Keywords
Computer Science and Digital Science
- A3.3. Data and knowledge analysis
- A3.4. Machine learning and statistics
- A5.2. Data visualization
- A5.3. Image processing and analysis
- A5.4. Computer vision
- A5.6. Virtual reality, augmented reality
- A5.9. Signal processing
- A6.1. Methods in mathematical modeling
- A6.2. Scientific computing, Numerical Analysis & Optimization
- A6.3. Computation-data interaction
- A8.3. Geometry, Topology
- A9. Artificial intelligence
- A9.2. Machine learning
- A9.3. Signal analysis
- A9.6. Decision support
- A9.7. AI algorithmics
Other Research Topics and Application Domains
- B2.2. Physiology and diseases
- B2.3. Epidemiology
- B2.4. Therapies
- B2.6. Biological and medical imaging
- B2.6.1. Brain imaging
- B2.6.2. Cardiac imaging
- B2.6.3. Biological Imaging
1 Team members, visitors, external collaborators
Research Scientists
- Nicholas Ayache [Team leader, Inria, Senior Researcher, HDR]
- James Benn [Inria, Starting Research Position, from Jul 2020]
- Hervé Delingette [Inria, Senior Researcher, HDR]
- Marco Lorenzi [Inria, Researcher, HDR]
- Xavier Pennec [Inria, Senior Researcher, HDR]
- Maxime Sermesant [Inria, Researcher, HDR]
Post-Doctoral Fellows
- Irene Balelli [Inria, from Feb 2020]
- Marie Deprez [Univ Côte d'Azur, from Feb 2020]
- Marta Nunez Garcia [Univ de Bordeaux]
- Dimbihery Rabenoro [Inria, from May 2020]
PhD Students
- Clement Abi Nader [Univ Côte d'Azur]
- Luigi Antelmi [Univ Côte d'Azur]
- Benoit Audelan [Univ Côte d'Azur]
- Tania Marina Bacoyannis [Inria]
- Jaume Banus Cobo [Univ Côte d'Azur]
- Paul Blanc-Durand [Hôpital Henri Mondor, HDR]
- Nicolas Cedilnik [Inria]
- Mikael Chelli [Centre hospitalier universitaire de Nice]
- Hind Dadoun [Inria]
- Gaetan Desrues [Inria]
- Yann Fraboni [Inria]
- Nicolas Guigui [Inria]
- Dimitri Hamzaoui [Univ Côte d'Azur]
- Josquin Harrison [Inria, from Oct 2020]
- Etrit Haxholli [Inria]
- Shuman Jia [Inria, until Mar 2020]
- Florent Jousse [Quantificare, CIFRE]
- Victoriya Kashtanova [Inria, from Mar 2020]
- Julian Krebs [Inria, until Aug 2020]
- Buntheng Ly [Univ Côte d'Azur]
- Elodie Maignant [Inria, from Oct 2020]
- Morten Pedersen [Inria, from Jul 2020]
- Santiago Smith Silva Rincon [Univ Côte d'Azur, from Feb 2020]
- Hari Sreedhar [Univ Côte d'Azur, from Dec 2020]
- Yann Thanwerdas [Univ Côte d'Azur]
- Paul Tourniaire [Inria]
- Clair Vandersteen [Centre hospitalier universitaire de Nice, from Feb 2020]
- Zihao Wang [Inria]
- Wen Wei [Inria, from Feb 2020 until May 2020]
- Yingyu Yang [Inria, from Sep 2020]
Technical Staff
- Wen Wei [Inria, Engineer, Sep 2020]
Interns and Apprentices
- Josquin Harrison [Inria, from Mar 2020 until Aug 2020]
- Julien Moreira [Univ Côte d'Azur, until Aug 2020]
- Andrea Senacheribbe [Inria, from Sep 2020]
- Yingyu Yang [Inria, from April 2020 to August 2020 ]
Administrative Assistant
- Isabelle Strobant [Inria]
Visiting Scientist
- Gerard Marti [Universitat Pompeu Fabra, from Sep 2020 until Oct 2020]
External Collaborators
- Valeria Manera [CHU Nice, CoBTeK]
- Marco Milanesio [Univ Côte d'Azur]
- Pamela Moceri [CHU Nice]
- Philippe Robert [CHU Nice, CoBTeK]
2 Overall objectives
2.1 Description
Our long-term goal is to contribute to the development of what we call the e-patient (digital patient) for e-medicine (digital medicine).
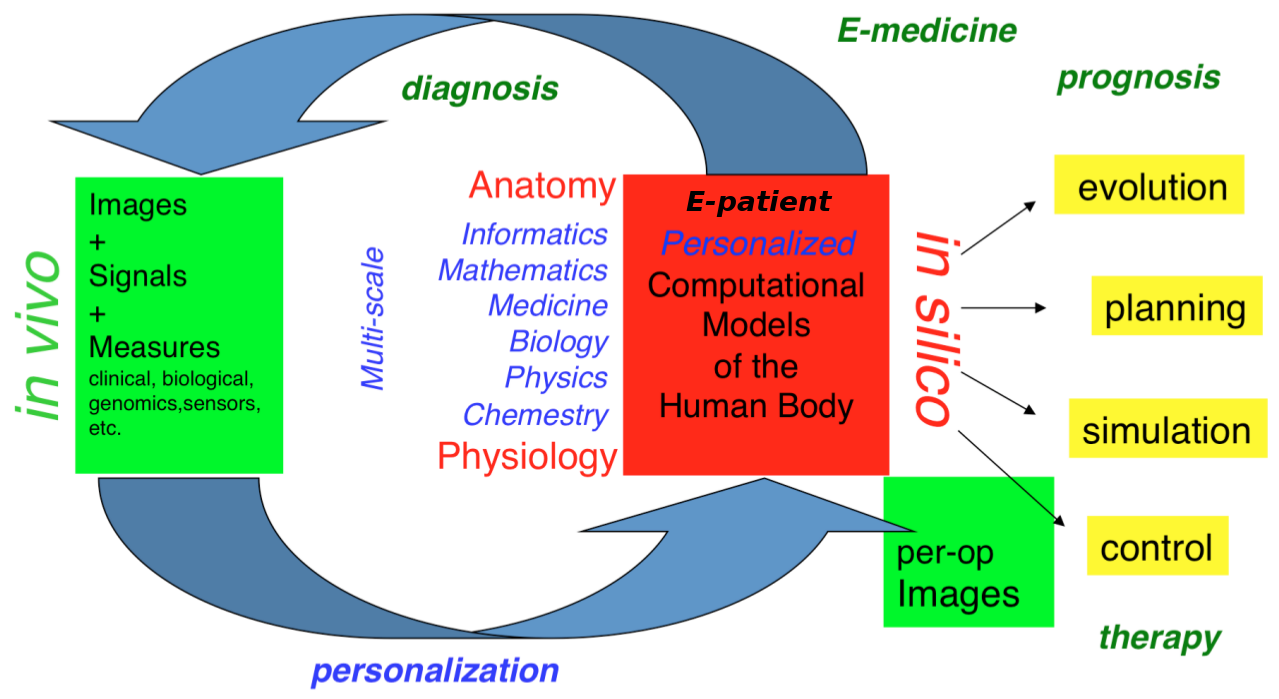
- the e-patient (or digital patient) is a set of computational models of the human body able to describe and simulate the anatomy and the physiology of the patient's organs and tissues, at various scales, for an individual or a population. The e-patient can be seen as a framework to integrate and analyze in a coherent manner the heterogeneous information measured on the patient from disparate sources: imaging, biological, clinical, sensors, ...
- e-medicine (or digital medicine) is defined as the computational tools applied to the e-patient to assist the physician and the surgeon in their medical practice, to assess the diagnosis/prognosis, and to plan, control and evaluate the therapy.
The models that govern the algorithms designed for e-patients and e-medicine come from various disciplines: computer science, mathematics, medicine, statistics, physics, biology, chemistry, etc. The parameters of those models must be adjusted to an individual or a population based on the available images, signals and data. This adjustment is called personalization and usually requires solving difficult inverse problems. The overall picture of the construction of the personalized e-patient for e-medicine was presented at the College de France through an inaugural lecture and a series of courses and seminars (fr), concluded by an international workshop.
2.2 Organisation
The research organization in our field is often built on a virtuous triangle. On one vertex, academic research requires multidisciplinary collaborations associating informatics and mathematics to other disciplines: medicine, biology, physics, chemistry ... On a second vertex, a clinical partnership is required to help defining pertinent questions, to get access to clinical data, and to clinically evaluate any proposed solution. On the third vertex, an industrial partnership can be introduced for the research activity itself, and also to transform any proposed solution into a validated product that can ultimately be transferred to the clinical sites for an effective use on the patients.
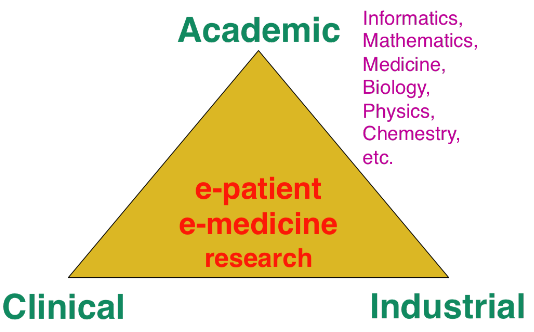
Keeping this triangle in mind, we choose our research directions within a virtuous circle: we look at difficult problems raised by our clinical or industrial partners, and then try to identify some classes of generic fundamental/theoretical problems associated to their resolution. We also study some fundamental/theoretical problems per se in order to produce fundamental scientific advances that can help in turn to promote new applications.
3 Research program
3.1 Introduction
Our research objectives are organized along 5 scientific axes:
- Biomedical Image Analysis & Machine Learning
- Imaging & Phenomics, Biostatistics
- Computational Anatomy, Geometric Statistics
- Computational Physiology & Image-Guided Therapy
- Computational Cardiology & Image-Based Cardiac Interventions
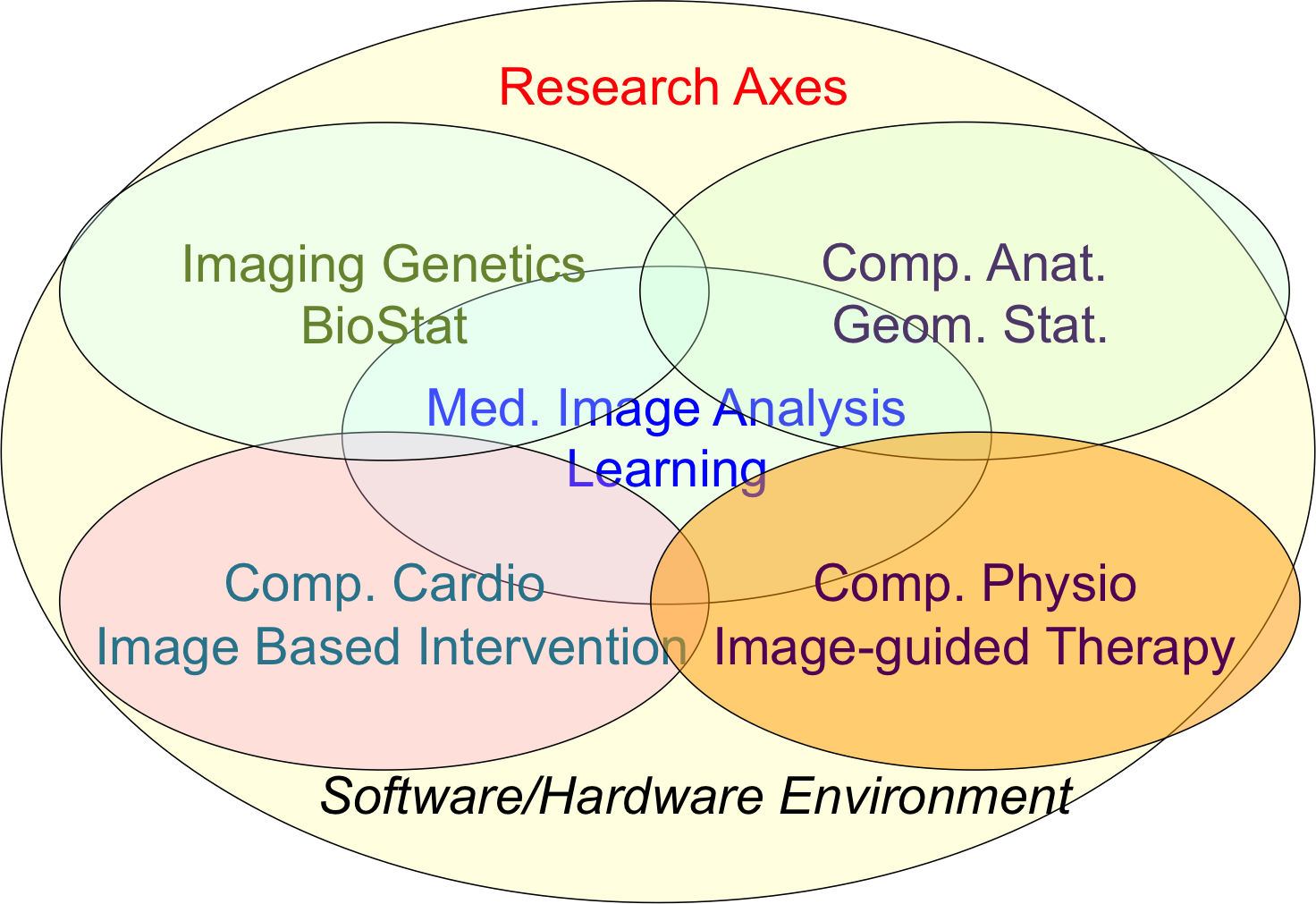
Epione's five main research axes
For each scientific axis, we introduce the context and the long term vision of our research .
3.2 Biomedical Image Analysis & Machine Learning
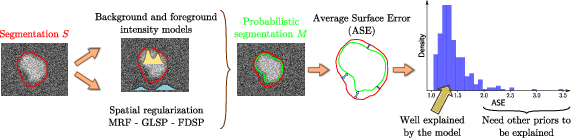
The long-term objective of biomedical image analysis is to extract, from biomedical images, pertinent information for the construction of the e-patient and for the development of e-medicine. This relates to the development of advanced segmentation and registration of images, the extraction of image biomarkers of pathologies, the detection and classification of image abnormalities, the construction of temporal models of motion or evolution from time-series of images, etc.
In addition, the growing availability of very large databases of biomedical images, the growing power of computers and the progress of machine learning (ML) approaches have opened up new opportunities for biomedical image analysis.
This is the reason why we decided to revisit a number of biomedical image analysis problems with ML approaches, including segmentation and registration problems, automatic detection of abnormalities, prediction of a missing imaging modality, etc. Not only those ML approaches often outperform the previous state-of-the-art solutions in terms of performances (accuracy of the results, computing times), but they also tend to offer a higher flexibility like the possibility to be transferred from one problem to another one with a similar framework. However, even when successful, ML approaches tend to suffer from a lack of explanatory power, which is particularly annoying for medical applications. We also plan to work on methods that can interpret the results of the ML algorithms that we develop.
3.3 Imaging & Phenomics, Biostatistics
The human phenotype is associated with a multitude of heterogeneous biomarkers quantified by imaging, clinical and biological measurements, reflecting the biological and patho-physiological processes governing the human body, and essentially linked to the underlying individual genotype. In order to deepen our understanding of these complex relationships and better identify pathological traits in individuals and clinical groups, a long-term objective of e-medicine is therefore to develop the tools for the joint analysis of this heterogeneous information, termed Phenomics, within the unified modeling setting of the e-patient.
To date the most common approach to the analysis of the joint variation between the structure and function of organs represented in medical images, and the classical -omics modalities from biology, such as genomics or lipidomics, is essentially based on the massive univariate statistical testing of single candidate features out of the many available. This is for example the case of genome-wide association studies (GWAS) aimed at identifying statistically significant effects in pools consisting of up to millions of genetics variants. Such approaches have known limitations such as multiple comparison problems, leading to underpowered discoveries of significant associations, and usually explain a rather limited amount of data variance . Although more sophisticated machine learning approaches have been proposed, the reliability and generalization of multivariate methods is currently hampered by the low sample size relatively to the usually large dimension of the parameters space.
To address these issues this research axis investigates novel methods for the integration of this heterogeneous information within a parsimonious and unified multivariate modeling framework. The cornerstone of the project consists in achieving an optimal trade-off between modeling flexibility and ability to generalize on unseen data by developing statistical learning methods informed by prior information, either inspired by "mechanistic" biological processes, or accounting for specific signal properties (such as the structured information from spatio-temporal image time series). Finally, particular attention will be paid to the effective exploitation of the methods in the growing Big Data scenario, either in the meta-analysis context, or for the application in large datasets and biobanks.
Federated learning in multi-centric studies. The current research scenario is characterised by medium/small scale (typically from 50 to 1000 patients) heterogeneous datasets distributed across centres and countries. The straightforward extension of learning algorithms successfully applied to big data problems is therefore difficult, and specific strategies need to be envisioned in order to optimally exploit the available information. To address this problem, we focus on learning approaches to jointly model clinical data localized in different centres. This is an important issue emerging from recent large-scale multi-centric imaging-genetics studies in which partners can only share model parameters (e.g. regression coefficients between specific genes and imaging features), as represented for example by the ENIGMA imaging-genetics study, led by the collaborators at University of Southern California. This problem requires the development of statistical methods for federated model estimation, in order to access data hosted in different clinical institutions by simply transmitting the model parameters, that will be in turn updated by using the local available data. This approach is extended to the definition of stochastic optimization strategies in which model parameters are optimized on local datasets, and then summarized in a meta-analysis context. Finally, this project studies strategies for aggregating the information from heterogeneous datasets, accounting for missing modalities due to different study design and protocols. The developed methodology finds important applications within the context of Big Data, for the development of effective learning strategies for massive datasets in the context of medical imaging (such as with the UK biobank), and beyond.
3.4 Computational Anatomy, Geometric Statistics
Computational anatomy is an emerging discipline at the interface of geometry, statistics and image analysis which aims at developing algorithms to model and analyze the biological shape of tissues and organs. The goal is not only to establish generative models of organ anatomies across diseases, populations, species or ages but also to model the organ development across time (growth or aging) and to estimate their variability and link to other functional, genetic or structural information. Computational anatomy is a key component to support computational physiology and is evidently crucial for building the e-patient and to support e-medicine.
Pivotal applications include the spatial normalization of subjects in neuroscience (mapping all the anatomies into a common reference system) and atlas to patient registration to map generic knowledge to patient-specific data. Our objectives will be to develop new efficient algorithmic methods to address the emerging challenges described below and to generate precise specific anatomical model in particular for the brain and the heart.
The objects of computational anatomy are often shapes extracted from images or images of labels (segmentation). The observed organ images can also be modeled using registration as the random diffeomorphic deformation of an unknown template (i.e. an orbit). In these cases as in many other applications, invariance properties lead us to consider that these objects belong to non-linear spaces that have a geometric structure. Thus, the mathematical foundations of computational anatomy rely on statistics on non-linear spaces.
