Keywords
Computer Science and Digital Science
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.1.4. Multiscale modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.6. Optimization
- A6.2.7. High performance computing
- A6.2.8. Computational geometry and meshes
- A6.3. Computation-data interaction
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.3.4. Model reduction
- A6.3.5. Uncertainty Quantification
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1.1.2. Molecular and cellular biology
- B1.1.8. Mathematical biology
- B2.2.1. Cardiovascular and respiratory diseases
- B2.2.6. Neurodegenerative diseases
- B2.4.1. Pharmaco kinetics and dynamics
- B2.6.2. Cardiac imaging
1 Team members, visitors, external collaborators
Research Scientists
- Jacques Henry [Inria, Emeritus, HDR]
- Amel Karoui [Univ de Bordeaux, Researcher, from Oct 2020]
- Michael Leguèbe [Inria, Researcher]
- Mark Potse [Univ de Bordeaux, Researcher]
- Nejib Zemzemi [Inria, Researcher]
Faculty Members
- Yves Coudiere [Team leader, Univ de Bordeaux, Professor, HDR]
- Mostafa Bendahmane [Univ de Bordeaux, Associate Professor, HDR]
- Jerome Fehrenbach [Univ Paul Sabatier, Associate Professor, from Sep 2020, HDR]
- Lisl Weynans [Univ de Bordeaux, Associate Professor, HDR]
Post-Doctoral Fellows
- Mohamadou Malal Diallo [Univ de Bordeaux]
- Eduardo Hernandez Montero [Universidad Adolfo Ibanez - Santiago Chili, until Oct 2020]
PhD Students
- Andony Arrieula [Inria]
- Oumayma Bouhamama [Univ de Bordeaux]
- Syed Hassaan Ahmed Bukhari [Université de Saragosse - Espagne]
- Narimane Gassa [Univ de Bordeaux, from Sep 2020]
- Amel Karoui [Univ de Bordeaux, until Oct 2020]
- Niami Nasr [Univ de Bordeaux, from Oct 2020]
- Bachar Tarraf [Inria]
Technical Staff
- Mehdi Juhoor [Inria, Engineer]
- Pauline Migerditichan [Inria, Engineer]
- Louise-Amélie Schmitt [Inria, Engineer, from Jun 2020 until Sep 2020]
Interns and Apprentices
- Nosseiba Ben Salem [Inria, from Mar 2020 until Aug 2020]
- Seong Eun Choi [Inria, from May 2020 until Jun 2020]
- Leila El Ouaazizi [Inria, from Jun 2020 until Jul 2020]
- Niami Nasr [Univ de Bordeaux, from Mar 2020 until Aug 2020]
- Adele Perrin [Inria, from Feb 2020 until Jul 2020]
- Taina Portecop [Inria, from May 2020 until Jul 2020]
- Harinjara Ratrimoarison [Univ de Bordeaux, from Nov 2020]
Administrative Assistant
- Nathalie Robin [Inria]
Visiting Scientists
- Abir Amri [Ecole Nationale d’Ingénieur de Tunis - Tunisie, from Oct 2020]
- Khouloud Kordoghli [Ecole Nationale d’Ingénieur de Tunis - Tunisie, from Oct 2020]
2 Overall objectives
The Carmen team develops and uses models and numerical methods to simulate the electrophysiology of the heart from the molecular to the whole-organ scale, and its relation to measurable signals inside the heart and on the body surface. It aims at:
- improving understanding of normal and pathological cardiac electrophysiology,
- improving the efficiency and accuracy of numerical models,
- exploitation of all available electrical signals for diagnosis, in particular for prediction of life-threatening cardiac arrhythmias.
The numerical models used and developed by the team incorporate the gating dynamics of the ion channels in the cardiac cell membranes and the heterogeneities and coupling processes on the cellular scale into macroscopic reaction-diffusion models. At the same time we use reduced models to solve the inverse problems related to non-invasive electrical imaging of the heart.
The fields involved in our research are: ordinary and partial differential equations (PDE), inverse problems, numerical analysis, high-performance computing, image segmentation, and mesh construction.
A main goal of the team is to contribute to the work packages
defined in the IHU LIRYC (http://
We cooperate with physiologists and cardiologists on several projects. The team is building new models and powerful simulation tools that will help to understand the mechanisms behind cardiac arrhythmias and to establish personalized and optimized treatments. A particular challenge consists in making the simulations reliable and accessible to the medical community.
3 Research program
3.1 Complex models for the propagation of cardiac action potentials
The contraction of the heart is coordinated by a complex electrical activation process which relies on about a million ion channels, pumps, and exchangers of various kinds in the membrane of each cardiac cell. Their interaction results in a periodic change in transmembrane potential called an action potential. Action potentials in the cardiac muscle propagate rapidly from cell to cell, synchronizing the contraction of the entire muscle to achieve an efficient pump function. The spatio-temporal pattern of this propagation is related both to the function of the cellular membrane and to the structural organization of the cells into tissues. Cardiac arrythmias originate from malfunctions in this process. The field of cardiac electrophysiology studies the multiscale organization of the cardiac activation process from the subcellular scale up to the scale of the body. It relates the molecular processes in the cell membranes to the propagation process and to measurable signals in the heart and to the electrocardiogram, an electrical signal on the torso surface.
Several improvements of current models of the propagation of the action potential are being developed in the Carmen team, based on previous work 35 and on the data available at IHU LIRYC:
- Enrichment of the current monodomain and bidomain models 35, 46 by accounting for structural heterogeneities of the tissue at an intermediate scale. Here we focus on multiscale analysis techniques applied to the various high-resolution structural data available at the LIRYC.