Geometric Statistics aim at studying this abstracted problem at the theoretical level. Our goal is to advance the fundamental knowledge in this area, with potential applications to new areas outside of medical imaging. Beyond the now classical Riemannian spaces, we aim at developping the foundations of statistical estimation on affine connection spaces (e.g. Lie groups), quotient and stratified metric spaces (e.g. orbifolds and tree spaces). In addition to the curvature, one of the key problem is the introduction of singularities at the boundary of the regular strata (non-smooth and non-convex analysis).
A second objective is to develop parametric and non-parametric dimension reduction methods in non-linear space. An important issue is to estimate efficiently not only the model parameters (mean point, subspace, flag) but also their uncertainty. We also want to quantify the influence of curvature and singularities on non-asymptotic estimation theory since we always have a finite (and often too limited) number of samples. A key challenge in developing such a geometrization of statistics will not only be to unify the theory for the different geometric structures, but also to provide efficient practical algorithms to implement them.
A third objective is to learn the geometry from the data. In the high dimensional but low sample size (small data) setting which is the common situation in medical data, we believe that invariance properties are essential to reasonably interpolate and approximate. New apparently antagonistic notions like approximate invariance could be the key to this interaction between geometry and learning.
Beyond the traditional statistical survey of the anatomical shapes that is developed in computational anatomy above, we intend to explore other application fields exhibiting geometric but non-medical data. For instance, applications can be found in Brain-Computer Interfaces (BCI), tree-spaces in phylogenetics, Quantum Physics, etc.
3.5 Computational Physiology & Image-Guided Therapy
Computational Physiology aims at developing computational models of human organ functions, an important component of the e-patient , with applications in e-medicine and more specifically in computer-aided prevention, diagnosis, therapy planning and therapy guidance. The focus of our research is on descriptive (allowing to reproduce available observations), discriminative (allowing to separate two populations), and above all predictive models which can be personalized from patient data including medical images, biosignals, biological information and other available metadata. A key aspect of this scientific axis is therefore the coupling of biophysical models with patient data which implies that we are mostly considering models with relatively few and identifiable parameters. To this end, data assimilation methods aiming at estimating biophysical model parameters in order to reproduce available patient data are preferably developed as they potentially lead to predictive models suitable for therapy planning.
Previous research projects in computational physiology have led us to develop biomechanical models representing quasi-static small or large soft tissue deformations (e.g. liver or breast deformation after surgery), mechanical growth or atrophy models (e.g. simulating brain atrophy related to neurodegenerative diseases), heat transfer models (e.g. simulating radiofrequency ablation of tumors), and tumor growth models (e.g. brain or lung tumor growth).
To improve the data assimilation of biophysical models from patient data, a long term objective of our research will be to develop joint imaging and biophysical generative models in a probabilistic framework which simultaneously describe the appearance and function of an organ (or its pathologies) in medical images. Indeed, current approaches for the personalization of biophysical models often proceed in two separate steps. In a first stage, geometric, kinematic or/ functional features are first extracted from medical images. In a second stage, they are used by personalization methods to optimize model parameters in order to match the extracted features. In this process, subtle information present in the image which could be informative for biophysical models is often lost which may lead to limited personalization results. Instead, we propose to develop more integrative approaches where the extraction of image features would be performed jointly with the model parameter fitting. Those imaging and biophysical generative models should lead to a better understanding of the content of images, to a better personalization of model parameters and also better estimates of their uncertainty.
3.6 Computational Cardiology & Image-Based Cardiac Interventions
Computational Cardiology has been an active research topic within the Computational Anatomy and Computational Physiology axes of the previous Asclepios project, leading to the development of personalized computational models of the heart designed to help characterizing the cardiac function and predict the effect of some device therapies like cardiac resynchronisation or tissue ablation . This axis of research has now gained a lot of maturity and a critical mass of involved scientists to justify an individualized research axis of the new project Epione, while maintaining many constructive interactions with the 4 other research axes of the project. This will develop all the cardiovascular aspects of the e-patient for cardiac e-medicine.
The new challenges we want to address in computational cardiology are related to the introduction of new levels of modeling and to new clinical and biological applications. They also integrate the presence of new sources of measurements and the potential access to very large multimodal databases of images and measurements at various spatial and temporal scales.
4 Application domains
The main applications of our research are in the field of healthcare and more precisely the domain of digital medicine and biomedical data analysis. The axes of research presented above are related to many branches of medicine including cardiology, oncology, urology, neurology, otology, pneumology, radiology, surgery, dermatology, nuclear medicine. Within those branches, the applications cover the following different stages of medicine : prevention, diagnosis, prognosis, treatment.
5 Highlights of the year
5.1 Awards
Nicholas Ayache received the International Steven Hoogendijk Award 2020. An official ceremony for this prestigious award should take place in June 2021 in the Rotterdam City Hall.
Nicolas Guigui obtained the 2nd Prize at the 4th edition of the Pierre Laffitte Prize (October 23th).
5.2 Habilitations
Marco Lorenzi defended his Habilitation à Diriger des Recherces (HDR), entitled “Modeling Pathological Processes from Heterogeneous and High-Dimensional Biomedical Data” (January 10th, 2020).
6 New software and platforms
6.1 New software
6.1.1 CardiacSegmentationPropagation
- Keywords: 3D, Segmentation, Cardiac, MRI, Deep learning
- Functional Description: Training of a deep learning model which is used for cardiac segmentation in short-axis MRI image stacks.
- Publication: hal-01753086
- Authors: Qiao Zheng, Hervé Delingette, Nicolas Duchateau, Nicholas Ayache
- Contact: Qiao Zheng
6.1.2 CardiacMotionFlow
- Keywords: 3D, Deep learning, Cardiac, Classification
- Functional Description: Creation of a deep learning model for the motion tracking of the heart, extraction of characteristic quantities of the movement and shape of the heart to classify a sequence of cine-MRI cardiac images in terms of the types of pathologies (infarcted heart, dilated , hypertrophied, abnormality of the right ventricle).
- Authors: Qiao Zheng, Hervé Delingette, Nicholas Ayache
- Contact: Qiao Zheng
6.1.3 MedINRIA
- Keywords: Visualization, DWI, Health, Segmentation, Medical imaging
- Scientific Description: MedInria aims at creating an easily extensible platform for the distribution of research algorithms developed at Inria for medical image processing. This project has been funded by the D2T (ADT MedInria-NT) in 2010, renewed in 2012. A fast-track ADT was awarded in 2017 to transition the software core to more recent dependencies and study the possibility of a consortium creation.The Empenn team leads this Inria national project and participates in the development of the common core architecture and features of the software as well as in the development of specific plugins for the team's algorithm.
- Functional Description: MedInria is a free software platform dedicated to medical data visualization and processing.
-
URL:
https://
med. inria. fr - Contact: Olivier Commowick
- Participants: Maxime Sermesant, Olivier Commowick, Théodore Papadopoulo
- Partners: HARVARD Medical School, IHU - LIRYC, NIH
6.1.4 GP-ProgressionModel
- Name: GP progression model
- Keywords: Data modeling, Data visualization, Data integration, Machine learning, Biostatistics, Statistical modeling, Medical applications, Evolution, Brain, Uncertainly, Uncertainty quantification, Alzheimer's disease, Probability, Stochastic models, Stochastic process, Trajectory Modeling, Marker selection, Health, Statistic analysis, Statistics, Bayesian estimation
-
Functional Description:
Disease progression modeling (DPM) of Alzheimer's disease (AD) aims at revealing long term pathological trajectories from short term clinical data. Along with the ability of providing a data-driven description of the natural evolution of the pathology, DPM has the potential of representing a valuable clinical instrument for automatic diagnosis, by explicitly describing the biomarker transition from normal to pathological stages along the disease time axis.
In this software we reformulate DPM within a probabilistic setting to quantify the diagnostic uncertainty of individual disease severity in an hypothetical clinical scenario, with respect to missing measurements, biomarkers, and follow-up information. The proposed formulation of DPM provides a statistical reference for the accurate probabilistic assessment of the pathological stage of de-novo individuals, and represents a valuable instrument for quantifying the variability and the diagnostic value of biomarkers across disease stages.
This software is based on the publication:
Probabilistic disease progression modeling to characterize diagnostic uncertainty: Application to staging and prediction in Alzheimer's disease. Marco Lorenzi, Maurizio Filippone, Daniel C. Alexander, Sebastien Ourselin Neuroimage. 2019 Apr 15,190:56-68. doi: 10.1016/j.neuroimage.2017.08.059. Epub 2017 Oct 24. HAL Id : hal-01617750 https://
hal. archives-ouvertes. fr/ hal-01617750/ - Release Contributions: - New interface and output - Completely based on Pytorch
-
URL:
https://
gitlab. inria. fr/ epione/ GP_progression_model_V2 - Publication: hal-01617750
- Contact: Marco Lorenzi
- Participant: Marco Lorenzi
6.1.5 Music
- Name: Multi-modality Platform for Specific Imaging in Cardiology
- Keywords: Medical imaging, Cardiac Electrophysiology, Computer-assisted surgery, Cardiac, Health
- Functional Description: MUSIC is a software developed by the Asclepios research project in close collaboration with the IHU LIRYC in order to propose functionalities dedicated to cardiac interventional planning and guidance. This includes specific tools (algorithms of segmentation, registration, etc.) as well as pipelines. The software is based on the MedInria platform.
-
URL:
https://
team. inria. fr/ asclepios/ software/ music/ - Contact: Maxime Sermesant
- Participants: Florent Collot, Mathilde Merle, Maxime Sermesant
- Partner: IHU- Bordeau
6.1.6 SOFA
- Name: Simulation Open Framework Architecture
- Keywords: Real time, Multi-physics simulation, Medical applications
- Functional Description: SOFA is an Open Source framework primarily targeted at real-time simulation, with an emphasis on medical simulation. It is mostly intended for the research community to help develop new algorithms, but can also be used as an efficient prototyping tool. Based on an advanced software architecture, it allows the creation of complex and evolving simulations by combining new algorithms with algorithms already included in SOFA, the modification of most parameters of the simulation (deformable behavior, surface representation, solver, constraints, collision algorithm etc.) by simply editing an XML file, the building of complex models from simpler ones using a scene-graph description, the efficient simulation of the dynamics of interacting objects using abstract equation solvers, the reuse and easy comparison of a variety of available methods.
- News of the Year: The new version v20.06 has been released including new elements on SoftRobots + ModelOrderReduction integration, in addition to an improved architecture and lots of cleans and bugfixes.
-
URL:
http://
www. sofa-framework. org - Contact: Hugo Talbot
- Participants: Christian Duriez, François Faure, Hervé Delingette, Stéphane Cotin, Hugo Talbot, Maud Marchal
- Partner: IGG
6.1.7 geomstats
- Name: Computations and statistics on manifolds with geometric structures
- Keywords: Geometry, Statistic analysis
-
Scientific Description:
Geomstats is an open-source Python package for computations and statistics on manifolds. The package is organized into two main modules: “geometry“ and “learning“.
The module `geometry` implements concepts in differential geometry, and the module `learning` implements statistics and learning algorithms for data on manifolds.
The goal is to provide an easily accessible library for learning algorithms on Riemannian manifolds.
- Functional Description: Geomstats is a Python package that performs computations on manifolds such as hyperspheres, hyperbolic spaces, spaces of symmetric positive definite matrices and Lie groups of transformations. It provides efficient and extensively unit-tested implementations of these manifolds, together with useful Riemannian metrics and associated Exponential and Logarithm maps. The corresponding geodesic distances provide a range of intuitive choices of Machine Learning loss functions. We also give the corresponding Riemannian gradients. The operations implemented in geomstats are available with different computing backends such as numpy, tensorflow and keras. Geomstats manifold computations have are integrated into keras deep learning framework thanks to GPU-enabled implementations.
- Release Contributions: Major update of the library with full documentation and code restructuring before publication in JMLR.
- News of the Year: In 2020, two coding sprints were organized in order to restructure the library, to fully document it and to add tutorials. This allowed to publish papers describing the library in the SciPy conference and in the Journal of Machine Learning Research (JMLR) and to develop new applications of statistical learning on geometric objects.
-
URL:
https://
github. com/ geomstats/ - Publications: hal-02536154, hal-02908006
- Contact: Nicolas Guigui
- Participants: Nicolas Guigui, Xavier Pennec, Yann Thanwerdas, Nina Miolane
- Partner: Stanford Department of Statistics
6.1.8 MC-VAE
- Name: Multi Channel Variational Autoencoder
- Keywords: Machine learning, Artificial intelligence, Medical applications, Dimensionality reduction, High Dimensional Data, Unsupervised learning, Heterogeneity
- Scientific Description: Interpretable modeling of heterogeneous data channels is essential in medical applications, for example when jointly analyzing clinical scores and medical images. Variational Autoencoders (VAE) are powerful generative models that learn representations of complex data. The flexibility of VAE may come at the expense of lack of interpretability in describing the joint relationship between heterogeneous data. To tackle this problem, this software extends the variational framework of VAE to introduce sparsity of the latent representation, as well as interpretability when jointly account for latent relationships across multiple channels. In the latent space, this is achieved by constraining the variational distribution of each channel to a common target prior. Parsimonious latent representations are enforced by variational dropout. Experiments on synthetic data show that our model correctly identifies the prescribed latent dimensions and data relationships across multiple testing scenarios. When applied to imaging and clinical data, our method allows to identify the joint effect of age and pathology in describing clinical condition in a large scale clinical cohort.
-
Functional Description:
This software implements the work published in the paper "Sparse Multi-Channel Variational Autoencoder for the Joint Analysis of Heterogeneous Data" presented at the conference ICML 2019 (Long Beach, California, USA).
The software extends classical variational autoencoders by identifying a joint latent code associated to heterogeneous data represented in different channels. The software is implemented in Python and is based on Pytorch. It can be applied to any kind of data arrays, and provides functions for optimisation, visualisation and writing of the modelling results.
- Release Contributions: First release
- News of the Year: Method presented in the International Conference on Machine Learning (ICML 2019).
-
URL:
https://
gitlab. inria. fr/ epione_ML/ mcvae - Contact: Luigi Antelmi
- Participants: Luigi Antelmi, Marco Lorenzi, Nicholas Ayache
- Partner: CoBteK
6.1.9 SOFA-CardiacReduction
- Keywords: Simulation, 3D modeling, Model Order Reduction, Cardiac
- Scientific Description: Modification of a finite element deformation model : meshless approach and frame-based description, reduction in the number of affine degrees of freedom and integration points.
- Functional Description: This SOFA plugin is intented to build a reduced model for deformable solids (especially cardiac simulations).
- Contact: Gaetan Desrues
- Participants: Gaetan Desrues, Hervé Delingette, Maxime Sermesant
6.1.10 Fed-BioMed
- Name: A general software framework for federated learning in healthcare
- Keywords: Federated learning, Medical applications, Machine learning, Distributed Applications, Deep learning
- Scientific Description: While data in healthcare is produced in quantities never imagined before, the feasibility of clinical studies is often hindered by the problem of data access and transfer, especially regarding privacy concerns. Federated learning allows privacy-preserving data analyses using decentralized optimization approaches keeping data securely decentralized. There are currently initiatives providing federated learning frameworks, which are however tailored to specific hardware and modeling approaches, and do not provide natively a deployable production-ready environment. To tackle this issue, Fed-BioMed proposes an open-source federated learning frontend framework with application in healthcare. Fed-BioMed framework is based on a general architecture accommodating for different models and optimization methods
-
Functional Description:
The project is based on: - the development of a distributed software architecture - the establishment of a server instance from which remote experiments are triggered on the clients sites - distribution of the software to the client's sites, allowing to run machine learning models on the local data. Model parameters are then trasmitted to the server for federated aggregation
Fed-BioMed finds application in all projects based on the development of learning models for multi-centric studies.
- Release Contributions: Fed-BioMed is based on Python and Pytorch. The software was recently revised (under an ADT) to adopt the libraries Pygrid and Pysyft.
-
News of the Year:
- Ongoing ADT (2 coding sprints in 2020, 1 in 2021).
- Software presented in the article:
Silva, S., Altmann, A., Gutman, B., & Lorenzi, M. (2020). Fed-BioMed: A General Open-Source Frontend Framework for Federated Learning in Healthcare. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning (pp. 201-210). Springer, Cham.
-
URL:
https://
fedbiomed. gitlabpages. inria. fr/ - Publication: hal-02966789v1
- Authors: Marco Lorenzi, Santiago Smith Silva Rincon, Marc Vesin, Tristan Cabel, Irene Balelli, Andréa Senacheribbe, Carlos Zubiaga Pena, Jonathan Levy
- Contact: Marco Lorenzi
6.1.11 EchoFanArea
- Name: delineation of the border of the fan in ultrasound
- Keywords: Ultrasound fan area, Deep learning, Statistics, Image processing
- Functional Description: This software allows the delimitation of the acquisition cone in ultrasound imaging and the inpainting of annotations (lines, characters) inside the cone. It allows both the perfect de-identification of the images but also to standardize the content of the images. It relies on a parametric probabilistic approach to generate a training dataset with region of interest (ROI) segmentation masks. This data will then be used to train a deep U-Net network to perform the same task in a supervised manner, thus considerably reducing the calculation time of the method, one hundred and sixty times faster. These images are then processed with existing filling methods to remove annotations present within the signal area.
-
URL:
https://
gitlab. inria. fr/ hdadoun/ pre-process-US - Publication: hal-03127809
- Authors: Hind Dadoun, Hervé Delingette, Nicholas Ayache, Anne-Laure Rousseau
- Contact: Hind Dadoun
- Partner: Nhance
6.1.12 ProMFusion
- Keywords: Image segmentation, Data fusion
- Functional Description: Code related to the paper "Robust Fusion of Probability Maps" by Benoît Audelan, Dimitri Hamzaoui, Sarah Montagne, Raphaële Renard-Penna and Hervé Delingette. The proposed approach allows to fuse probability maps in a robust manner with a spatial regularization of the consensus.
-
URL:
https://
gitlab. inria. fr/ epione/ promfusion - Publication: hal-02934590
- Contact: Benoit Audelan
- Participants: Benoit Audelan, Hervé Delingette, Dimitri Hamzaoui
6.1.13 ClinicSim
- Name: Disease progression modeling for clinical intervention simulation
- Keywords: Data modeling, Clinical analysis, Clinical trial simulator, Alzheimer's disease, Pytorch, Variational Autoencoder, Ordinary differential equations
- Scientific Description: Recent failures of clinical trials in Alzheimer’s Disease underline the critical importance of identifying optimal intervention time to maximize cognitive benefit. While several models of disease progression have been proposed, we still lack quantitative approaches simulating the effect of treatment strategies on the clinical evolution. In this work, we present a data-driven method to model dynamical relationships between imaging and clinical biomarkers. Our approach allows simulating intervention at any stage of the pathology by modulating the progression speed of the biomarkers, and by subsequently assessing the impact on disease evolution.
- Functional Description: A machine-learning framework allowing to simulate the impact of intervention on the long-term progression of imaging and clinical biomarkers from collections of healthcare data
- Contact: Marco Lorenzi
- Participants: Clément Abi Nader, Marco Lorenzi, Nicholas Ayache, Philippe Robert
- Partner: Université Côte d'Azur (UCA)
6.1.14 SOFA-CardiacReduction
- Keywords: Simulation, 3D modeling, Model Order Reduction, Cardiac
- Functional Description: This SOFA plugin is intented to build a reduced model for deformable solids (especially cardiac simulations).