- Coupling of the tissues from the different cardiac compartments and conduction systems. Here, we develop models that couple 1D, 2D and 3D phenomena described by reaction-diffusion PDEs.
These models are essential to improve our in-depth understanding of cardiac electrical dysfunction. To this aim, we use high-performance computing techniques in order to numerically explore the complexity of these models.
We use these model codes for applied studies in two important areas of cardiac electrophysiology: atrial fibrillation 39 and sudden-cardiac-death (SCD) syndromes 7, 642. This work is performed in collaboration with several physiologists and clinicians both at IHU Liryc and abroad.
3.2 Simplified models and inverse problems
The medical and clinical exploration of the cardiac electric signals is based on accurate reconstruction of the patterns of propagation of the action potential. The correct detection of these complex patterns by non-invasive electrical imaging techniques has to be developed. This problem involves solving inverse problems that cannot be addressed with the more compex models. We want both to develop simple and fast models of the propagation of cardiac action potentials and improve the solutions to the inverse problems found in cardiac electrical imaging techniques.
The cardiac inverse problem consists in finding the cardiac activation maps or, more generally, the whole cardiac electrical activity, from high-density body surface electrocardiograms. It is a new and a powerful diagnosis technique, which success would be considered as a breakthrough. Although widely studied recently, it remains a challenge for the scientific community. In many cases the quality of reconstructed electrical potential is not adequate. The methods used consist in solving the Laplace equation on the volume delimited by the body surface and the epicardial surface. Our aim is to:
- study in depth the dependance of this inverse problem on inhomogeneities in the torso, conductivity values, the geometry, electrode positions, etc.,
- improve the solution to the inverse problem by using new regularization strategies, factorization of boundary value problems, and the theory of optimal control.
Of course we will use our models as a basis to regularize these inverse problems. We will consider the following strategies:
- using complete propagation models in the inverse problem, like the bidomain equations, for instance in order to localize electrical sources,
- constructing families of reduced-order models using e.g. statistical learning techniques, which would accurately represent some families of well-identified pathologies,
- constructing simple models of the propagation of the activation front, based on eikonal or level-set equations, but which would incorporate the representation of complex activation patterns.
Additionaly, we will need to develop numerical techniques dedicated to our simplified eikonal/level-set equations.
3.3 Numerical techniques
We want our numerical simulations to be efficient, accurate, and reliable with respect to the needs of the medical community. Based on previous work on solving the monodomain and bidomain equations 37, 36, 45, 28, we will focus on:
- High-order numerical techniques with respect to the variables with physiological meaning, like velocity, AP duration and restitution properties,
- Efficient, dedicated preconditioning techniques coupled with parallel computing.
Existing simulation tools used in our team rely, among others, on mixtures of explicit and implicit integration methods for ODEs, hybrid MPI-OpenMP parallellization, algebraic multigrid preconditioning, and Krylov solvers. New developments include high-order explicit integration methods and task-based dynamic parallellism.
3.4 Cardiac Electrophysiology at the Microscopic Scale
Numerical models of whole-heart physiology are based on the approximation of a perfect muscle using homogenisation methods. However, due to aging and cardiomyopathies, the cellular structure of the tissue changes. These modifications can give rise to life-threatening arrhythmias. For our research on this subject and with cardiologists of the IHU LIRYC Bordeaux, we aim to design and implement models that describe the strong heterogeneity of the tissue at the cellular level and to numerically explore the mechanisms of these diseases.
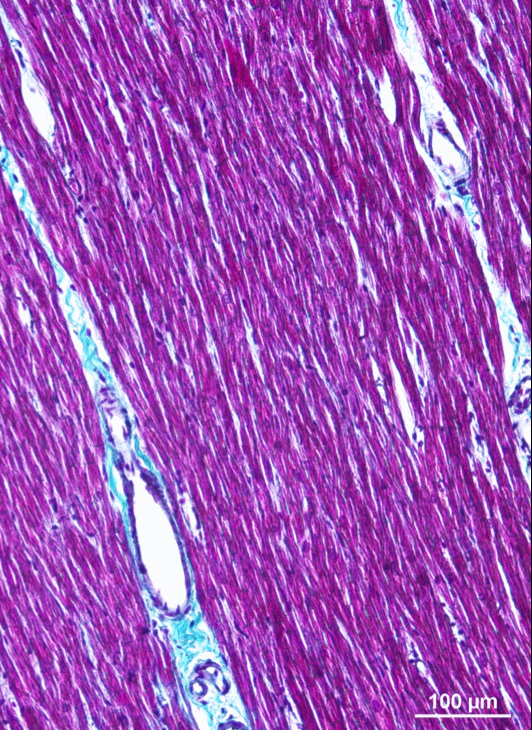
The literature on this type of model is still very limited 51. Existing models are two-dimensional 43 or limited to idealized geometries, and use a linear (purely resistive) behaviour of the gap-juction channels that connect the cells. We propose a three-dimensional approach using realistic cellular geometry (figure 1), nonlinear gap-junction behaviour, and a numerical approach that can scale to hundreds of cells while maintaining a sub-micrometer spatial resolution (10 to 100 times smaller than the size of a cardiomyocyte) 31, 30, 29. P-E. Bécue defended his PhD thesis on this topic in December 2018.
4 Application domains
4.1 Scientific context: the LIRYC
The University Hospital of Bordeaux (CHU de Bordeaux) is equipped with a specialized cardiology hospital, the Hôpital Cardiologique du Haut-Lévêque, where the group of Professor Michel Haïssaguerre has established itself as a global leader in the field of cardiac electrophysiology 41, 40, 33. Their discoveries in the area of atrial fibrillation and sudden cardiac death syndromes are widely acclaimed, and the group is a national and international referral center for treatment of cardiac arrhythmia. Thus the group also sees large numbers of patients with rare cardiac diseases.
In 2011 the group has won the competition for a 40 million euro
Investissements d'Avenir grant for the establishment of IHU
Liryc, an institute that combines clinical, experimental, and
numerical research in the area of cardiac arrhythmia
(http://
The Carmen team was founded to partner with IHU Liryc. We bring applied mathematics and scientific computing closer to experimental and clinical cardiac electrophysiology. In collaboration with experimental and clinical researchers at Liry we work to enhance fundamental knowledge of the normal and abnormal cardiac electrical activity and of the patterns of the electrocardiogram, and we develop new simulation tools for training, biological, and clinical applications.
4.2 Basic experimental electrophysiology
Our modeling is carried out in coordination with the experimental teams from IHU Liryc. It help to write new concepts concerning the multiscale organisation of the cardiac action potentials that will serve our understanding in many electrical pathologies. For example, we model the structural heterogeneities at the cellular scale 32, and at an intermediate scale between the cellular and tissue scales.
At the atrial level, we apply our models to understand the mechanisms of complex arrythmias and the relation with the heterogeneities at the insertion of the pulmonary veins. We will model the heterogeneities specific to the atria, like fibrosis or fatty infiltration 4839. These heterogeneities ara thought to play a major role in the development of atrial fibrillation.
At the ventricular level, we focus on (1) modeling the complex coupling between the Purkinje network and the ventricles, which is supposed to play a major role in sudden cardiac death, and (2) modeling the heteogeneities related to the complex organization and disorganization of the myocytes and fibroblasts, which is important in the study of infarct scars for instance.
4.3 Clinical electrophysiology
Treatment of cardiac arrhythmia is possible by pharmacological means, by implantation of pacemakers and defibrillators, and by curative ablation of diseased tissue by local heating or freezing. In particular the ablative therapies create challenges that can be addressed by numerical means. Cardiologists would like to know, preferably by noninvasive means, where an arrhythmia originates and by what mechanism it is sustained.
We address this issue in the first place using inverse models, which attempt to estimate the cardiac activity from a (high-density) electrocardiogram. A new project aims at performing this estimation on-site in the catheterization laboratory and presenting the results, together with the cardiac anatomy, on the screen that the cardiologist uses to monitor the catheter positions 44.
An important prerequisite for this kind of interventions and for inverse modeling is the creation of anatomical models from imaging data. The Carmen team contributes to better and more efficient segmentation and meshing through the IDAM project.
5 Highlights of the year
5.1 Awards
- 2 european projects founded: (Microcard, SimCardioTest)
- 1 project MSCA-ITN lauched (PersonalizeAF)
- The year 2020 was marked by the covid crisis, and its impact on society and its overall activity. The world of research was also greatly affected: Faculty members have seen their teaching load increase significantly; PhD students and post-docs have often had to deal with a worsening of their working conditions, as well as with reduced interactions with their supervisors and colleagues; most scientific collaborations have been greatly affected, with many international activities cancelled or postponed to dates still to be defined.
6 New software and platforms
6.1 New software
6.1.1 CEPS
- Name: Cardiac ElectroPhysiology Simulation
- Keywords: Simulation, Health, Mesh, Cardiac, 3D, Cardiac Electrophysiology
- Scientific Description: As compared to other existing softwares, CEPS aims at providing a more general framework of integration for new methods or models and a better efficiency in parallel. CEPS is designed to run on massively parallel architectures, and to make use of state-of-the-art and well known computing libraries to achieve realistic and complex heart simulations. CEPS also includes software engineering and validation tools.
- Functional Description: CEPS is a modular high-performance computing software for performing numerical simulations in cardiac electrophysiology. It is based on modules : - management of geometries represented by meshes in 3D, 2D or 1D (volumes, surfaces, trees), - model simulation of cellular electrophysiology, - calculating the tissue propagation of the action potentials in the cardiac geometries, - calculation of extracardiac potentials, - time approximation methods in order 2, 3 and 4 specific to electrocardiography.