- Contact: Hervé Delingette
7 New results
7.1 Medical Image Analysis & Machine Learning
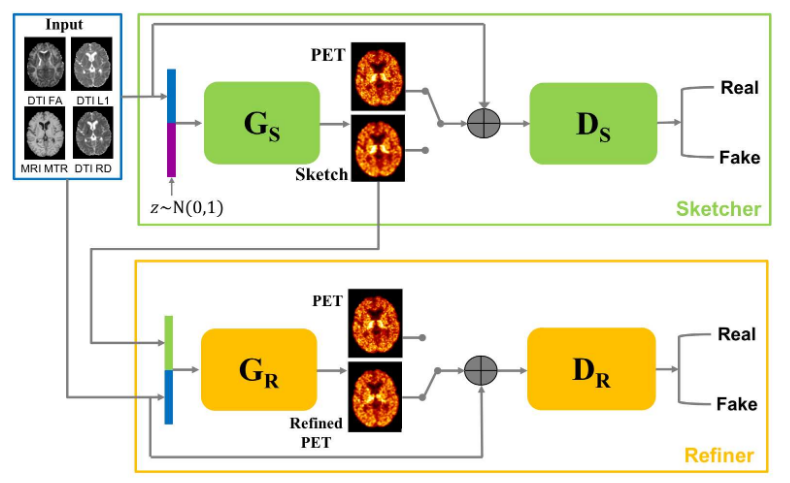
7.1.1 Predicting PET-derived demyelination from multimodal MRI using sketcher-refiner adversarial training for multiple sclerosis
Keywords: Multiple Sclerosis, MRI, PET, GANs.
Participants: Wen Wei, Nicholas Ayache, Olivier Colliot.
By using multiparametric MRI, we proposed to use a 3D FCNN to predict FLAIR MRI which is used clinically for the detection of white matter (WM) lesions. In addition, we proposed Sketcher-Refiner GANs to predict PET-derived demyelination from multiparametric MRI 14 with the following contributions:
- Learning the complex relationship between myelin content and multimodal MRI data;
- Comparing quantitatively our approach to other state-of-the-art techniques;
- Proposing visual attention saliency maps to better interpret the neural networks;
- Comparing different combinations of MRI modalities and features to assess which is the optimal input;
This work was presented during the thesis defense of Wen Wei 40.
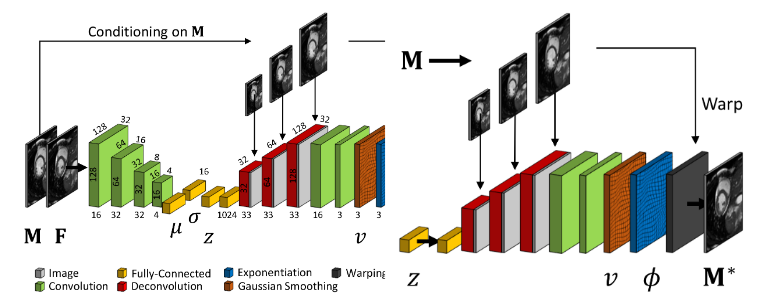
7.1.2 Learning a Probabilistic Model for Diffeomorphic Registration and Motion Modeling
This work is funded by Siemens Healthineers, Princeton, NJ, USA
Keywords: deformable registration, probabilistic motion modeling, artificial intelligence, latent variable model, deformation transport.
Participants: Julian Krebs, Hervé Delingette, Tommaso Mansi, Nicholas Ayache.
We developed a probabilistic approach for multi-scale deformable motion tracking from 2D and 3D cardiac MRI image sequences using conditional variational autoencoder.
This includes:
- A probabilistic formulation of the registration problem through unsupervised learning of an encoded deformation model.
- A generative motion model using explicit time-dependent temporal convolutional networks (Fig. 5).
- Demonstration on cardiac cine-MRI for cardiac motion tracking, simulation, transport and temporal super-resolution.
This work was presented during the thesis defense of Julian Krebs 38 and was the topic of 2 patents 54, 52.
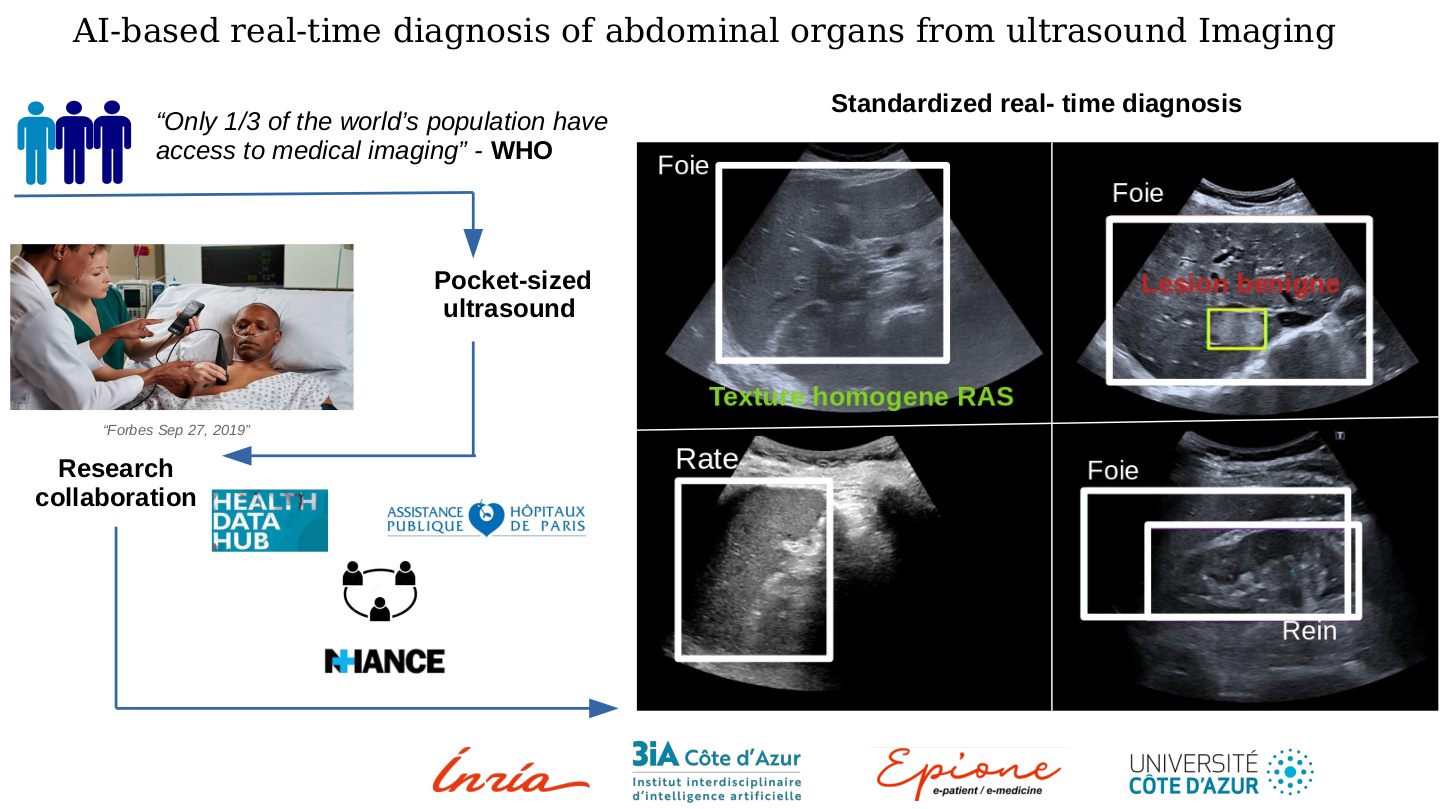
7.1.3 AI-Based Real Time Diagnosis of Abdominal Ultrasound
This work is done in collaboration with Dr Anne-Laure Rousseau (AP-HP)
Keywords: Machine Learning, Ultrasound, Computer-Aided Diagnosis.
Participants: Hind Dadoun, Hervé Delingette, Nicholas Ayache.
We worked on the use of Deep Learning-based algorithms for object detection algorithms, in order to:
- Localize and recognize abdominal organs.
- Detect, localize and characterize Focal Liver Lesions in Abdominal Ultrasound.
To support those objectives, we also proposed a preprocessing step to detect the location of the fan in ultrasound images, combining Deep Learning and Bayesian Learning 18 (Figure 6).
We also worked in collaboration with the Health Data Hub and the Data Warehouse of AP-HP to create a large dataset of abdominal ultrasound images, with more than 60,000 images.
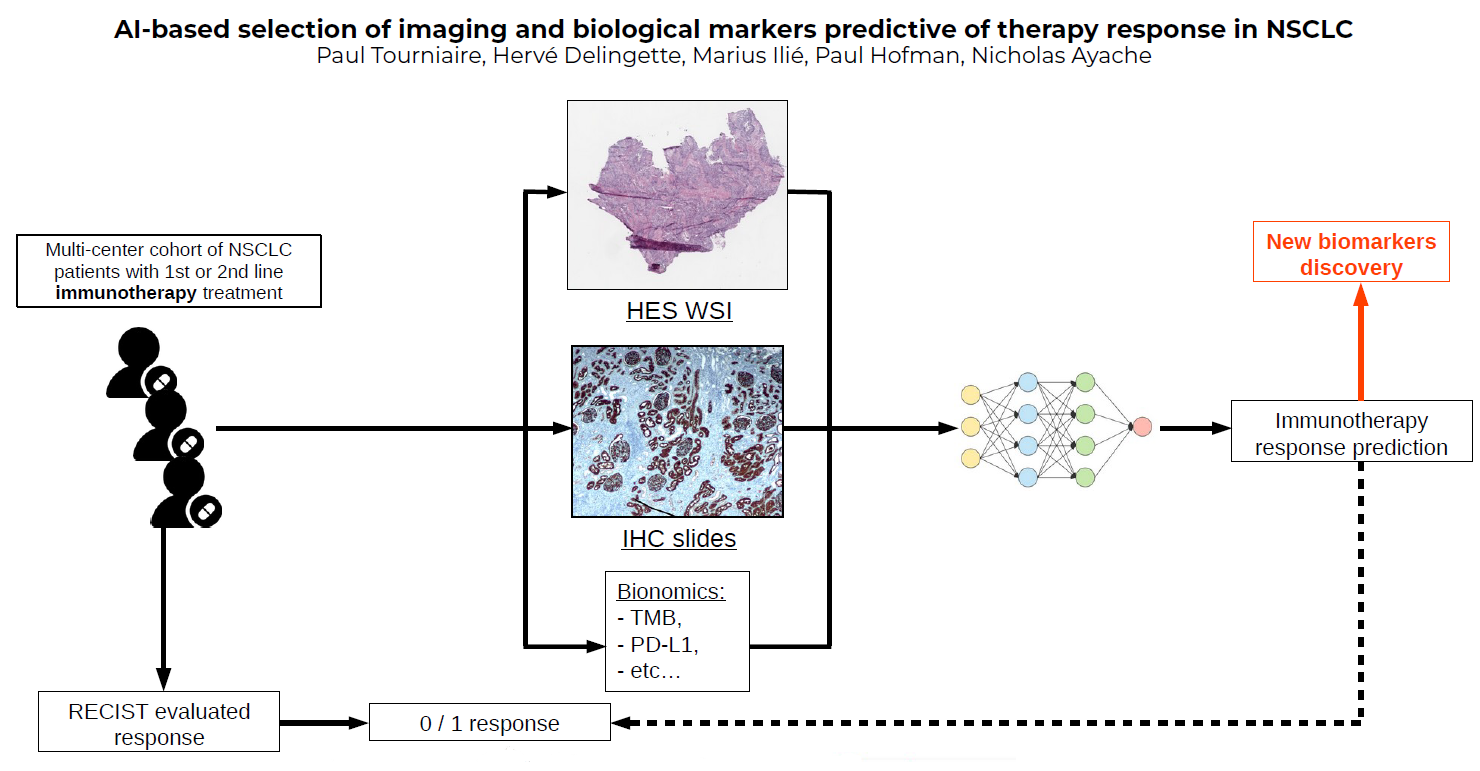
7.1.4 AI-based selection of imaging and biological markers predictive of therapy response in NSCLC
This work has been supported by the French government, through the 3IA Côte d’Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002
Keywords: Immunotherapy, multi-omics analysis, machine learning..
Participants: Paul Tourniaire, Hervé Delingette, Nicholas Ayache, Marius Ilié, Paul Hofman.
- This works aims to predict immunotherapy response for patients with non small cell lung cancer (NSCLC) based on RECIST criteria for the outcome.
- It is based on multiple instance learning and neural networks to extract predictive biomarkers of the response from histological whole slide images.
- We rely on a multi-centric cohort to test the model's robustness (Figure 7).
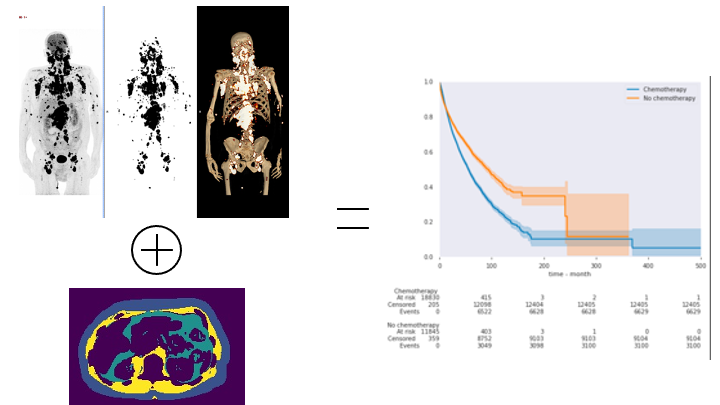
7.1.5 Leveraging artificial intelligence for Nuclear Medicine
We thank the Fond Henri Mondor for their financial support. 1
Keywords: PET/CT ; deep learning ; nuclear medicine ; anthropometry ; anomaly detection.
Participants: Paul Blanc-Durand, Hervé Delingette.
Nuclear medicine is an hybrid imaging technique that combines the morphological information from CT or MRI and metabolical information from PET. PET allows to easily detect tumors 2 and CT can be used for both a better characterization of the detected anomalies and the extraction of additional information about the nutritional status of patients 3. Combining those information in a multidimensional setting will help us to identify patients at risk of complications or to select those that will not benefit from chemotherapies (Figure 8).
7.1.6 Joint Biological & Imaging markers for the Diagnosis of severe lung diseases
Keywords: Image segmentation, Segmentation quality control, data fusion.
Participants: Benoit Audelan, Dimitri Hamzaoui, Hervé Delingette, Nicholas Ayache.
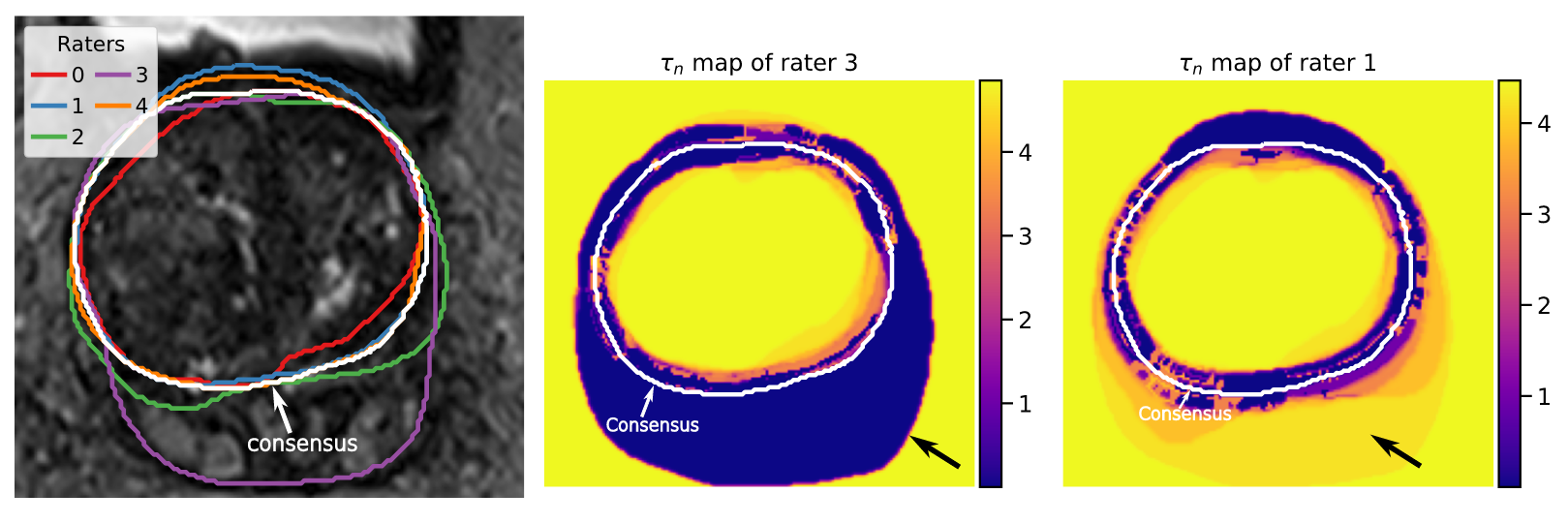
Monitoring the quality of image segmentation is critical to many clinical applications. This quality assessment can be carried out by a human expert when the number of cases is limited. However, it becomes onerous when dealing with large image databases, so partial automation of this process is required. We propose an unsupervised image segmentation quality control framework, based on a smoothness and intensity probabilistic model 1. The workflow is presented in Fig. 9. The method was tested on several datasets containing various types of images and segmented structures, showing its ability to isolate atypical cases and therefore to perform quality control assessment.
Another investigated topic is the fusion of continuous probability maps. When working on a collection of probability maps produced by several algorithms or human raters, the challenge is to weight properly the combination of maps in order to reflect the agreement among raters, the presence of outliers and the spatial uncertainty in the consensus. To this end, we expand the classical continuous STAPLE algorithm by introducing Student's t-distributions allowing local estimates of raters’ performances 16. In Fig. 10, the rater 3 is an outlier at the bottom of the image with some over-segmentation. is the scale variable that modulates the local contribution of each rater to the consensus. We can see that rater 3 exhibits smaller tau values at the bottom than rater 1 and is therefore less contributing to the consensus in this area. Moreover, the introduction of bias and spatial priors leads to proper rater bias estimates and a control over the smoothness of the consensus map.