-
URL:
https://
gforge. inria. fr/ projects/ ceps/ - Authors: Mehdi Juhoor, Nejib Zemzemi, Charlie Douanla Lontsi, Marc Fuentes, Yves Coudière, Florian Caro
- Contacts: Michael Leguebe, Yves Coudière, Nejib Zemzemi
- Participants: Mehdi Juhoor, Nejib Zemzemi, Antoine Gerard, Charlie Douanla Lontsi, Pierre-Elliott Bécue, Marc Fuentes, Yves Coudière, Michael Leguebe, Andjela Davidovic, Pauline Migerditichan, Florian Caro
- Partners: Université de Bordeaux, Fondation Bordeaux Université, CHU de Bordeaux, Inria
6.1.2 OptimDBS
- Name: Optimizing the Deep Brain Stimulation
- Keywords: Image analysis, Deep brain stimulation, Statistical learning
- Functional Description: Targeting software for deep brain stimulation
-
URL:
https://
gitlab. inria. fr/ optimdbs/ optimdbs-medinria/ -/ wikis/ home - Authors: Nejib Zemzemi, Louise-Amelie Schmitt
- Contact: Nejib Zemzemi
- Participants: Nejib Zemzemi, Louise-Amelie Schmitt, Emmanuel Cuny, Julien Engelhardt
- Partner: CHU de Bordeaux
6.1.3 MUSIC - Carmen plugins
- Name: Carmen plugins for multi-modality imaging in Cardiology
- Keywords: Image segmentation, Mesh generation, Image filter, Numerical simulations, Cardiac Electrophysiology, Inverse problem, Finite element modelling, Visualization, 3D interaction, Registration
- Scientific Description: Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
- Functional Description: Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
- Publications: hal-01923927, hal-01923763, hal-01400889
- Authors: Mehdi Juhoor, Pauline Migerditichan, Mathilde Merle, Julien Castelneau, Florent Collot, Nejib Zemzemi, Pauline Migerditichan, Pauline Migerditichan
- Contacts: Nejib Zemzemi, Yves Coudière, Mehdi Juhoor, Pauline Migerditichan
- Participants: Hubert Cochet, Florent Collot, Mathilde Merle, Maxime Sermesant, Julien Castelneau, Mehdi Juhoor, Pauline Migerditichan, Nejib Zemzemi, Yves Coudière
- Partners: Université de Bordeaux, IHU - LIRYC
6.2 New platforms
CEMPACK
CEMPACK is a new collection of software that was previously
archived in different places. It includes the high-performance
simulation code Propag and a suite of software for the creation of
geometric models, preparing inputs for Propag, and analysing its
outputs. In 2017 the code was collected in an archive
on Inria's GitLab platform, and a public website was created
for documentation (http://
-
Propag-5.1
Applied modeling studies performed by the Carmen team in collaboration with IHU Liryc and foreign partners 749, 39, 38, 34 rely on high-performance computations on the national supercomputers Irene, Occigen, and Turing. The Propag-5 code is optimized for these systems. It is the result of a decades-long development first at the Université de Montréal in Canada, then at Maastricht University in the Netherlands, and finally at the Institute of Computational Science of the Università della Svizzera italiana in Lugano, Switzerland. Since 2016 most of the development on Propag has been done by M. Potse at the Carmen team 50. The code scales excellently to large core counts and, as it is controlled completely with command-line flags and configuration files, it can be used by non-programmers. It also features:
- a plugin system for membrane models,
- a completely parallel workflow, including the initial anatomy input and mesh partitioning, which allows it to work with meshes of more than nodes,
- a flexible output scheme allowing hundreds of different state variables and transient variables to be output to file, when desired, using any spatial and temporal subsampling,
- a configurable, LUSTRE-aware parallel output system in which groups of processes write HDF5/netCDF files, and
- CWEB documentation of the entire code base.
The code has been stable and reliable for several years. It can be considered the workhorse for our HPC work until CEPS takes over.
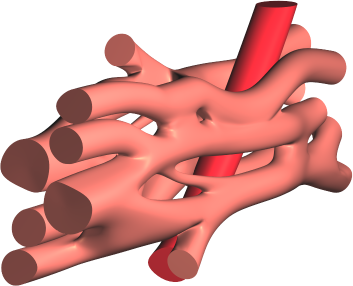
- Gepetto The Gepetto suite, named after a famous model maker, transforms a surface mesh of the heart into a set of (semi-)structured meshes for use by the Propag software or others. It creates the different fiber orientations in the model, including the transmurally rotating ventricular fibers and the various bundle structures in the atria (figure 2), and creates layers with possibly different electrophysiological properties across the wall. A practically important function is that it automatically builds the matching heart and torso meshes that Propag uses to simulate potentials in the torso (at a resolution of 1 mm) after projecting simulation results from the heart model (at 0.1 to 0.2 mm) on the coarser torso mesh 47. Like Propag, the Gepetto software results from a long-term development that started in Montreal, Canada, around 2002. The code for atrial fiber structure was developed by our team.
-
Blender plugins
Blender (https://
www. blender. org) is a free software package for the production of 3-D models, renderings, and animations, comparable to commercial software such as Cinema4D. CEMPACK includes a set of plugins for Blender that facilitate the production of anatomical models and the visualization of measured and simulated data. It uses the MMG remeshing library, which is developed by the CARDAMOM team at Inria Bordeaux.