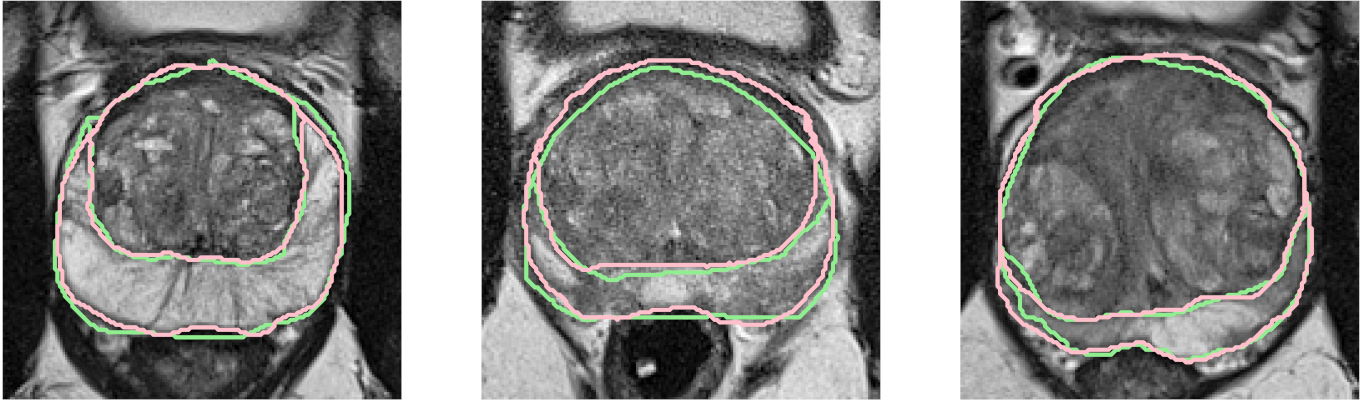
7.1.7 AI-Based Diagnosis of Prostate Cancer from Multiparametric MRI
This work is funded through the DS4H doctoral school, and is in collaboration with Pr Renard-Penna and Dr Montagne, from AP-HP.
Keywords: Image segmentation, Prostate Cancer, Variability.
Participants: Dimitri Hamzaoui, Benoît Audelan, Hervé Delingette, Nicholas Ayache.
The objective of our work is to build a deep learning based model for the zonal segmentation of the prostate from T2-weighted MRI 25 (Figure 11), to facilitate the construction of an atlas of the prostate for further applications, such as detection and characterization of tumoral lesions.
We also worked on the characterization of the inter-rater variability of zonal segmentation of the prostate according to their characteristics, and on an automatized method to characterize variability of segmentations between several raters 16.
7.1.8 A Deep Learning based Fast Signed Distance Map Generation
This work is funded by the Provence-Alpes-Côte-d'Azur region, the Université Côte d’Azur and Oticon Medical within the CIMPLE research project.
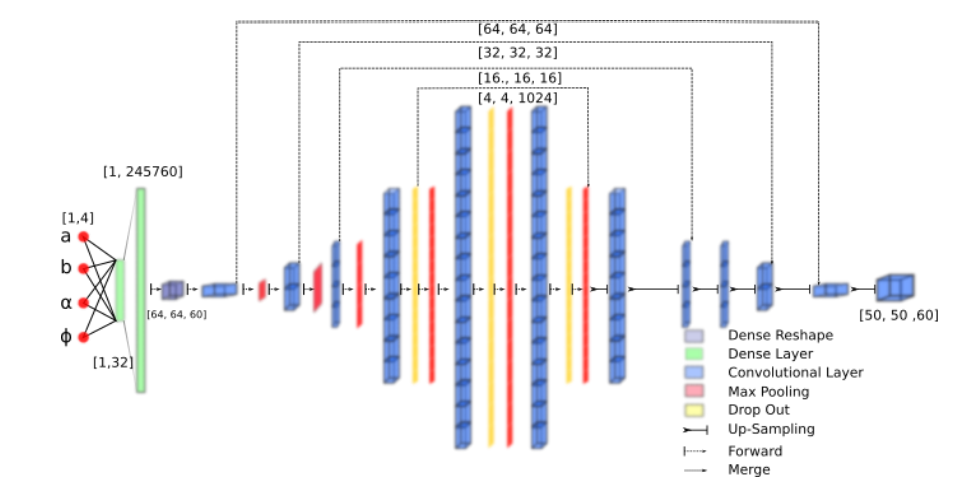
Keywords: Signed Distance Map, Cochlea Shape Model.
Participants: Zihao Wang, Clair Vandersteen, Thomas Demarcy, Dan Gnansia, Charles Raffaelli, Nicolas Guevara, Hervé Delingette.
We propose a neural network to generate Signed Distance Map (SDM) from shape parameters 24.- We propose the SDM Neural Network (Fig. 12) that generates a cochlea signed distance map depending on four input parameters.
- We show that the deep learning approach leads to a 60 fold improvement in terms of computation time compared to more classical SDM generation methods.
7.1.9 Quasi-Symplectic Langevin Variational Autoencoder.
This work is funded by the Provence-Alpes-Côte-d'Azur region, the Université Côte d’Azur and Oticon Medical through CIMPLE research project.
Keywords: Stochastic Inference, VAE, Statistical Learning.
Participants: Zihao Wang, Hervé Delingette.
We propose a new flow-based Bayesian inference framework by introducing the quasi-symplectic Langevin dynamic for building a stochastic Evidence Lower Bound 47. The proposed approach can reduce significantly the GPU memory requirement for training this deterministic dynamic flow-based inference method.7.2 Imaging & Phenomics, Biostatistics
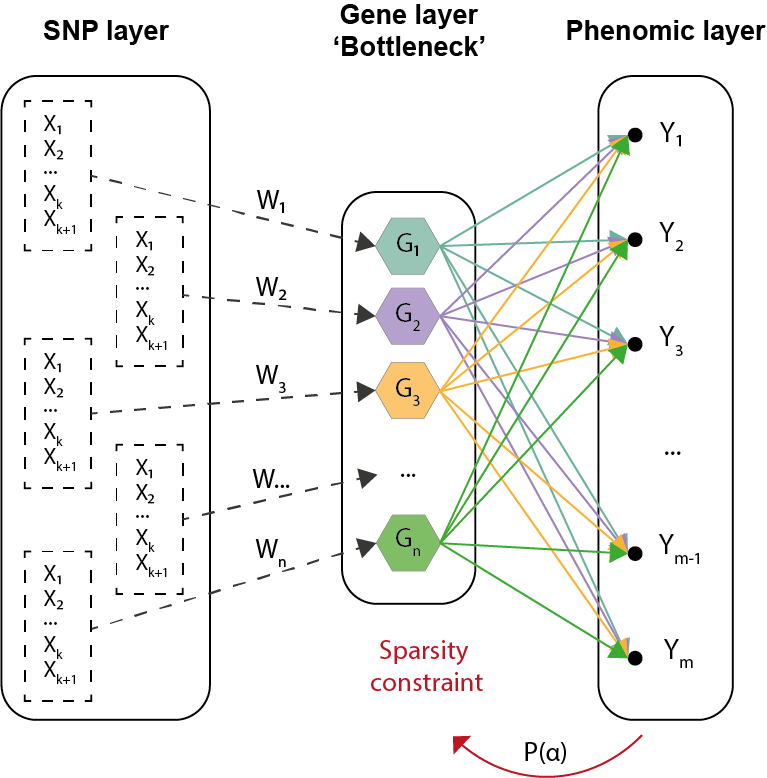
7.2.1 Development of biologically inspired constrained Bayesian neural network for the integration of multi-omics data
Work funded by UCA Neuromod institut
Keywords: Deep Learning, Multi-omics, Alzheimer's disease, Biological constraints.
Participants: Marie Deprez, Marco Lorenzi, Maxime Sermesant.
This project aims to integrate multi-omics data using deep learning methods to improve understanding of complex biological conditions and identify key risk factors in diseases (Alzheimer's Disease, Atrial Fibrillation).
- Work inspired from system biology dogma which uses the combined analysis of multiple biological read-outs to get a better understanding of a biological system as a whole;
- Development of a Bayesian neural network with biologically inspired constraint to associate/integrate multi-omics data (genetic information grouped by genes which relate to phenotype, Figure 13);
- Introduction of a sparsity constraint to pinpoint key features which have a statistical influence on the output layer (phenomic features).
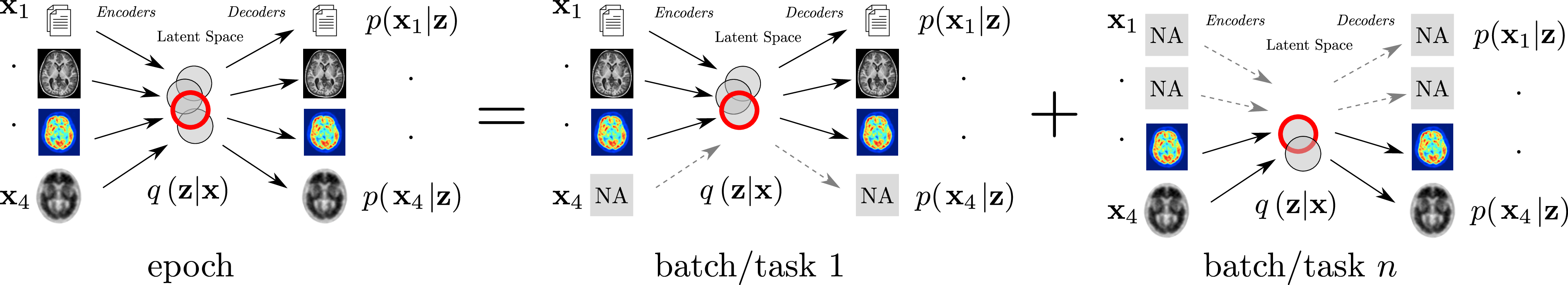
7.2.2 Combining Multi-Task Learning and Multi-Channel Variational Auto-Encoders to Exploit Datasets with Missing Observations - Application to Multi-Modal Neuroimaging Studies in Dementia
Acknowledgments: IDEX UCAJEDI MNC3 Project (ANR-15-IDEX-01); 3IA (ANR-19-P3IA-0002); the OPAL infrastructure from Université Côte d'Azur.
Keywords: Multi Task Learning, Missing Data, Multimodal Data Analysis..
Participants: Luigi Antelmi, Nicholas Ayache, Philippe Robert, Federica Ribaldi, Valentina Garibotto, Giovanni B. Frisoni, Marco Lorenzi.
The aim of this work is to build scalable learning models for the joint analysis of heterogeneous biomedical data, to improve diagnosis, treatment, and monitoring of neurological and psychiatric diseases, in collaboration with the Institut Claude Pompidou (CHU of Nice), within the MNC3 initiative (Médecine Numérique: Cerveau, Cognition, Comportement). Data comes from collections of brain imaging, biological and clinical evaluations available in current large-scale databases such as ADNI and local clinical cohorts, where the joint analysis is impaired by data missingness, that is non-overlapping sets of modalities across subjects (e.g. imaging data, clinical scores, biological measurements) and across datasets. The main contributions and results of this work 41 are:
- a multi-task generative latent-variable model (Fig.14) where the common variability across datasets stems from the estimation of a shared latent representation across heterogeneous data modalities;
- to consistently analyze high-dimensional and heterogeneous information with missing data;
7.2.3 Statistical Learning of Heterogeneous Data in Large-Scale Clinical Databases
Acknowledgments: IDEX UCAJEDI MNC3 Project (ANR-15-IDEX-01); 3IA (ANR-19-P3IA-0002); the OPAL infrastructure from Université Côte d'Azur.
Keywords: Ordinary Differential Equations, Generative models, Alzheimer's Disease, Disease Progression Modelling.
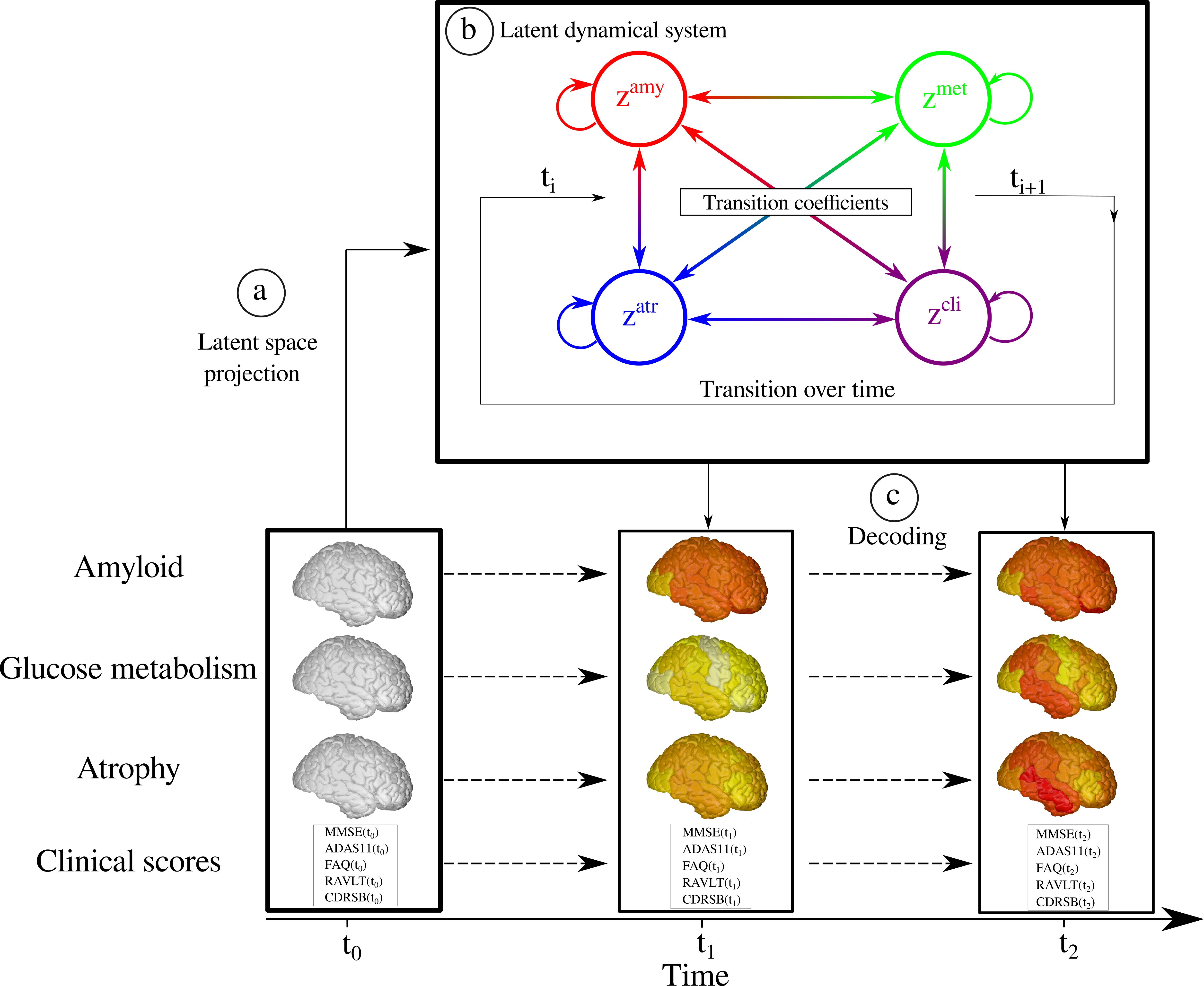
Participants: Clement Abi Nader, Nicholas Ayache, Philippe Robert, Marco Lorenzi.
Alzheimer's disease is a neurodegenerative disorder whose dynamics still remain partially unknown. The aim of this work is to develop computational models to better understand Alzheimer's disease progression, based on the analysis of imaging and clinical data. Lately, a collaboration with the LANVIE laboratory from the Geneva University Hospitals (HUG) has allowed to test the developed model in a clinical context.
- Development of statistical models for describing the evolution of Alzheimer's disease.
- Recent work focuses on simulating the effect of drug intervention on the progression of Alzheimer's disease 8.
- The method relies on probabilistic generative model, coupled with a system of Ordinary Differential Equations (ODE) to model Alzheimer's disease (Figure 15).
7.2.4 Bayesian latent variables model for time-series analysis
This work is funded by Inria-SAM
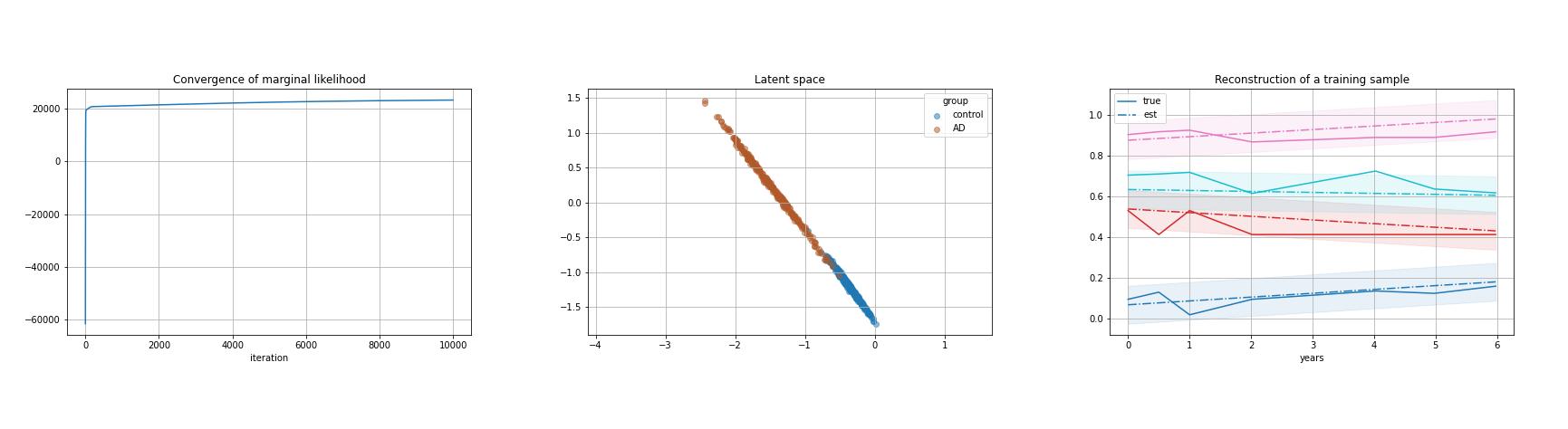
Keywords: Time series, Latent variables, Bayesian modelling, PPCA.
Participants: Andrea Senacheribbe, Marco Lorenzi, Irene Balelli.
This project aims to develop a latent variable model for the analysis of time series data, based on the Bayesian framework of Probabilistic Principal Component Analysis (PPCA). During this work:
- we analytically formulated the model in its centralised version
- we implemented the model in Python, paying particular attention on the computational efficiency
- we tested the model on synthetically generated data
- we tested the model on clinical scores and MRI brain volumes of Alzheimer's patients from the ADNI dataset, see Figure 16
7.2.5 Exploring latent dynamical models for failure prediction in time-series of high-dimensional and heterogeneous data
The work was funded by a contract with the startup MyDataModels, and by 3IA Côte d'Azur
Keywords: Anomaly Detection, Time-Series, High-Dimensional, Neural ODE, Machine Learning, EEG.
Participants: Etrit Haxholli, Marco Lorenzi.
Anomaly detection is the identification of rare observations which differ significantly from the majority of the data. The goal of this project is to provide a novel general algorithm for real-time anomaly detection in time-series of high-dimensional and heterogeneous data. Our model will be applied to the analysis of epilepsy in Electroencephalography (EEG) data-sets, such as the one provided by our collaborators, Dr Massimo Mantegazza (CNRS) and Dr Fabrizio Capitano (CNRS).
7.3 Computational Anatomy & Geometric Statistics
7.3.1 Uncertainty estimation in a geometric framework
This work is funded by ERC AdG G-Statistics.
Keywords: PCA, Central limit theorems on manifolds..
Participants: Dimbihery Rabenoro, Xavier Pennec.
This project aims to study the uncertainty of the estimation of the mean and higher dimensional sub-spaces in extensions of PCA to manifolds.
7.3.2 Lie group invariant Riemannian metrics on covariance and correlation matrices
This work was partially supported by the EU H2020 ERC AdV grant G-Statistics (No 786854), by the UCAJEDI IDEX (ANR-15-IDEX-01) and by the 3IA Côte d’Azur (ANR-19-P3IA-0002).
Keywords: Covariance matrices, correlation matrices, symmetric positive definite matrices, Riemannian geometry, group actions.
Participants: Yann Thanwerdas, Xavier Pennec.
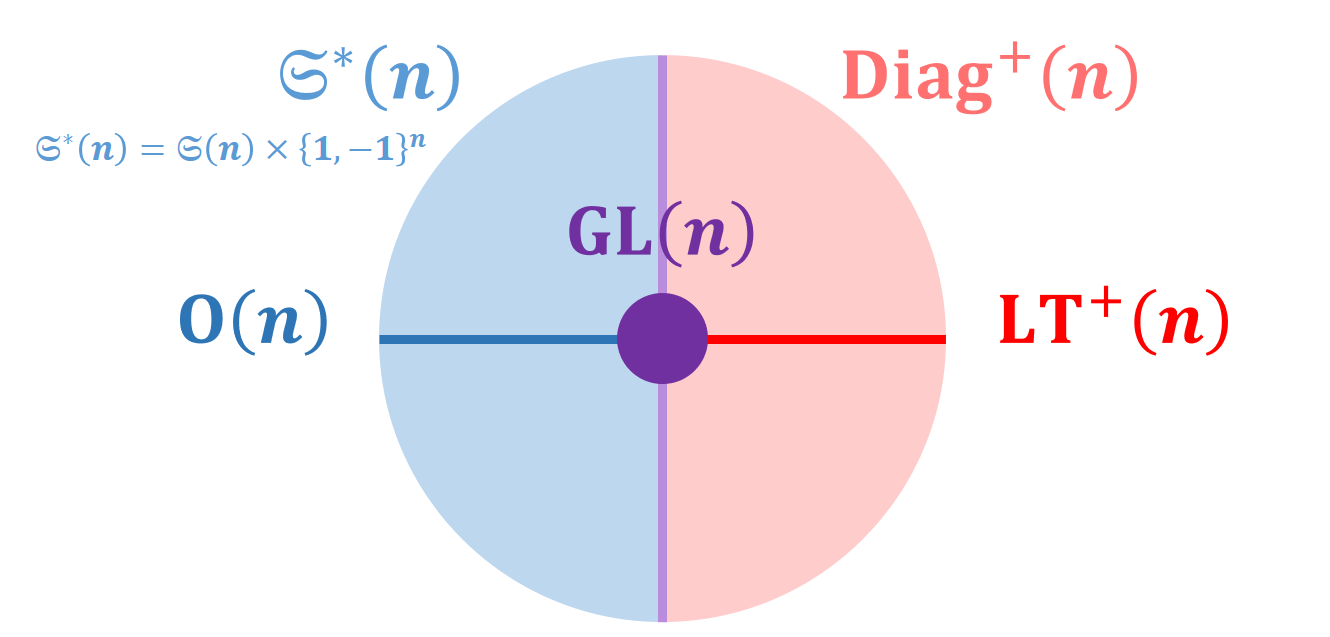
Covariance and correlation matrices are Symmetric Positive Definite matrices which have been used in many fields of medical data analysis. We introduced new structures on these spaces based on invariance under Lie group actions (Figure 18):
- characterization of -invariant metrics
- characterization of permutation-invariant Euclidean metrics
- fundamental Riemannian quotient operations of the quotient-affine metric
- families of Lie-Cholesky metrics and quotient-Lie-Cholesky metrics.
7.3.3 Numerical accuracy of parallel transport with pole ladder
This work was partially supported by the EU H2020 ERC AdV grant G-Statistics (No 786854) and by the 3IA Côte d’Azur (ANR-19-P3IA-0002).
Keywords: Shape Analysis, parallel transport, LDDMM, numerical accuracy.
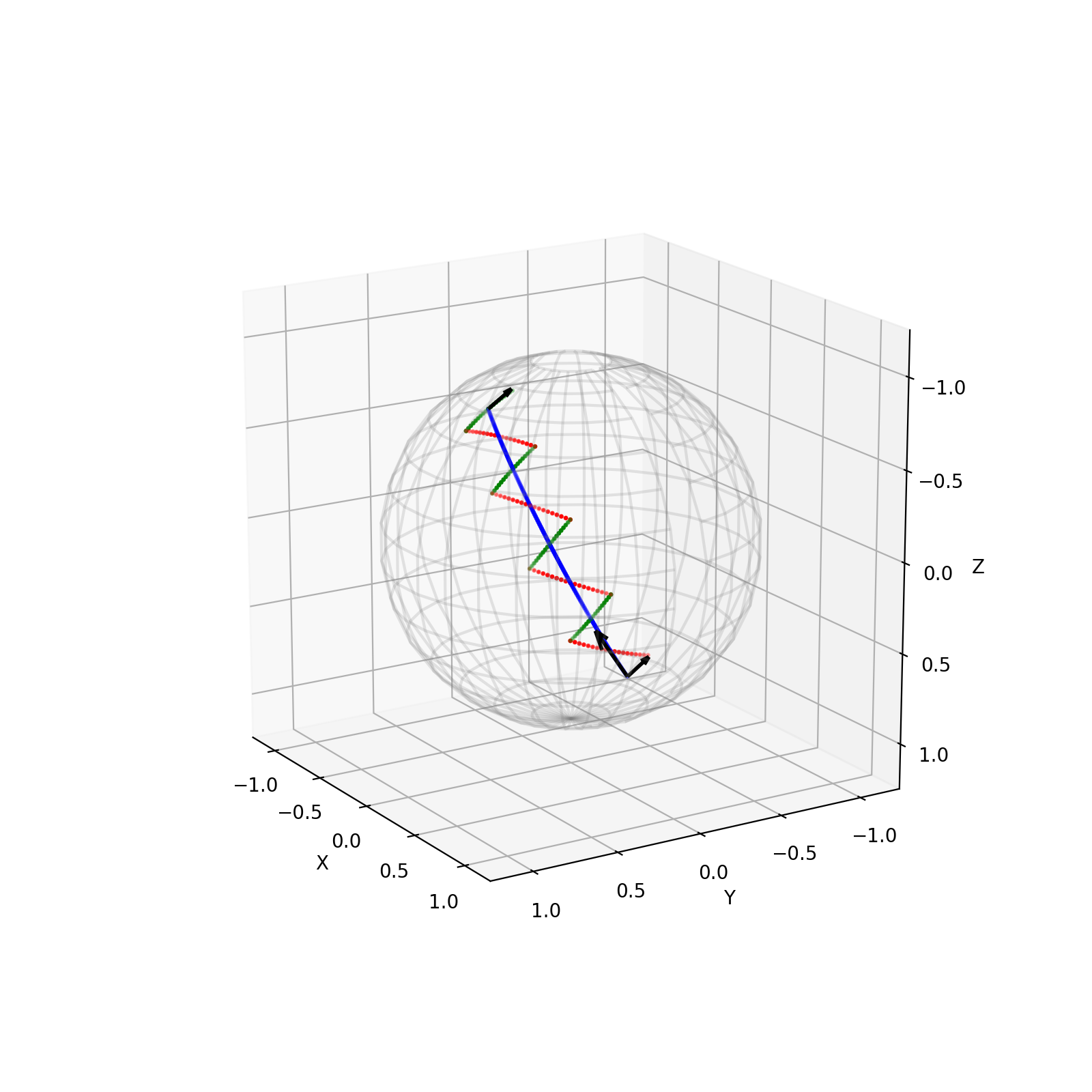
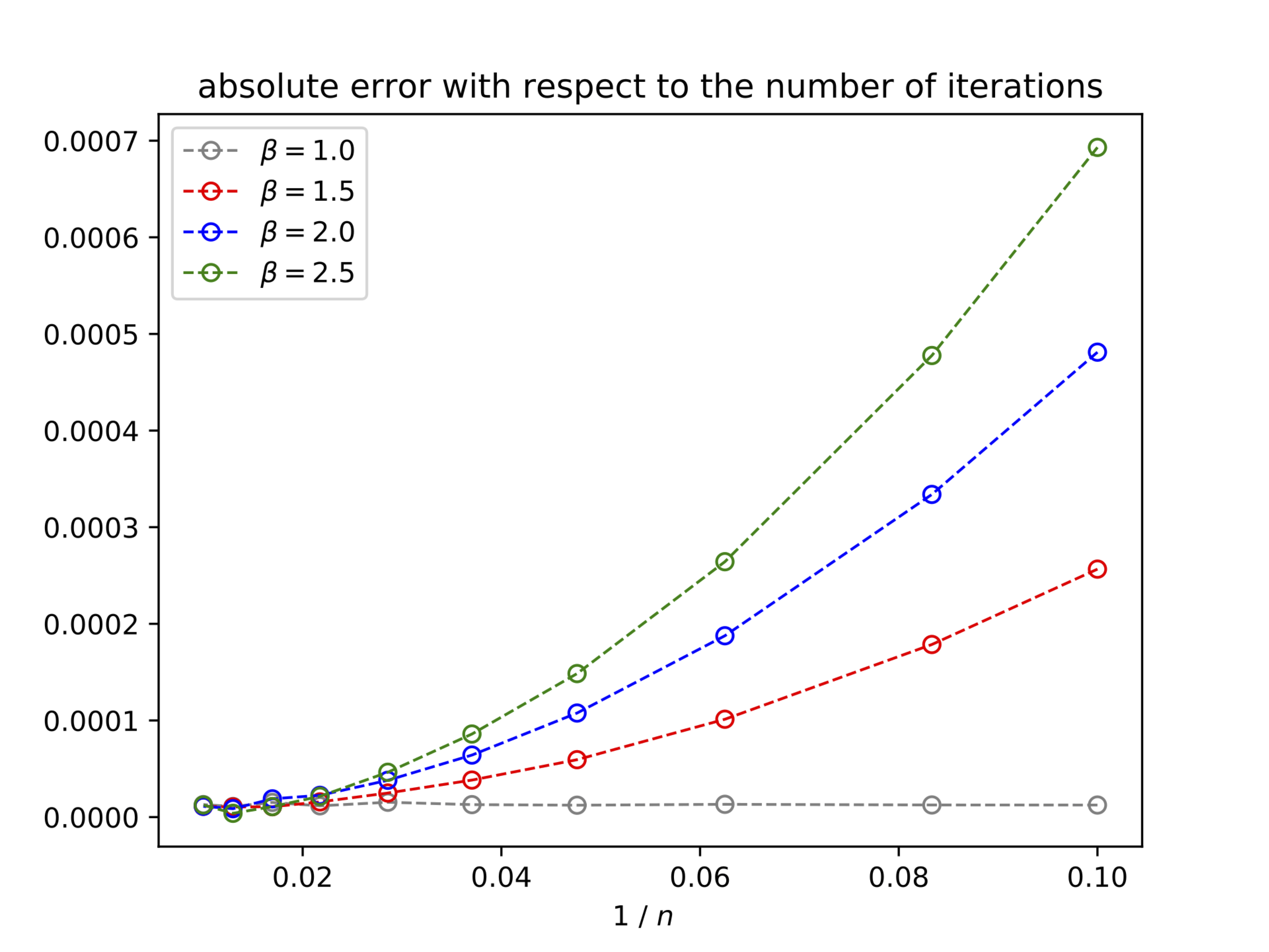
Participants: Nicolas Guigui, Xavier Pennec.
The work of 43 focused on the Pole Ladder algorithm, an approximation of parallel transport on manifolds based on the construction of geodesic parallelograms (Figure 19). We proved that this algorithm converged with a quadratic speed in the number of steps, with a rate proportional to the Riemannian curvature. This method is more efficient than other state-of-the-art methods by an order of magnitude. Illustrations on the 2-sphere and the special Euclidean group with an anisotropic metric show that the theoretical errors we have established are measured with a high accuracy (Figure 20).
7.3.4 Advances in geometric statistics
This work was partially supported by the EU H2020 ERC AdV grant G-Statistics (No 786854) and by the 3IA Côte d’Azur (ANR-19-P3IA-0002).
Keywords: Lie group, Riemannian manifolds, statistical models, dimension reduction.
Participants: Xavier Pennec, Nicolas Guigui, Emmanuel Chevalier, Nina Miolane, Alice Le Brigant.
In 2020, an important collective effort was done on the development of the open-source Python package Geomstats that implements the main operations and algorithms of geometric statistics. The package encompasses for the first time very complex notions of Riemannian geometry into a consistent object-oriented API that makes it easy for mathematicians to add their code. From an applied perspective, the use of the algorithms does not require a deep understanding of the mathematics behind - which is an evident strength of the package. The package received an excellent feedback so far 20, 9.
Thanks to the new computing possibilities offered by this package, we collaborated to illustrate several new statistical models: with E. Chevalier, a statistical model of wrapped densities for bi-invariant statistics on the group of rigid motions of a Euclidean space was studied 4 ; with Alice Le Brigant, the information geometry of distributions was studied in 45, with applications to the clustering of medical data.
On a more abstract level, foundations for a geometric view of statistics were established in 44. A summary of the generalization of principal component analysis to Riemannian manifolds using barycentric subspaces was published in the book chapter 32.
7.3.5 Statistical shape analysis of faces for computer aided dermatology and plastic surgery
Supported by the company Quantificare through a CIFRE funding (ANRT no 2019/0101).
Keywords: Gaussian Processes, non rigid registration.
Participants: Florent Jousse, Xavier Pennec, Hervé Delingette, Matilde Gonzalez.
The objective of this work is to model complex face deformations such as natural aging, facial expressions, surgical interventions and posture motions to improve the 3D reconstruction of faces and to normalize their analysis. It includes the development of non-rigid registration methods of textured meshes and their statistical modeling.
7.4 Computational Cardiology & Image-Based Cardiac Interventions
7.4.1 Re-entrant Ventricular Tachycardia Simulation from Non-invasive Data
This work is funded by the IHU Liryc, Bordeaux.
Keywords: computed tomography, cardiac electrophysiology, image-based modelling, ventricular tachycardia.
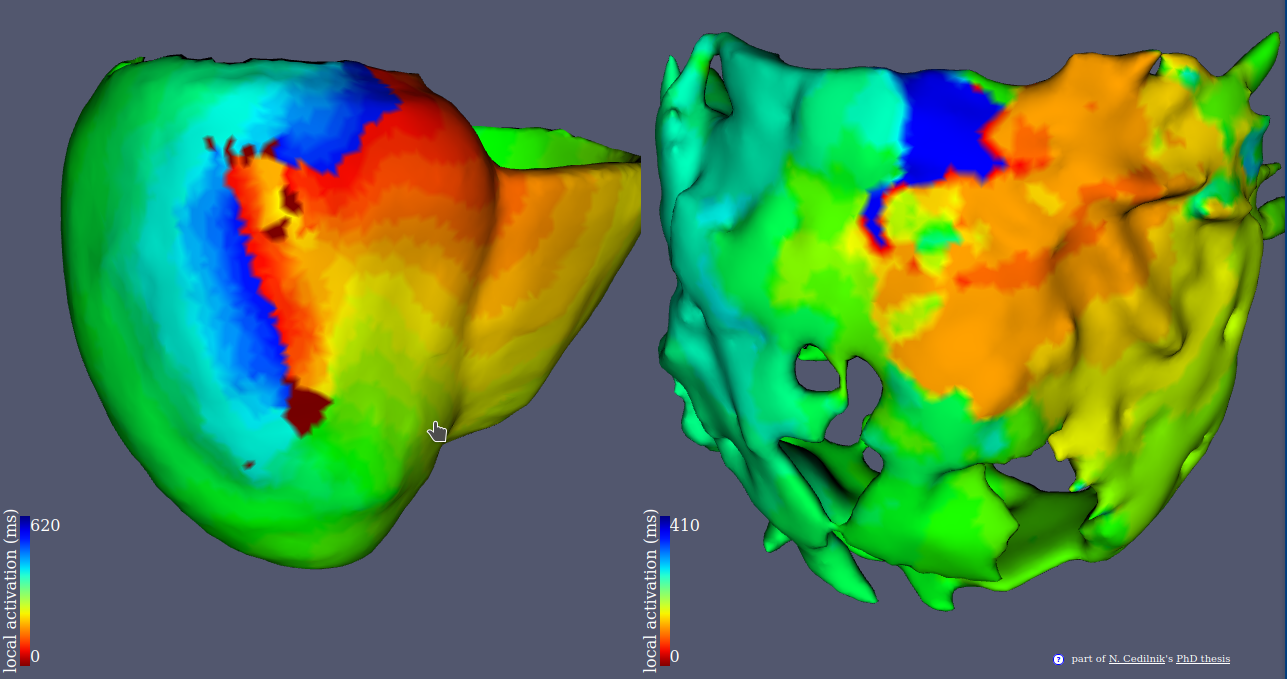
Participants: Nicolas Cedilnik, Pierre Jaïs, Frédéric Sacher, Hubert Cochet, Maxime Sermesant.
This project aims at improving ventricular tachycardia ablation procedures planning with in silico electro-anatomical exploration of the arrhythmogenic substrate using cadiac imaging as a support for model personalisation.
- We conducted a large scale study of the relationship between imaging and electrophysiological features using in-vivo human data (Figure 20).
- We used our findings to parametrise a reaction diffusion model of activation wave propagation.
- We showed that this personalisation framework was able to reproduce re-entrant patterns that match intra cardiac.
This work was presented during Nicolas Cedilnik's PhD defense in front of an international jury 37.