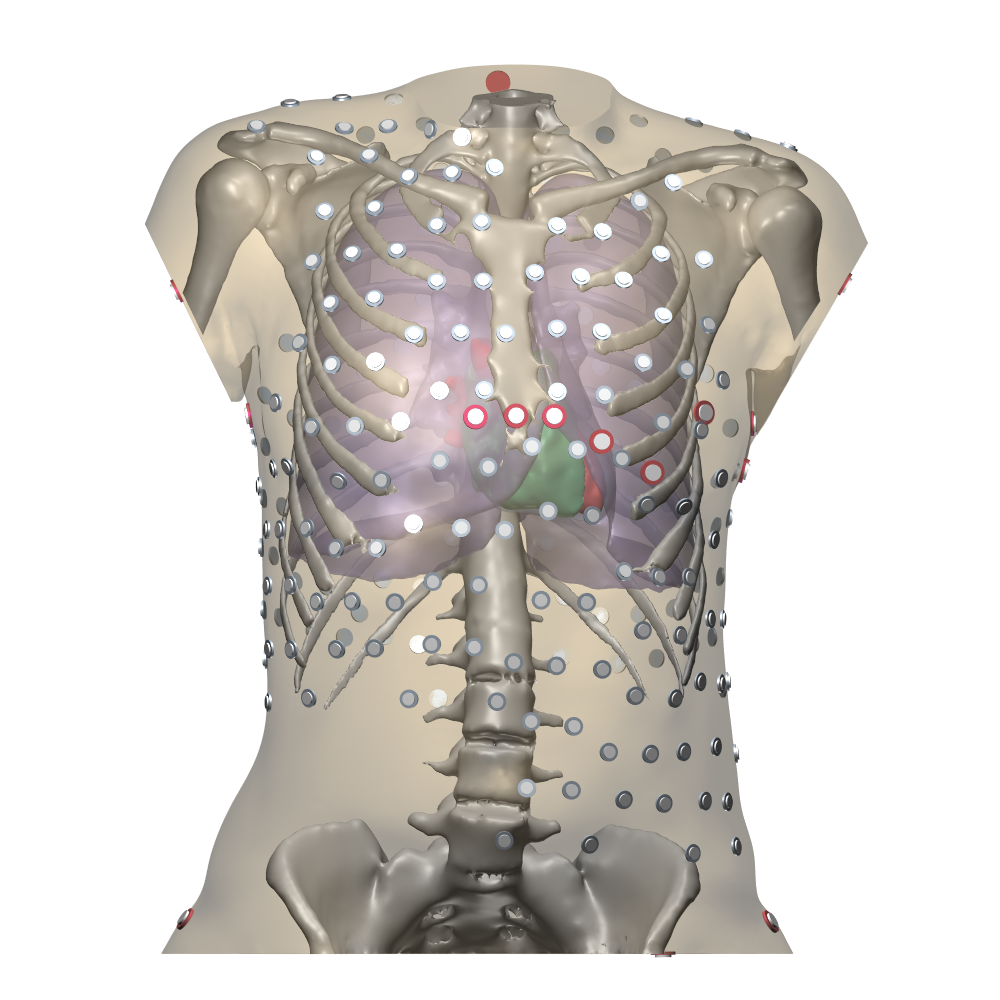
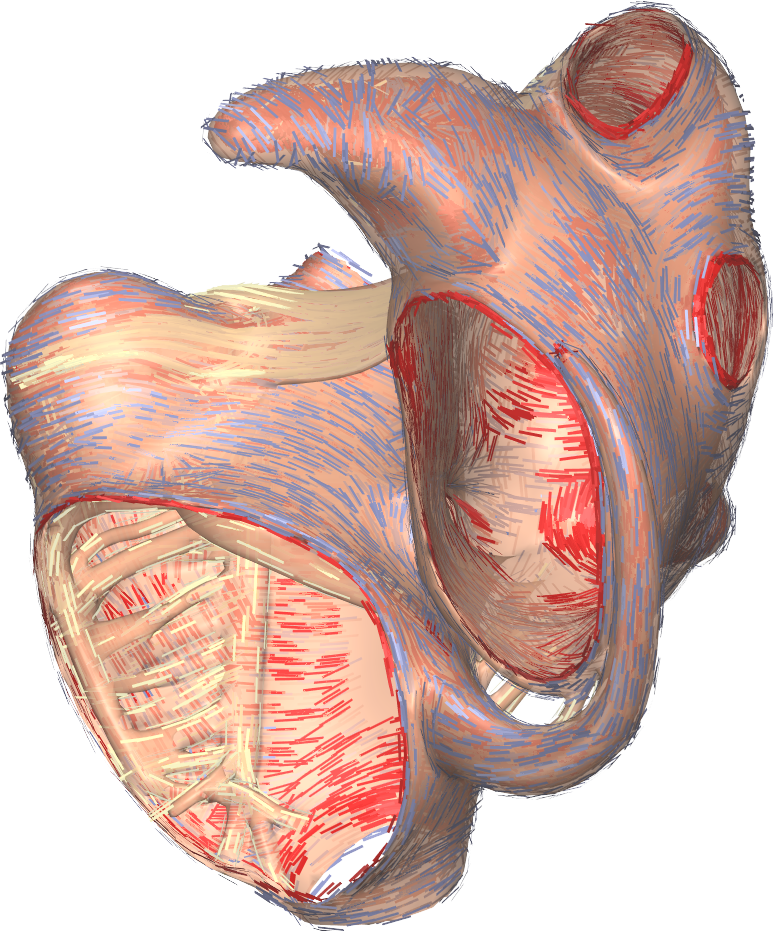
7 New results
7.1 Partial differential equations analysis
- Theoretical analysis of Purkinje/myocardium coupling, with as final objective the theoretical and numerical analysis of coupling parameters (Ph.D. project of Khouloud Kordoghli),
- Theoretical analysis of parameters identification problems: in particular the parameters of the ionic model (conductances of main ionic channels, sodium, calcium and potassium) (Ph.D. thesis of Yassine Abidi defended in 2020, and beginning Ph.D. project of Abir Amri).