7.4.2 Deep Learning Formulation of ECGI for Data-driven Integration of Spatiotemporal Correlations and Imaging Information
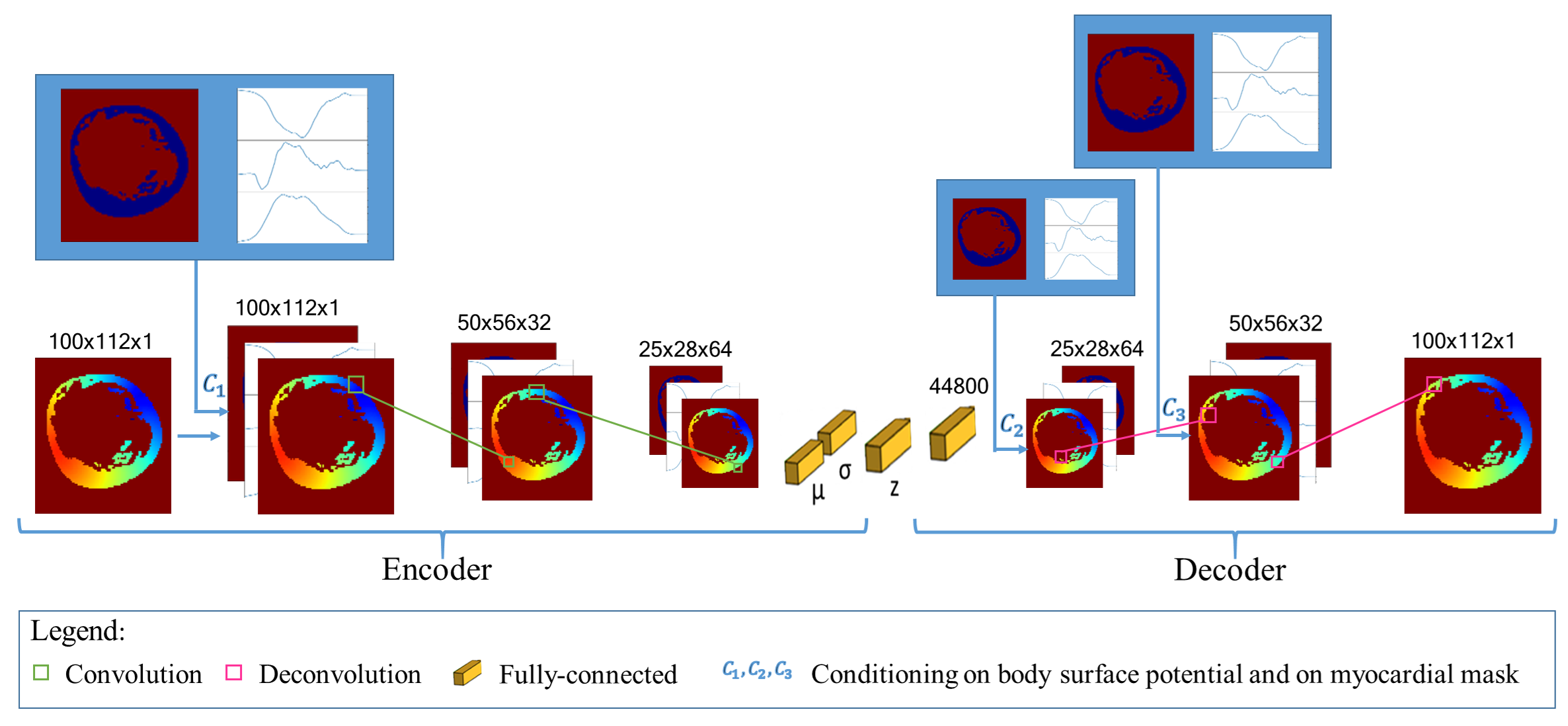
This work is funded within the ERC Project ECSTATIC with the IHU Liryc, in Bordeaux.
Keywords: Deep Learning, Electrocardiographic Imaging, Inverse problem of ECG, Electrical simulation, Generative Model..
Participants: Tania-Marina Bacoyannis, Hubert Cochet, Maxime Sermesant.
Electrocardiographic Imaging (ECGI) is a promising tool to map the electrical activity of the heart non-invasively using Body Surface Potentials (BSP). However, it is still challenging due to the mathematically ill-posed nature of the inverse problem to solve. Novel approaches leveraging progress in artificial intelligence could alleviate these difficulties. To this end, we extended to 3D our previous work2, where a novel Deep Learning method based on Conditional Variational Autoencoder was designed to solve this problem in 2D (Figure 22).
Next, we will test it on real data provided by the IHU Liryc.
7.4.3 Deep learning and cardiac electrical activity modelling using 3D non-invasive cardiac imaging: for the application of Sudden Cardiac Death prevention and ventricular arrhythmia treatment.
This project is funded by University Côte d'Azur and under the co-direction of Maxime Sermesant (Inria) and Hubert Cochet (IHU Liryc).
Keywords: Sudden Cardiac Death, Ventricular Arrhythmia, Cardiac Imaging, Cardiac Modelling, Deep Learning,.
Participants: Buntheng Ly, Hubert Cochet, Maxime Sermesant.
The aim of this project is to build an automatic pipeline for ventricular arrhythmia prediction, in order to develop tools for Sudden Cardiac Death prevention (Figure 23). To this end, we use Deep Learning and cardiac simulation to extract relevant features from 3D non-invasive cardiac imaging, including CT and MR imaging. Furthermore, we also wish to identify high risk features by utilizing Explainable Learning.
This is a joint project with IHU Liryc, as the medical collaborator. It also exploits a large imaging and clinical database collected by the Liryc institute.

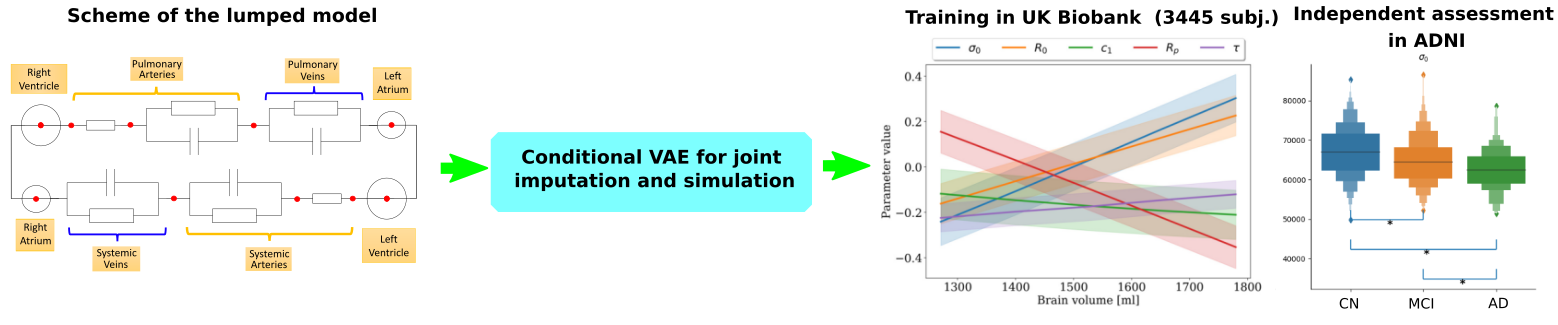
7.4.4 Discovering the link between cardiovascular pathologies and neurodegeneration through biophysical and statistical models of cardiac and brain images
This project is funded by Université Côte d'Azur (UCA)
Keywords: Lumped models - Biophysical simulation - Statistical learning.
Participants: Jaume Banus Cobo, Maxime Sermesant, Marco Lorenzi, Oscar Camara Rey.
The project aims at developing a computational model of the relationship between cardiac function and brain damage from large‐scale clinical databases of multi‐modal and multi‐organ medical images (Figure 24). The model is based on advanced statistical learning tools for discovering relevant imaging features related to cardiac dysfunction and brain damage. However, the limited cardiac data available in datasets focused on neurodegenerative diseases hinders the study of the role of cardiac function in these diseases. Hence, we developed a joint imputation framework based on variational inference and Gaussian process (GP) regression. The GP emulates the cardiac dynamics of a mechanistic model and constraints the data imputation conditioned on the available brain information 17.
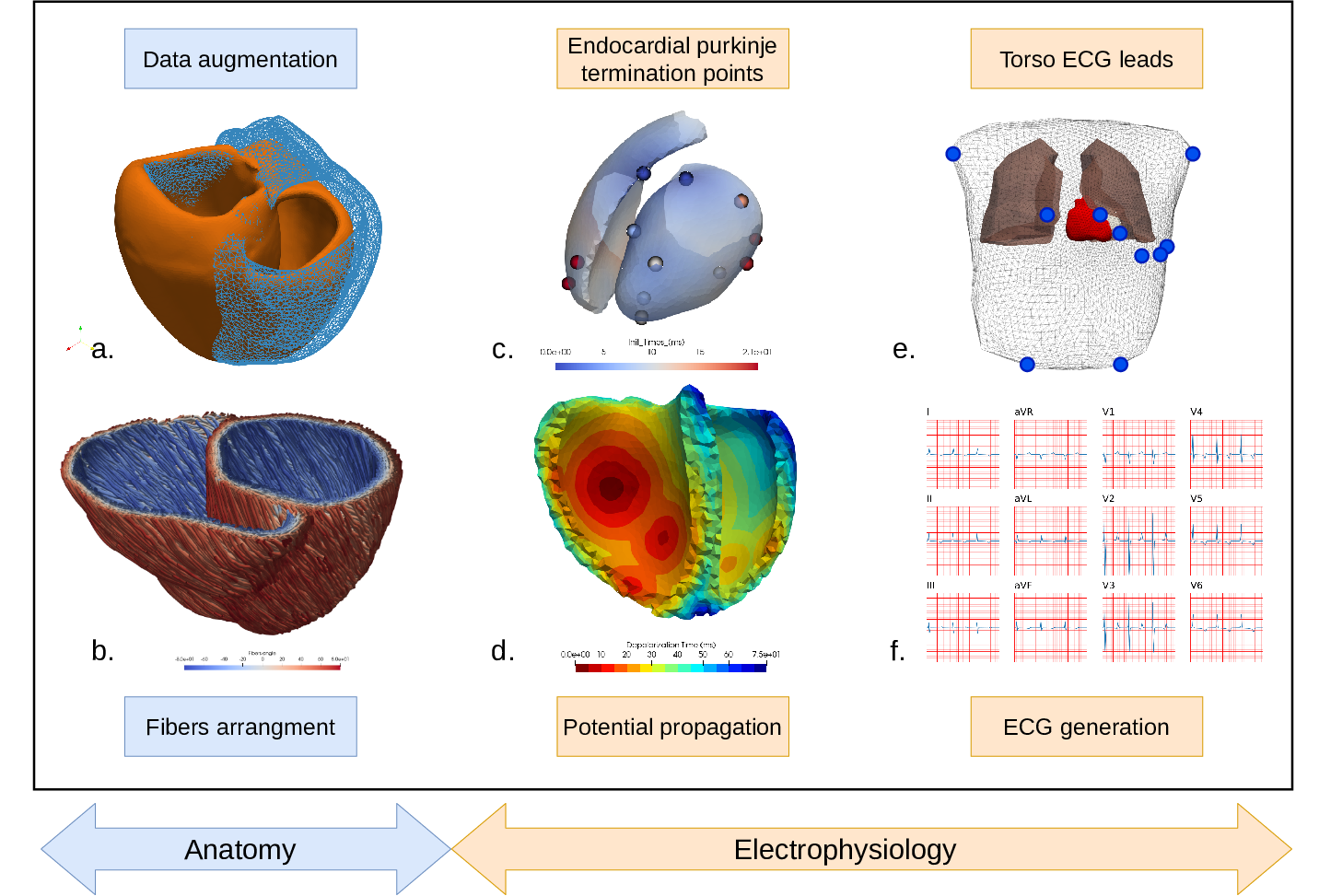
7.4.5 3D electromechanical cardiac modelling for heart failure patients stratification and prediction of cardiac resynchronisation therapy response
Keywords: Cardiac electrophysiology, Patient-specific modeling.
Participants: Gaëtan Desrues, Serge Cazeau, Thierry Legay, Hervé Delingette, Delphine Feuerstein, Maxime Sermesant.
Patient-specific 3D models can help in improving patient selection, therapy optimisation and interventional guidance. The aim of this project is to study cardiac desynchronisation and the resynchronisation therapy, based on personalised models of the heart.
Given a patient-specific geometry of the heart (often extracted from MRI or CT), muscle fibre orientations are generated in the myocardium (Fig.25b). Some algorithms have been developed to introduce deformations to the generated meshes, allowing to simulate various pathological cases (Fig.25a).
At each cardiac cycle, the mechanical contraction of the heart is driven by the electrical activation. The electrical signal is conducted along the His bundles and the Purkinje fibres to the endocardium (Fig.25c). Then, the anisotropic propagation of the activation potential is computed with an Eikonal model (Fig.25d). After a short period of time (duration of the QRS), all the myocardial cells are activated and contract until the repolarisation wave arrives.
Finally, using a dipole formulation, it is possible to record the electrical potential at the 10 leads placed on a virtual torso (Fig.25e) and to plot the corresponding ECG (Fig.25f). This electrocardiogram is a tool for cardiologists to detect cardiac desynchronisation, or more generally, cardiac pathologies.
This electrical activation can then be used to simulate cardiac contraction.
7.4.6 Segmentation and motion tracking for 2D Echocardiography
This work is funded by Inria
Keywords: UNet Segmentation, Polyaffine Registration.
Participants: Yingyu Yang, Maxime Sermesant.
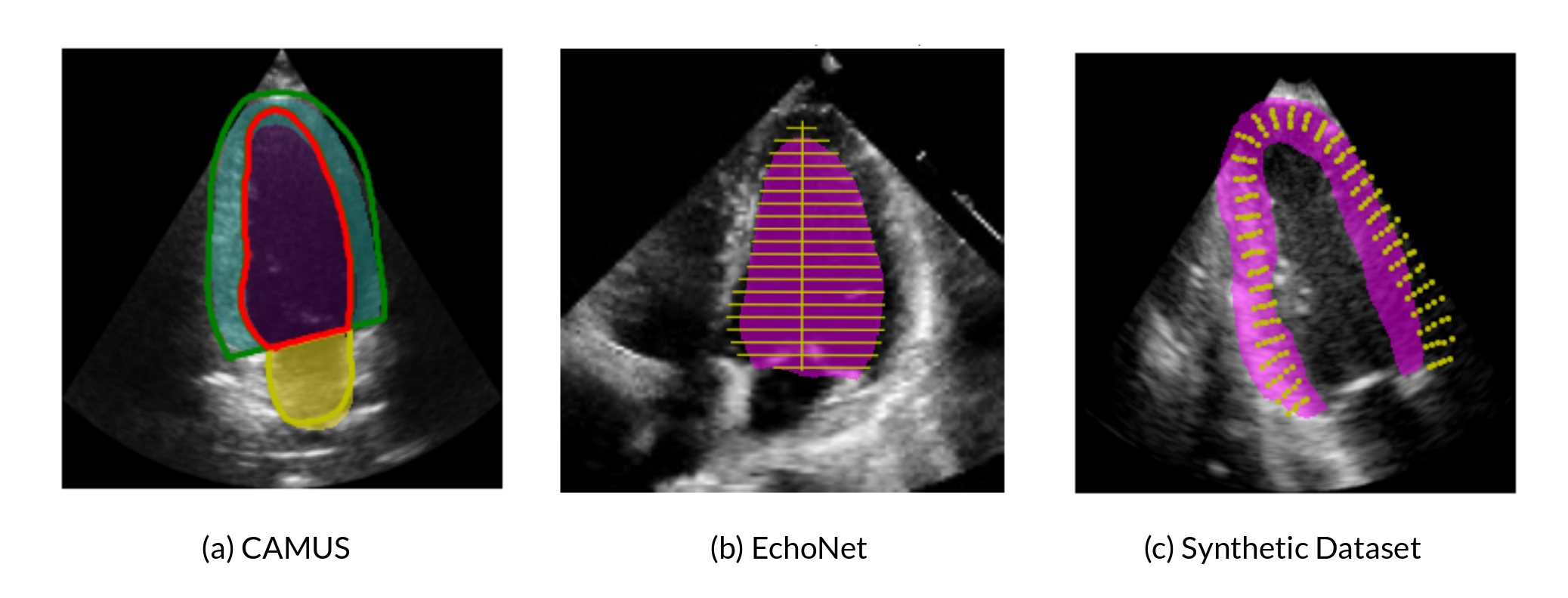
- 2D cross dataset segmentation: Using a adapted UNet along with plausible augmentation techniques during training phase (rotation, cropping, contrast & brightness adjustment, noise modification etc) on CAMUS dataset, we only achieved the state-of-art segmentation accuracy on the test data from the same dataset, but also gained much improvement on the other unseen dataset (EchoNet), with only 2.5% loss dice coefficient comparing with the state-of-art segmentation performance on this dataset (Figure 26).
- 2D Echocardiography motion tracking: We first did exploration of polyaffine log-demons and polyaffine block matching method on 2D synthetic echocardiography. Polyaffine log-demons demonstrated better tracking performance, while it demands longer computation time. We turned to deep learning method with the intention to reduce computation time and a better tracking result (work in progress).
7.4.7 Predicting thrombosis from Left-Atrium CT-Scan
This work is funded by PARIS EU Project, and is in collaboration with the IHU Liryc of Bordeaux and the Pompeu Fabra University in Barcelona
Keywords: Diffeomorphic Registration, Graph Representation, Latent Variable Models.
Participants: Josquin Harrison, Hubert Cochet, Oscar Camara Rey, Xavier Pennec, Maxime Sermesant.
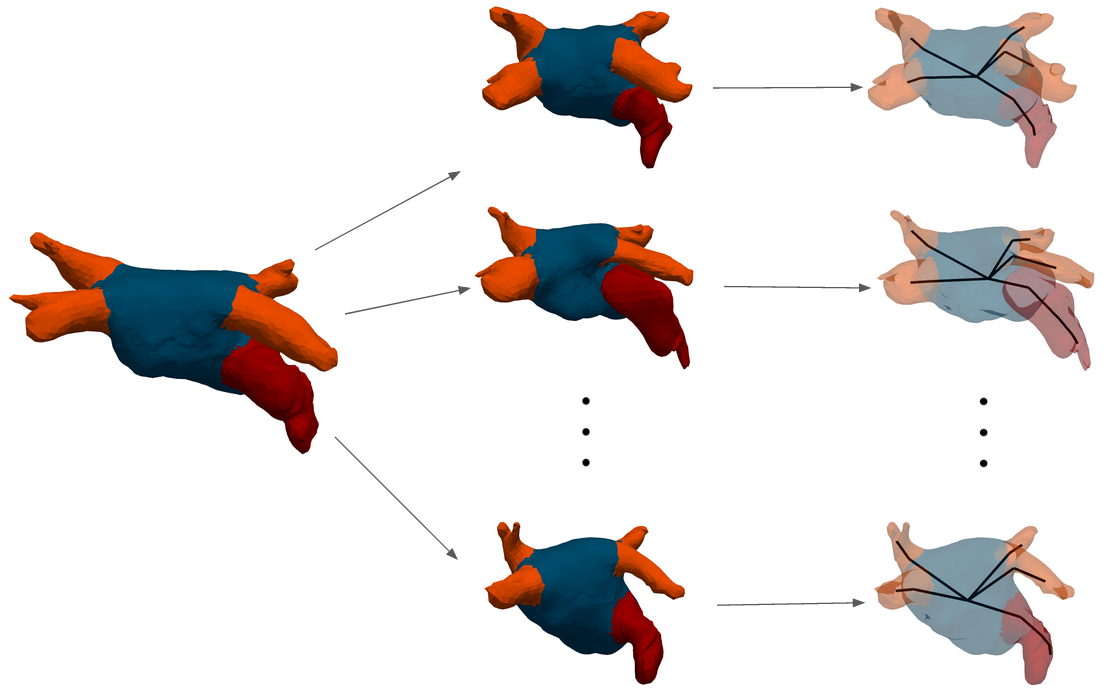
The objective of this project is to find image-based biomarkers in order to predict the risk of thrombus-related stroke for atrial fibrillation patients. 3D shapes of the left atrium are analysed, in combination with detailed blood flow simulations (Figure 27). A retrospective database of 300 patients from Bordeaux University Hospital is currently analysed.
This work is part of the European project PARIS, in collaboration with Simula, Oslo, Norway and Hambourg University Hospital, Germany.
7.5 Multi-centric data and Federated Learning
7.5.1 Bayesian federated learning framework for heterogeneous multi-view data
This project received financial support by the French government through the Agence Nationale de la Recherche (ANR), project Fed-BioMed ref. num. ANR-19-CE45-0006.
Keywords: Federated Learning, Hierarchical Generative Model, Heterogeneity.
Participants: Irene Balelli, Santiago Silva, Marco Lorenzi.
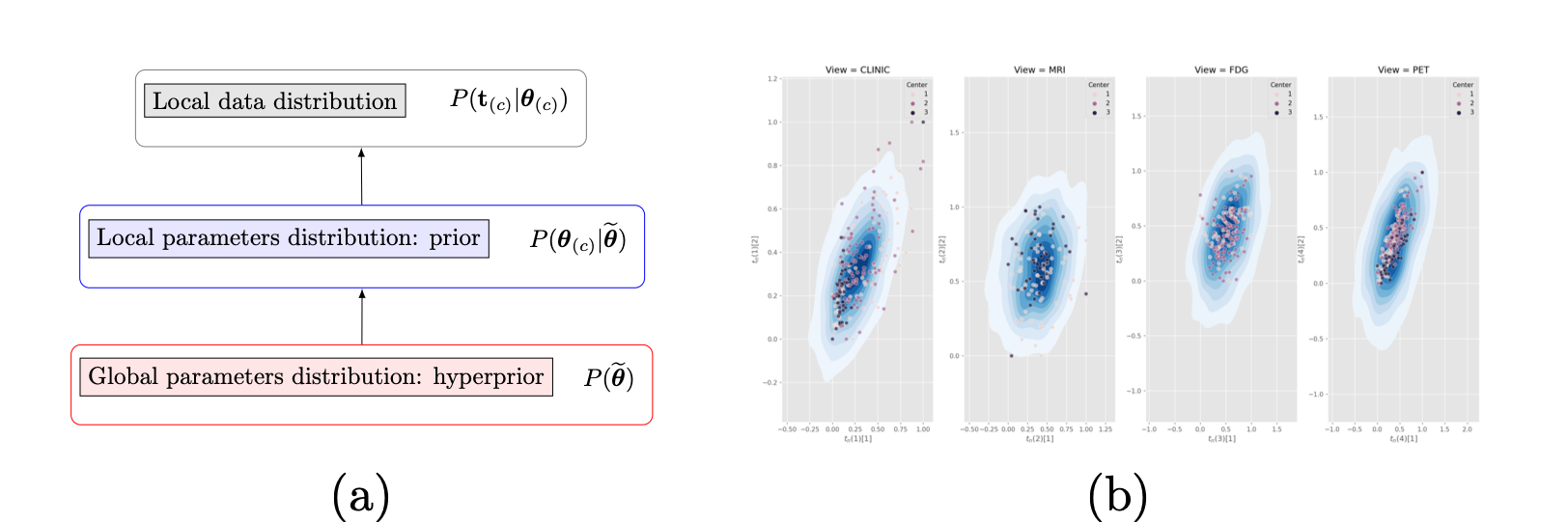
We developed a novel federated learning framework based on a hierarchical generative model (Fig. 28 (a)) for dimensionality reduction of heterogeneous multi-view data. The Bayesian approach deployed for parameter estimation allows to handle heterogeneous distribution of datasets across centers, and missing views, while providing an interpretable model of data variability. We applied our method to multi-modal medical imaging data and clinical scores from distributed datasets of patients affected by Alzheimer's disease from ADNI. The results prove the robustness of our method when data is distributed in iid and non-iid manners, and the high-quality of data reconstruction (Fig. 28 (b)), outperforming competitive methods.- A novel Bayesian federated learning paradigm
- Hierarchical generative model for dimensionality reduction of multi-view heterogeneous decentralized data
- High quality data reconstruction even in the presence of missing views
7.5.2 Fed-BioMed: A general open-source frontend framework for federated learning in healthcare
Acknowledgments: IDEX UCAJEDI BoostUrCareer; Agence Nationale de la Recherche (ANR), project Fed-BioMed ref. num. ANR-19-CE45-0006.
Keywords: Federated, healthcare, medical imaging.
Participants: Santiago Silva, Andre Altmann, Boris Gitman, Marco Lorenzi.
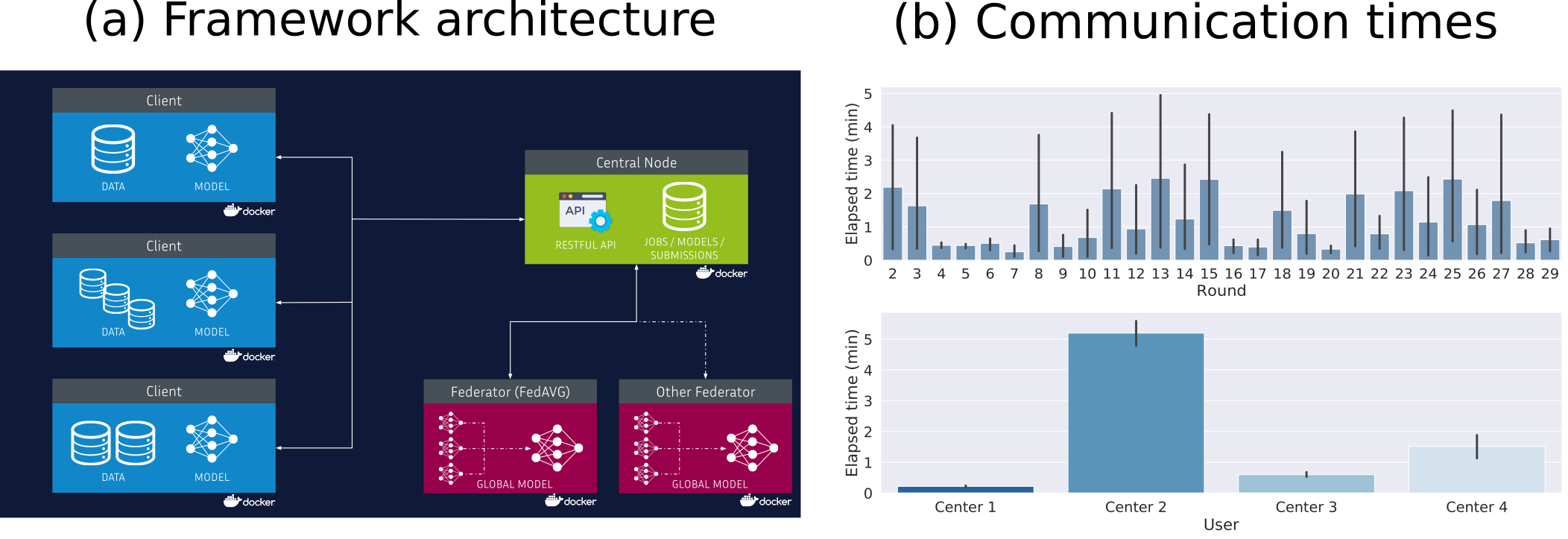
This project aims to propose solutions and methods for federated learning in healthcare by respecting privacy concerns such as the General Data Protection Regulation (GDPR) and keeping data securely decentralized. Continuing our previous proposal for federated variability analysis on high dimensional data, we proposed an open-source framework for federated learning in healthcare that was tested on neuroimaging data in a real-world scenario (Figure 29). This work was presented and published in the 1st MICCAI Workshop on "Distributed And Collaborative Learning" 2020 23. This project is available at https://
7.5.3 Free-rider attacks in Federated Learning
This work has been done as part of a CIFRE PhD on Bias in Federated Learning with Accenture Labs Sophia Antipolis, and has been partly funded by the Agence Nationale de la Recherche (ANR), project Fed-BioMed ref. num. ANR-19-CE45-0006.
Keywords: free-riders, federated learning, FedAvg, FedProx, Ornstein-Uhlenbeck process.
Participants: Yann Fraboni, Marco Lorenzi.
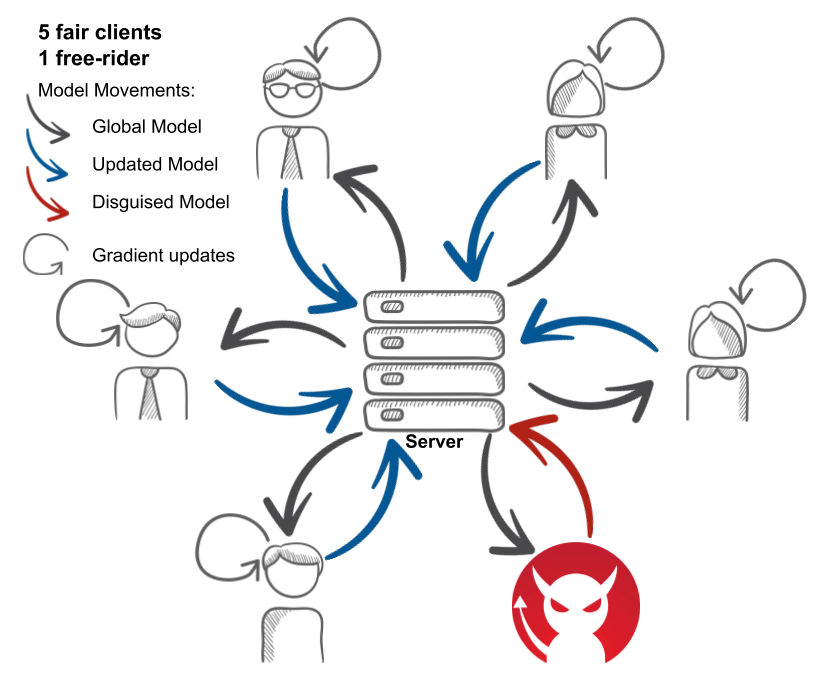
Federated Learning (FL), enables different clients to jointly learn a global model without sharing their data. We investigate the feasibility of free-riders on FL. Such attackers are clients pretending to have data to participate to the learning process and aiming at obtaining the jointly learnt model. We propose in our work two strategies for free-riders to achieve their goal without changing the global model performances that we prove both theoretically and experimentally. 42
8 Bilateral contracts and grants with industry
8.1 Bilateral contracts with industry
8.1.1 Siemens HealthCare
Siemens Healthcare, Medical Imaging Technologies, Princeton, NJ (U.S.A). is funding the PhD work of Julian Krebs which aims at developing robust medical image registration methods
8.1.2 Quantificare
The company Quantificare is funding the PhD of Florent Jousse through a CIFRE grant, on the statistical analysis of shapes, deformations and appearance of anatomical surfaces for computer-aided dermatology and plastic surgery. The primary purpose is to model complex face deformations such as natural aging, facial expressions, surgical interventions and posture motions.
8.1.3 Oticon Medical
Oticon Medical, Vallauris, France, is co-funding the PhD work of Zihao Wang which aims at developing robust medical image algorithms for cochlea image segmentation.
8.1.4 Accenture Labs
Accenture Labs, Sophia Antipolis, France, is funding the CIFRE PhD work of Yann Fraboni, which aims at investigating the problem of bias and fairness in federated learning applications.
8.1.5 MyDataModels
MyDataModels, startup based in Sophia Antipolis, France, funded the internship of Etrit Haxholli, with the goal of investigating novel approaches for anomaly detection in time series data.
8.1.6 Spin-off company inHEART
inHEART3 is a spin-off of the Epione team and IHU Liryc founded in 2017. inHEART provides a service to generate detailed anatomical and structural meshes from medical images, that can be used during ablation interventions. inHEART received 2 awards, one from Aquitaine region and one i-LAB from the BPI. It raised 3.2 million euros in 2020. It currently employs 19 people.
9 Partnerships and cooperations
9.1 International initiatives
9.1.1 Inria International Labs
GeomStats
- Title: Geometric Statistics in Computational Anatomy: Non-linear Subspace Learning Beyond the Riemannian Structure
- Duration: 2018 - 2020
- Coordinator: Xavier Pennec
-
Partners:
- Stanford (USA), Department of Statistics, Pr. Susan Holmes.
- Inria contact: Xavier Pennec
-
Website:
http://
www-sop. inria. fr/ asclepios/ projects/ GeomStats/ - Summary: The scientific goal of the associated team is to develop the field of geometric statistics with key applications in computational anatomy. Computational anatomy is an emerging discipline at the interface of geometry, statistics, image analysis and medicine that aims at analysing and modelling the biological variability of the organs shapes at the population level. An important application in neuroimaging is the spatial normalization of subjects that is necessary to compare anatomies and functions through images in populations with different clinical conditions. Following the developments of the last 3 years of the associated team GeomStat, the new research directions have been broken into three axes. The first axis aims at continuing the progresses in theoretical and applied Geometric statistics, with a first theme studying the impact of curvature on the estimation with a finite sample, and a second axis extending the current work on Barycentric Subspace Analysis (BSA), notably with algorithms. The second axis aims at developing a hierarchical atlas of the brain anatomy based on the stratification of the space of image orbits under diffeomorphisms. The third axis explores three important applications of low-dimensional subspace learning in manifolds using BSA in neuroscience: the approximation of EEG signals for Brain-Computer Interfaces (BCI); the acceleration and robustification of Tensor Distribution Functions (TDF) estimation in diffusion images; and the efficient inference in spaces of rank-deficient symmetric matrices for imaging-genetics from multi-centric databases.
9.1.2 Inria international partners
Department of Computer Science, University of Copenhagen, DK
Xavier Pennec is collaborating with Pr. Stefan Sommer on stochastic and sub-Riemannian approached to statistics, in the framework of his ERC G-statistics and of the jointly advised PhD thesis of Morten Akhoj Pedersen.
Laboratory of Physics of Fluids, University of Twente, NL
Herve Delingette is collaborating with Assistant Professor Guillaume Lajoinie, on the topics of Deep Learning for ultrasound imaging in the framework of the BoostUrCareer Cofund program and the thesis of Hari Sreedhar.
University College London (UCL), London, UK
Marco Lorenzi is collaborator of the COMputational Biology in Imaging and geNEtics (COMBINE) group within the Centre for Medical Image Computing (CMIC) of UCL, and with the UCL Institute of Ophtalmology. His collaboration is on the topic of spatio-temporal analysis of medical images, with special focus on brain imaging analysis and biomarker development. He is also collaborating with the “Progression Over Neurodegenerative Disorders” (POND) group (Prof. Daniel Alexander) for developing new computational models and techniques for learning characteristic patterns of disease progression using large longitudinal clinical data sets, with special focus on dementias.
Imaging Genetics Center (IGC), University of Southern California (USC), CA, USA and Illinois Institute of Technology (IIT, IL, USA)
Marco Lorenzi is currently collaborator of IGC and IIT for the investigation of the complex relationship between brain atrophy and genetics in Alzheimer's disease, in particular for demonstrating the effectiveness of multivariate statistical models in providing a meaningful description of the relationship between genotype and brain phenotype.
Laboratory of Neuroimaging of Aging (LANVIE), Faculty of Medicine, Geneva University Hospitals (HUG)
Marco Lorenzi collaborates with the LANVIE laboratory led by Prof. Giovanni B. Frisoni. The collaboration consists in developing and translating novel approaches for disease progression modeling in neurodegenerative disorders, such as Alzheimer's disease. St Thomas' Hospital, King's College London, United Kingdom
Other International Hospitals
Collaborations with several other European hospitals have been established through the European projects VP2HF, MD PAEDIGREE, SysAFib and with BarcelonaBeta research centre for Alzheimer.
9.2 European initiatives
9.2.1 FP7 & H2020 Projects
ERC G-Statistics
- Title: Geometric Statistics
- Type: ERC
- Program: H2020
- Duration: 2018-2023
- Inria contact: Xavier Pennec
- Coordinator: Inria
-
Summary:
G-Statistics aims at exploring the foundations of statistics on non-linear spaces with applications in the Life Siences. Invariance under gauge transformation groups provides the natural structure explaining the laws of physics. In life sciences, new mathematical tools are needed to estimate approximate invariance and establish general but approximate laws. Rephrasing Poincaré: a geometry cannot be more true than another, it may just be more convenient, and statisticians must find the most convenient one for their data. At the crossing of geometry and statistics, G-Statistics aims at grounding the mathematical foundations of geometric statistics and to exemplify their impact on selected applications in the life sciences.
So far, mainly Riemannian manifolds and negatively curved metric spaces have been studied. Other geometric structures like quotient spaces, stratified spaces or affine connection spaces naturally arise in applications. G-Statistics will explore ways to unify statistical estimation theories, explaining how the statistical estimations diverges from the Euclidean case in the presence of curvature, singularities, stratification. Beyond classical manifolds, particular emphasis will be put on flags of subspaces in manifolds as they appear to be natural mathematical object to encode hierarchically embedded approximation spaces.
In order to establish geometric statistics as an effective discipline, G-Statistics will propose new mathematical structures and characterizations of their properties. It will also implement novel generic algorithms and illustrate the impact of some of their efficient specializations on selected applications in life sciences. Surveying the manifolds of anatomical shapes and forecasting their evolution from databases of medical images is a key problem in computational anatomy requiring dimension reduction in non-linear spaces and Lie groups. By inventing radically new principled estimations methods, we aim at illustrating the power of the methodology and strengthening the “unreasonable effectiveness of mathematics” for life sciences.
ERC ECSTATIC
- Title: Electrostructural Tomography – Towards Multiparametric Imaging of Cardiac Electrical Disorders
- Type: ERC
- Program: H2020
- Duration: 2017 - 2022
- Inria contact: Maxime Sermesant
- Coordinator: U. Bordeaux
-
Summary:
Cardiac electrical diseases are directly responsible for sudden cardiac death, heart failure and stroke. They result from a complex interplay between myocardial electrical activation and structural heterogeneity. Current diagnostic strategy based on separate electrocardiographic and imaging assessment is unable to grasp both these aspects. Improvements in personalized diagnostics are urgently needed as existing curative or preventive therapies (catheter ablation, multisite pacing, and implantable defibrillators) cannot be offered until patients are correctly recognized.
ECSTATIC aims at achieving a major advance in the way cardiac electrical diseases are characterized and thus diagnosed and treated, through the development of a novel non-invasive modality (Electrostructural Tomography), combining magnetic resonance imaging (MRI) and non-invasive cardiac mapping (NIM) technologies.
The approach will consist of: (1) hybridising NIM and MRI technologies to enable the joint acquisition of magnetic resonance images of the heart and torso and of a large array of body surface potentials within a single environment; (2) personalising the inverse problem of electrocardiography based on MRI characteristics within the heart and torso, to enable accurate reconstruction of cardiac electrophysiological maps from body surface potentials within the 3D cardiac tissue; and (3) developing a novel disease characterisation framework based on registered non-invasive imaging and electrophysiological data, and propose novel diagnostic and prognostic markers.
This project will dramatically impact the tailored management of cardiac electrical disorders, with applications for diagnosis, risk stratification/patient selection and guidance of pacing and catheter ablation therapies. It will bridge two medical fields (cardiac electrophysiology and imaging), thereby creating a new research area and a novel semiology with the potential to modify the existing classification of cardiac electrical diseases.
9.3 National initiatives
Consulting for Industry
- Marco Lorenzi was a scientific consultant for the company MyDataModels (Sophia Antipolis), and for the company Flexper (Sophia Antipolis).
- Maxime Sermesant is a scientific advisor for the company inHEART (Bordeaux).