7.2 Numerical analysis and development of numerical methods
- Publication of an article about Rush-Larsen time-stepping methods of high order for stiff problems in cardiac electrophysiology (following Ph.D. thesis of C. Douanla Lontsi),
- Numerical analysis of a numerical scheme (positive cell vertex Godunov scheme) for the Beeler-Reuter model of cardiac activity,
-
Convergence proof of the numerical scheme for the virtual element method for a nonlocal
FitzHugh–Nagumo model of cardiac electrophysiology (Mostafa Bendahmane in collaboration with Veronica Anaya and David Mora (university Bip-Bio, Concepcion, Chile),
- Proof of concept of AI based methods for the development of new ECGi algorithms: second part of Amel Karoui Ph.D. thesis,
- Develoment of ECGI algorithms for transmural reconstruction in presence of scars (Malal Diallo, part of ERC Ecstatic of H. Cochet),
- Develoment of a real time algorithm to predict accurately the exit site of a ventricular extrasystole based on its 12-lead ECG. The algorithm is “trained” using ECGs obtained by pacing at a few sites whose locations are documented with an electroanatomical mapping system,
- Validation of the « patchwork method », a ECGI method selecting among several numerical method the one minimizing the residuals on the torso, on simulated data without/with noise. (Ph.D thesis of Oumayma Bouhamama, co-supervised with Laura Bear from Signal Processing team),
-
Development of a new method on cartesian grids to solve the inverse problem of Electrical Impe-
dance Tomography in 2 dimensions (Ph.D thesis of Niami Nasr, co-supervised by J. Dardé, IMT Toulouse),
- Development of an eikonal model to solve the propagation of the front on the heart surface, in collaboration with J. Fehrenbach (IMT, Toulouse),
- Numerical study of parameters identification problems: in particular the parameters of the ionic model (conductances of main ionic channels, sodium, calcium and potassium) (Ph.D. thesis of Yassine Abidi defended in 2020, and beginning Ph.D. project of Abir Amri).
7.3 Modelling
- Modelling of heterogeneities in the electrical conduction parameters (follow up of Andjela Davidovic Ph.D thesis). It enables to include in a model informations about the microstructure available from high resolution imaging modalities, such as fibrosis or diverse types of inclusions,
-
Model of cardiac mitochondria that can be coupled to cardiac electrophysiology models, para-
meter identification results. It is an interdisciplinairy work between Liryc teams (modelling and metabolism) and other laboratories (ISM Bordeaux and LASS Toulouse), Ph.D. thesis of Bachar Tarraf,
- Simulations of atrial fibrillation induced by rapid pacing have shown an extreme sensitivity to model parameters. When the refractory period is reduced, and the pacing frequency is well adjusted to the refractory period, it turns out to be almost always possible to induce arrhythmia, even in a structurally normal model. Normal anatomical features such as thick muscle bundles and abrupt changes in fiber orientation play an important role in these phenomena.
7.4 High performance computing
We have participated in a Grand Challenge call of TGCC, the computing center of the CEA, on the occasion of the installation of a new partition of the national supercomputer Irène Joliot-Curie. This competitive call for proposals has given us 20 million compute hours and a preferential access to this new cluster computer with more than 260 thousand compute cores, allowing us to test our simulation code on a scale 1000 times larger than our routine simulations. The test results were positive, showing that the code remains efficient at this scale and that we can essentially make our computations as large as the machine can store in its memory. This gives us confidence for our projects on future supercomputers at an even larger scale.
8 Partnerships and cooperations
8.1 International initiatives
8.1.1 Inria International Labs
SPICY
- Title: Stochastic forward and inverse Problems In Cardiac electrophysiologY
- Duration: 2020 -
- Coordinator: Mostafa Bendahmane
- Partner: LAMSIN (LIRIMA)
- Inria contact: Mostafa Bendahmane
8.1.2 Participation in other international programs
Ecos-Sud funds
For 3 years, collaboration with University bio-Bio, Chile, PI: Mostafa Benhdamane for Bordeaux University/Liryc.
8.2 International research visitors
8.2.1 Visits of international scientists
In Fall 2020, visits of Khouloud Kordoghli et Abir Amri (Ph.D. student).
8.3 European initiatives
8.3.1 Collaborations in European programs, except FP7 and H2020
SimCardioTest
Simulation of Cardiac Devices and Drugs for in-silico Testing and Certification in Accelerating the uptake of computer simulations for testing medicines and medical devices – TOPIC ID:SC1-DTH-06-2020, himself in Digital transformation in Health and Care (H2020-SC1-DTH-2018-2020), which is in WP Health, demographic change and wellbeing, 857 875 € for UB/Liryc. Main costs: PhD, engineers, experiments with animals, experiments with torso tank, PI: M. Sermesant from Inria, PI for Bordeaux University/Liryc: Yves Coudière.
EuroHPC MICROCARD
5.8M€ (1.6 M€ for UBx) with 10 European partners, both academic and industrial. The project aims to build a software code that will be able to simulate the heart cell-by-cell on future “exascale” supercomputers. PI: Mark Potse for Bordeaux University/Liryc.
MSCA/ITN personalizeAF:
an interdisciplinary research and training project involving several countries, several scientific fields. The aim of the project is the individual-specific characterization of AF substrate in order to increase the treatment efficiency, 278k euros, for UBx (IHU-LIRYC), PI: Nejib Zemzemi for Bordeaux University/Liryc.
8.4 National initiatives
8.4.1 ANR Exacard
Granted by ANR in July 2018, it is a collaborative project with computer scientists from Labri that targets hexascale computing in cardiac electrophysiology, it serves as a sandbox to prepare the project EuroHPC Microcard. PI for Bordeaux University/Liryc: Yves Coudière.
8.4.2 ANR MITOCARD
The MITOCARD project (Electrophysiology of Cardiac Mitochondria), coordinated by S. Arbault (Université de Bordeaux, ISM), was granted by the ANR in July 2017. The objective of MITOCARD is to improve understanding of cardiac physiology by integrating the mitochondrial properties of cell signaling in the comprehensive view of cardiac energetics and rhythm pathologies. It was recently demonstrated that in the heart, in striking contrast with skeletal muscle, a parallel activation by calcium of mitochondria and myofibrils occurs during contraction, which indicates that mitochondria actively participate in Ca2+ signaling in the cardiomyocyte. We hypothesize that the mitochondrial permeability transition pore (mPTP), by rhythmically depolarizing inner mitochondrial membrane, plays a crucial role in mitochondrial Ca2+ regulation and, as a result, of cardiomyocyte Ca2+ homeostasis. Moreover, mitochondrial reactive oxygen species (ROS) may play a key role in the regulation of the mPTP by sensing mitochondrial energetics balance. Consequently, a deeper understanding of mitochondrial electrophysiology is mandatory to decipher their exact role in the heart's excitation-contraction coupling processes. However, this is currently prevented by the absence of adequate methodological tools (lack of sensitivity or selectivity, time resolution, averaged responses of numerous biological entities). The MITOCARD project will solve that issue by developing analytical tools and biophysical approaches to monitor kinetically and quantitatively the Ca2+ handling by isolated mitochondria in the cardiomyocyte.
MITOCARD is a multi-disciplinary project involving 4 partners of different scientific fields: the CARMEN team as well as:
- ISM, the largest chemistry laboratory of the Université de Bordeaux, where the necessary measurement methods will be developed,
- Liryc, where mitochondria are studied at all levels of integration from the isolated mitochondrion to the intact heart,
- LAAS, the MiCrosystèmes d'Analyse (MICA) group at the Laboratory of Analysis and Architecture of Systems, which develops the biological microsensors for this project.
The project will:
- develop chips integrating 4 different electrochemical microsensors to monitor in real-time key mitochondrial signaling parameters: Ca2+, membrane potential, quinone reduction status, O2 consumption, and ROS production,
- develop microwell arrays integrating ring nanoelectrodes to trap single mitochondria within micrometric chambers and measure locally by combined fluorescence microscopy and electrochemical techniques intra- (by fluorescence) and extra-mitochondrial (electrochemistry) metabolites,
- develop a mathematical model of mitochondrial Ca2+ and ROS handling built on existing knowledge, new hypotheses, and the measured data.
The model may serve both to assess biological assumptions on the role of mitochondria in Ca2+ signaling and to integrate pathological data and provide clues for their global understanding.
8.5 Regional initiatives
Calm:
co-funding from Région Nouvelle Aquitaine-INRIA/Liryc to support the Ph.D. of Andony Arrieula on a real time algorithm to predict accurately the exit site of a ventricular extrasystole based on its 12-lead ECG, PI for Bordeaux University/Liryc: Mark Potse, collaboration with Pierre Jaïs.
9 Dissemination
9.1 Promoting scientific activities
9.1.1 Research administration
Yves Coudière is vice-head of the Mathematics Department.
9.2 Teaching - Supervision - Juries
9.2.1 Teaching
The 2 assistant professors and 1 professor of the team teach at several levels of the Bordeaux University programs in Mathematics and Neurosciences (respectively, 192, 192 and 96 h/year on average). The researchers also have a regular teaching activity, contributing to several courses in the Applied Mathematics at the Bachelor and Master levels (usually between 16 and 32 h/year).
The PhD student are used to teach between 32 and 64 h/year, usually courses of general mathematics in L1 or mathematics for biologists in L1 or L2.
Teaching responsibilities at the University of Bordeaux:
- Yves Coudière: Master MAS (Mathématiques Appliquées, Statistiques), parcours MNCHP (Modélisation Numérique Calcul Haute Performance),
- Yves Coudière: Licence Mathématique parcours ingénierie mathématique,
- Lisl Weynans: Mineure Mathématiques du parcours International de la Licence,
- Mostafa Bendahmane: Responsable de la mobilité internationale des étudiants de Licence MIASHS.
Courses (L for Bachelor level, M for Master level):
- Numerical analysis (L2)
- Programming for scientific computing with C++ (L3)
- Programming projects with Python (M1)
- Numerical approximation of PDEs: Finite Differences, Finite Elements, Finite Volumes (M1, M2)
- Supervision of programming projects (L3, M1)
- Mathematical modelling in L2 parcours medicine and physics
- Linear Algebra, Optimization under constraints (L2 and Essca school)
- Analysis, L2
- Computational Neurosciences, M2
- Neuropsychology and Psychophysiology, L3
- Graduate program EUR Digital Public Health, Bordeaux University, March 2019. Philosophy of science and the role of numerical models: introductory course in the module Modeling in life science
9.2.2 Supervision
- PhD in progress : Andony Arrieula, Oct. 2018, encadrant: Mark Potse
- PhD in progress: Oumayma Bouhamama, Oct. 2018 , encadrante: Lisl Weynans
- PhD in progress : Syed Hassaan Ahmed Bukhari, Oct. 2018, encadrant: Mark Potse
- PhD in progress : Amel Karoui, Oct 2017, encadrants: Mostafa Bendahmane and Nejib Zemzemi
- PhD in progress : Bachar Tarraf, Oct. 2018, encadrants: Yves Coudière and Michael Leguebe
- PhD in progress : Niami Nasr, Oct. 2020, encadrante: Lisl Weynans
- PhD in progress : Narimane Gassa, Oct. 2020, encadrants: Yves Coudi
ere and Nejib Zemzemi
10 Scientific production
10.1 Major publications
- 1 articleConvergence of discrete duality finite volume schemes for the cardiac bidomain modelNetworks and Heterogeneous Media622011, 195-240
- 2 articleA mathematical model of Purkinje-Muscle JunctionsMathematical Biosciences and Engineering842011, 915-930
- 3 articleExistence And Uniqueness Of The Solution For The Bidomain Model Used In Cardiac ElectrophysiologyNonlinear Anal. Real World Appl.1012009, 458-482URL: http://hal.archives-ouvertes.fr/hal-00101458/fr
- 4 articleA 2D/3D Discrete Duality Finite Volume Scheme. Application to ECG simulationInternational Journal on Finite Volumes612009, URL: http://hal.archives-ouvertes.fr/hal-00328251/fr
- 5 articleStability And Convergence Of A Finite Volume Method For Two Systems Of Reaction-Diffusion Equations In Electro-CardiologyNonlinear Anal. Real World Appl.742006, 916--935URL: http://hal.archives-ouvertes.fr/hal-00016816/fr
- 6 articleThe Early Repolarization Pattern; A Consensus PaperJournal of the American College of Cardiology662015, 470-477URL: http://dx.doi.org/10.1016/j.jacc.2015.05.033
- 7 article Reduced Sodium Current in the Lateral Ventricular Wall Induces Inferolateral J-Waves Front Physiol 7 365 August 2016
- 8 articlePreconditioning the bidomain model with almost linear complexityJournal of Computational Physics2311January 2012, 82--97URL: http://www.sciencedirect.com/science/article/pii/S0021999111005122
10.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Reports & preprints
Other scientific publications
10.3 Cited publications
- 28 articleConvergence of discrete duality finite volume schemes for the cardiac bidomain modelNetworks and Heterogeneous Media622011, 195-240
- 29 inproceedingsModélisation et simulation de l'électrophysiologie cardiaque à l'échelle microscopique43e Congrès National d'Analyse Numérique (CANUM)SMAIObernai, Alsace, FranceMay 2016, URL: http://smai.emath.fr/canum2016/resumesPDF/peb/Abstract.pdf
- 30 misc Theoretical and Numerical Study of Cardiac Electrophysiology Problems at the Microscopic Scale. Poster July 2016
- 31 inproceedingsA Three-Dimensional Computational Model of Action Potential Propagation Through a Network of Individual CellsComputing in Cardiology 2017Rennes, FranceSeptember 2017, 1-4
- 32 inproceedings Microscopic Simulation of the Cardiac Electrophysiology: A Study of the Influence of Different Gap Junctions Models Computing in Cardiology Maastricht, Netherlands September 2018
- 33 articleImpact of Septal Radiofrequency Ventricular Tachycardia Ablation; Insights From Magnetic Resonance ImagingCirculation1302014, 716-718
- 34 inproceedingsDo we need to enforce the homogeneous Neuman condition on the Torso for solving the inverse electrocardiographic problem by using the method of fundamental solution ?Computing in Cardiology 201643Computing in Cardiology 2016Vancouver, CanadaSeptember 2016, 425-428
- 35 articleOptimal monodomain approximations of the bidomain equations used in cardiac electrophysiologyMathematical Models and Methods in Applied Sciences246February 2014, 1115-1140
- 36 articleA 2D/3D Discrete Duality Finite Volume Scheme. Application to ECG simulationInternational Journal on Finite Volumes612009, URL: http://hal.archives-ouvertes.fr/hal-00328251/fr
- 37 articleStability And Convergence Of A Finite Volume Method For Two Systems Of Reaction-Diffusion Equations In Electro-CardiologyNonlinear Anal. Real World Appl.742006, 916--935URL: http://hal.archives-ouvertes.fr/hal-00016816/fr
- 38 articleSpatially Coherent Activation Maps for Electrocardiographic ImagingIEEE Transactions on Biomedical Engineering64May 2017, 1149-1156
- 39 inproceedings Epicardial Fibrosis Explains Increased Transmural Conduction in a Computer Model of Atrial Fibrillation Computing in Cardiology Vancouver, Canada September 2016
- 40 articleSudden cardiac arrest associated with early repolarizationN. Engl. J. Med.3582008, 2016--2023
- 41 articleSpontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veinsN. Engl. J. Med.3391998, 659-666
- 42 articleMechanism of Right Precordial ST-Segment Elevation in Structural Heart Disease: Excitation Failure by Current-to-Load MismatchHeart Rhythm72010, 238-248URL: http://dx.doi.org/10.1016/j.hrthm.2009.10.007
- 43 articleA microstructural model of reentry arising from focal breakthrough at sites of source-load mismatch in a central region of slow conductionAm. J. Physiol. Heart Circ. Physiol.3062014, H1341-1352
- 44 inproceedings A new ECG-based method to guide catheter ablation of ventricular tachycardia iMAging and eLectrical Technologies Uppsala, Sweden April 2018
- 45 articlePreconditioning the bidomain model with almost linear complexityJournal of Computational Physics2311January 2012, 82--97URL: http://www.sciencedirect.com/science/article/pii/S0021999111005122
-
46
articleA Comparison of monodomain and bidomain
-diffusion models for action potential propagation in the human heart'IEEE Transactions on Biomedical Engineering53122006, 2425-2435URL: http://dx.doi.org/10.1109/TBME.2006.880875 - 47 articleCardiac Anisotropy in Boundary-Element Models for the ElectrocardiogramMedical and Biological Engineering and Computing472009, 719--729URL: http://dx.doi.org/10.1007/s11517-009-0472-x
- 48 inproceedings Anatomically-induced Fibrillation in a 3D model of the Human Atria Computing in Cardiology Maastricht, Netherlands September 2018
- 49 misc Regional conduction slowing can explain inferolateral J waves and their attenuation by sodium channel blockers Poster September 2016
- 50 articleScalable and Accurate ECG Simulation for Reaction-Diffusion Models of the Human HeartFrontiers in Physiology9April 2018, 370
- 51 articleA Cell-Based Framework for Numerical Modeling of Electrical Conduction in Cardiac TissueFront. Phys.52017, 48