- Nicholas Ayache is a scientific consultant for the company Mauna Kea Technologies (Paris).
Institute 3IA Côte d'Azur
-
The 3IA Côte d'Azur https://
3ia. univ-cotedazur. eu/ is one of the four "Interdisciplinary Institutes of Artificial Intelligence" that were created in France in 2019. Its ambition is to create an innovative ecosystem that is influential at the local, national and international levels, and a focal point of excellence for research, education and the world of AI. - Epione is heavily involved in this institute since its 5 permanents researchers (N. Ayache, H. Delingette, M. Lorenzi, M. Sermesant and X.Pennec) are chair holders in this institute, and N. Ayache serves as scientific director. H. Delingette and N. Ayache are members of its scientific committee.
Funded projects
- Marco Lorenzi is principal investigator of the ANR JCJC project Fed-BioMed (2020-2023, 196k€).
- Hervé Delingette is among the main investigators of the DAICAP project (2020-2022, 300k€) selected by the Health Data Hub, the Grand Défi « Amélioration des diagnostics médicaux par l’Intelligence Artificielle », and Bpifrance in July 2020. That project aims to develop an algorithm able to produce a standardised MRI scan report to improve the early detection of prostate cancer.
Collaboration with national hospitals
The Epione project team collaborates with the following 3 French IHU (University Hospital Institute): the IHU-Strasbourg (Pr J. Marescaux and L. Soler) on image-guided surgery, the IHU-Bordeaux (Pr M. Haïssaguere and Pr P. Jaïs) on cardiac imaging and modeling and the IHU-Pitié Salpétrière (Dr. O. Colliot and S. Durrleman) on neuroimaging.
The Epione project team is involved in the following research projects with the Assistance Publique des Hôpitaux de Paris (AP-HP) : NHANCE project on abdominal ultrasound image analysis with Dr Anne-Laure Rousseau (Hospital St-Louis), PAIMRI project on prostate cancer detection with Pr Raphaele Renard-Penna (Hospital La Pitié Salpêtrière), CLARITI project on PET-CT anomaly detection with Pr Florent Besson (Hospital Kremlin Bicêtre), PET-CT lesion detection with Dr Paul Blanc-Durand (Hospital Henri Mondor).
In 2020, N. Ayache conducted for the Gustave Roussy Hospital (Paris) an expert study for the development of AI.
A collaboration with the Centre Hospitalier d'Antibes was launched in 2020 for providing support on the analysis and treatment of data collected throughout the COVID-19 outbreak. This latter project is in collaboration with Prof. Maureen Clerc (EPI Athena), and with the service d'Expérimentation et de Développement (SED) of Inria Sophia Antipolis.
We also have long term collaborations with the CHU Nice and Centre Antoine Lacassagne in Nice.
9.4 Regional initiatives
- Marco Lorenzi and Hervé Delingette received funding for a PhD salary from UCA under the European Program BoostUrCareer (Marie Sklodowska-Curie agreement 847581).
- Marco Lorenzi is principal investigator of the project Big Data for Brain Research, funded during 2017-20 by the Départment des Alpes Maritimes.
- Marco Lorenzi is principal investigator of the project MetaImaGen, funded by Idex UCA JEDI (2018-2020, 37k€).
- Hervé Delingette is the principal investigator of the LungMark project funded by Idex UCA JEDI (2018-2021).
- Hervé Delingette is the principal investigator of the CIMPLE project, funded by Idex UCA JEDI (2018-2021), the region PACA and Oticon Medical. The region PACA and Oticon Medical are co-funding the Phd of Zihao Wang.
- N. Ayache and P. Robert are principal investigators of the project MNC3 (Médecine Numérique, Cerveau, Cognition, Comportement) funded by Idex Jedi UCA (2017-2021, 450k€). M. Lorenzi (Inria) actively participates to the supervision of this project with the help of V. Manera (ICP).
- Maxime Sermesant is principal investigator of the project "The Digital Heart" and the innovation action "Digital Heart Phantom" with General Electrics, funded by Idex UCA JEDI. These projects gather the local cardiac research in academia, clinics and industry.
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
General chair, scientific chair
- X. Pennec organized a workshop on the Geometry of shapes (Math in the Cloud) regrouping about 20 people from June 29 to July 10, 2020. Originally planned in La minière de Vallauria (Alpes Maritimes, FR), the meeting became virtual due to the sanitary situation.
- M. Sermesant was a co-chair of the MICCAI 2020 Workshop Statistical Atlases and Computational Models of the Heart (STACOM 2020), which was held virtually in September 2020.
- Nicholas Ayache was a member of the organizing comittee of the 3rd conference "Medical imaging in the era of AI" held at ICM in 2020.
Member of organizing committees
- H. Delingette is a member of the organizing committee of the 2020 "SophI.A. summit" a scientific event that was held virtually and face to face in Sophia Antipolis from Nov. 15th til Nov. 25th.
Member of the conference program committees
- X. Pennec was a member of the program committee of the International Workshop on Differential Geometry in Computer Vision (Diff-CVML), the MICCAI workshop ShapeMe 2020, and of Information Processing in Medical Images (IPMI 2021).
Reviewer
- M. Lorenzi was a reviewer for the conferences Neural Information Processing Systems (NeurIPS 2020), International Conference on Machine Learning (ICML 2020), International Conferene on Artificial Intelligence and Statistics (AISTATS 2021), Medical Image Computing and Computer Aided Intervention (MICCAI 2020), International Conference on Learning Representations (ICLR 2021), Alzheimer's Association International Conference (AAIC 2020)
- M. Sermesant was a reviewer for Medical Image Computing and Computer Aided Intervention (MICCAI 2020), the MICCAI workshop STACOM and the Computing in Cardiology conference.
- H. Delingette was a reviewer for the Medical Image Computing and Computer Aided Intervention (MICCAI 2020), the European Conference on Computer Vision (ECCV'20), the Medical Imaging with Deep Learning (MIDL'20) conference, the International Symposium on Biomedical Imaging (ISBI'21).
10.1.2 Journal
Member of the editorial boards
- X. Pennec is a member of the editorial board of the journal Medical Image Analysis (MedIA, Elsevier), of the International Journal of Computer Vision (IJCV, Springer), and of the Journal of Mathematical Imaging and Vision (JMIV, Springer).
- M. Lorenzi is member of the editorial board of the journal Scientific Reports (Nature Publishing Group); he is also member of the Board of Statisticians of the Journal of Alzheimer’s Disease (IOS Press).
- H. Delingette is a member of the editorial board of the journal Medical Image Analysis (Elsevier).
- N. Ayache is the co-founder and the Co-Editor in Chief with J. Duncan (Professor at Yale) of Medical Image Analysis journal. This scientific journal was created in 1996 and is published by Elsevier.
- N. Ayache is a member of the editorial board of the following journals: Medical Image Technology (Japanese journal) and Journal of Computer Assisted Surgery (Wiley).
- I. Strobant is editorial coordinator for Medical Image Analysis, Elsevier (since october 2001).
Reviewer - reviewing activities
- X. Pennec was a reviewer for the following journals: Bernoulli, Annals of Statistics, Medical Image Analysis, Journal of the American Statistical Association (JASA),IEEE Transactions on Medical Imaging, Journal of Computational and Applied Mathematics, Information Geometry, Linear Algebra and its Applications (LAA).
- M. Lorenzi was a reviewer for the following journals: Journal of Alzheimer's Disease, Medical Image Analysis, IEEE Transactions on Medical Imaging, NeuroImage, International Journal of Computer Vision, Scientific Reports.
- M. Sermesant was a reviewer for the following journals: Journal of Machine Learning Research, Journal of the American College of Cardiology, IEEE Transactions on Medical Imaging, IEEE Transactions on Biomedical Engineering, Medical Image Analysis and Computers in Biology and Medecine.
- H. Delingette was a reviewer for the following journals: Medical Image Analysis (Elsevier), IEEE Transactions on Medical Imaging, Pattern Recognition, and Computer Vision and Image Understanding.
10.1.3 Invited talks
- N. Ayache gave a plenary invited lecture at the French Academy of Sciences on "AI and medical imaging: the role of models" in March 2020. He also gave a plenary talk at the opening ceremony of the winter school AI4health.
- X. Pennec was keynote speaker of the Computational Geometry Week (CG Week), Zürich, CH, June 22-26, 2020 and of the workshop Recent Advances in Statistical Analysis of Imaging Data organized by the American Statistical Association and FSU, USA, December 4-5 2020.
- M. Lorenzi gave an invited talk to the following events: European Glaucoma Society Congress 2020, Special session on Multi-view Methods for Imaging Genetics at the 26th Annual Meeting of the Organization for Human Brain Mapping, Monthly seminar of the Geneva Univeristy Hospital (CH).
- H. Delingette gave an invited talk to the following events: MICCAI workshop on Uncertainty for Safe Utilization of Machine Learning in Medical Imaging (UNSURE), 3e colloque sur l’imagerie médicale à l’heure de l’intelligence artificielle
10.1.4 Scientific expertise
- X. Pennec was a panel member for the Collaborative Research in Computational Neuroscience (CRCN) (joint call and evalutaion from NSF (US), NIH (US), ANR (FR), BMBF (DE), BSF (US-IL), NICT (JP)) and an expert for the Professor recruitment committee at the Medical University of Vienna (AT). He was also an evaluator for the Israel Science Foundation (ISF, IL), the European Research Concil (ERC CoG), the New Frontiers in Research Fund (NFRF, CRCC, CA), the French Agence Nationale de la Recherche (ANR).
- M. Lorenzi was reviewer of the funding agency ANR (Agence Nationale de la Recherche, France), of the Fonds de recherche du Québec en Santé (FRQS, CA), and of the Fondation France Alzheimer (France). He is providing scientific consulting for the company MyDataModels and Flexper through an Inria Tech research contract.
- H. Delingette is a member of the scientific committee of the institute 3IA Côte d'Azur and was reviewer of the funding agency ANR (Agence Nationale de la Recherche, France), an evaluator for the Israel Science Foundation (ISF, IL), the Fonds de recherche du Québec en Santé (FRQS, CA).
- N. Ayache is a member of the following scientific committees:
- 2021- Scientific Committee of the French Health Data Hub
- 2016 -: Scientific advisory committee for Région Ile de France (20 members),
- 2010 -: Scientific Advisory Boards in London (ICL,KCL,UCL), Oxford & Notthingham.
10.1.5 Research administration
- Nicholas Ayache is the scientific director of the 3IA Côte d'Azur since its creation in April 2019.
- Xavier Pennec is co-director of the Ecole doctorale STIC of Université Côte d'Azur. He is a member of the committee of EDSTIC, of the Doctoral follow-up Committee (CSD) at Inria Sophia Antipolis, and participated to the PhD fellowship granting committees of EDSTIC, Cofund at UCA, CORDI at Inria. He is a member of the "Comité de la Recherche Biomédicale en Santé Publique (CRBSP)" of the Nice University Hospital (CHU). At University Côte d'Azur /UCA JEDI, he is a member of the executive committee of the Academy 4 (Living systems Complexity and diversity), of the Scientific committee of the Academy 2 (Complex Systems), and of the Advanced Research Program Committee. At the national level, X. Pennec is a member of the International Chairs Selection Committee and of the Evaluation Committee of Inria. As such, he participated in 2020 to the promotion commitees for CRHC, DR1, DR0, to the PEDR committe and to the CRCN recruitment committee at Inria Rennes.
- Marco Lorenzi is a member of the local steering committee of the technological platforms (Comités Scientifiques de Pilotage des Plateformes) in charge of Cluster, Grid, Cloud, and HPC technologies. He is member of the Scientific Board of the UCA NeuroMod Institute, and he is part of the scientific committe for the 2021 AI4Health Winter School (Health Data Hub and 3IA).
- Hervé Delingette was a member of the local committee in charge of the scientific selection of visiting scientists (Comité NICE) and the local committee on the immersive platform until June 2020. He was the director of the Academy of excellence on “Networks, Information and Digital Society” and the deputy director of the “Ecole Universitaire de recherche” entitled "Digital Systems for Humans (DS4H)" at Université Côte d'Azur until Oct. 2020. He is a representive of Inria at the Federation Hospitalo-Universitaire Oncoage led by the CHU Nice. Since Oct 1st, he was nominated as one of the 4 program directors of the IDEX program UCA JEDI under the direction of the president of the Université Côte d'Azur. He is an administrator of the Groupement de Coopération Sanitaires (GCS) CARES involving the Université Côte d'Azur and the 3 local hospitals (CHU Nice, Centre Antoine Lacassagne, Fondation Lenval).
- Maxime Sermesant is an elected member of the Inria Sophia Antipolis research centre committee.
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
- Master: H. Delingette and X. Pennec, Introduction to Medical Image Analysis, 21h course (28.5 ETD), Master 2 MVA, ENS Saclay, France.
- X. Pennec is a member of the pedagogical council of the Computational biology master QCSBD, Univ. Côte d'Azur, France.
- Master: M. Lorenzi, Bayesian Learning, 30h course, Master Data Science, Univ. Côte d'Azur, France.
- Master: M. Lorenzi, Model Selection and Resampling Methods, 30h course, Master Data Science, Univ. Côte d'Azur, France.
10.2.2 Theses Defended
- Marco Lorenzi, Modeling Pathological Processes from Heterogeneous and High-Dimensional Biomedical Data. Université Côte d'Azur. Habilitation á Diriger des Recherches (HDR), Defended on January 10th, 2020.
- Wen Wei, Learning Brain Alterations in Multiple Sclerosis from Multimodal Neuroimaging Data,Université Côte d'Azur. Started in 2016. Co-directed by Nicholas Ayache and Olivier Colliot (ARAMIS). Defended on June the 5th, 2020.
- Julian Krebs, Robust image registration based on machine learning, Université Côte d'Azur. Started in 2016. Co-directed by Hervé Delingette and Nicholas Ayache. Defended on June the 15th, 2020.
- Nicolas Cedilnik, Image-based Personalised Models of Cardiac Electrophysiology for Ventricular Tachycardia Therapy Planning, Université Côte d'Azur. Started in 2017. Co-directed by Maxime Sermesant and Hubert Cochet (IHU Liryc). Defended on December the 14th, 2020.
10.2.3 PhD in progress
- Clément Abi-Nader, Statistical Learning of Heterogeneous Data in Large-Scale Clinical Databases, Université Côte d'Azur. Started in 2017. Co-directed by Philippe Robert (CoBTek) and Nicholas Ayache and supervised by Marco Lorenzi.
- Luigi Antelmi, Statistical learning on large databases of heterogeneous imaging, cognitive and behavioural data, Université Côte d'Azur. Started in 2017. Co-directed by Philippe Robert (CoBTek) and Nicholas Ayache and supervised by Marco Lorenzi.
- Benoît Audelan, Joint biological and imaging markers for the diagnosis of severe lung diseases. Started in 2018. Co-directed by Hervé Delingette and Nicholas Ayache.
- Tania-Marina Bacoyannis, Cardiac Imaging and Machine Learning for Electrostructural Tomography, Université Côte d'Azur. Started in 2017. Co-directed by Maxime Sermesant and Hubert Cochet (IHU Liryc).
- Jaume Banús Cobo, Heart & Brain: discovering the link between cardiovascular pathologies and neurodegeneration through biophysical and statistical models of cardiac and brain images, Université Côte d'Azur. Started in 2017. Directed by Maxime Sermesant and co-supervised by Marco Lorenzi.
- Florent Jousse, Analyse statistique de forme, de déformations et d'apparences de surfaces anatomiques pour l'aide à la dermatologie et à la chirurgie plastique. Started in 2019. Cifre fellowship with Quantificare. Co-directed by Xavier Pennec and Hervé Delingette.
- Buntheng Ly, Cardiac Image Analysis for Sudden Cardiac Death Prediction, Université Côte d'Azur. Started in 2019. Co-directed by Maxime Sermesant and Hubert Cochet (IHU Liryc).
- Nicolas Guigui, Statistical estimation on Riemannian and affine symmetric spaces with applications to the statistical survey of the brain anatomy, Université Côte d'Azur. Started in 2018. Directed by Xavier Pennec.
- Yann Thanwerdas, Statistical Dimension Reduction in Non-Linear Manifolds for Brain Shape Analysis, Connectomics & Brain-Computer Interfaces, Université Côte d'Azur. Started in 2019. Directed by Xavier Pennec.
- Zihao WANG, Cochlear Implantation Modeling, Planning & Evaluation. Started in 2018. Directed by Hervé Delingette.
- Hind Dadoun, AI-Based Real Time Diagnosis of Abdominal Ultrasound, Université Côte d'Azur. Started in December 2019. Co-directed by Nicholas Ayache and Hervé Delingette.
- Paul Tourniaire, AI-based selection of imaging and biological markers predictive of therapy response in lung cancer, Université Côte d'Azur. Started in 2019. Co-directed by Nicholas Ayache and Hervé Delingette.
- Dimitri Hamzaoui, AI-Based Diagnosis of Prostate Cancer from Multiparametric MRI, Université Côte d'Azur. Started in 2020. Co-directed by Nicholas Ayache and Hervé Delingette.
- Victoriya Kashtanova, Learning Cardiac 3D Electromechanical Dynamics with PDE-based Physiological Constraints for Data-Driven Personalized Predictions in Cardiology, Université Côte d'Azur . Started in 2020. Co-directed by Maxime Sermesant and Patrick Gallinari (Sorbonne University, LIP6, Paris).
- Hari Sreedhar, Learning-based detection and classification of thyroid nodules from ultrasound images, Université Côte d'Azur. Started in 2020. Co-directed by Hervé Delingette and Charles Raffaelli (CHU-Nice).
- Yingyu Yang, Artificial Intelligence for Automatic Cardiac Function Analysis: Multimodal and Biophysical Approach with Application to a Portable Imaging Device, Université Côte d'Azur. Started in 2020. Co-directed by Maxime Sermesant and Pamela Moceri (CHU-Nice)
- Josquin Harrison, Medical Imaging and learning for the prediction of strokes in the case of atrial fibrillation, Université Côte d'Azur. Started in 2020. Directed by Maxime Sermesant.
- Yann Fabroni, Bias in Federated Learning, Université Côte d'Azur. Started in 2020. Directed by Marco Lorenzi.
- Etrit Haxholli, Exploring latent dynamical models for failure prediction in time-series of high-dimensional and heterogeneous data, Université Côte d'Azur. Started in 2020. Directed by Marco Lorenzi.
- Santiago Smith Silva Rincon, Federated learning of biomedical data in large-scale networks of multicentric imaging-genetics information, Université Côte d'Azur. Started in 2020. Co-directed by Marco Lorenzi and Barbara Bardoni (INSERM)
- Elodie Maignan, Geometric learning of manifolds, Université Côte d'Azur. Started in 2020. Co-directed by Xavier Pennec and Alain Trouvé (ENS Paris-Saclay)
- Morten Pedersen, Lie group action, approximate invariance and Sub-Riemannian geometry in statistics, Université Côte d'Azur and University of Copenhagen. Started in 2020. Co-directed by Xavier Pennec and Stefan Sommer (Univ Copenhagen, DK).
- Gaëtan Desrues, 3D electromechanical cardiac modelling for heart failure patients stratification and prediction of cardiac resynchronisation therapy response, Université Côte d'Azur. Started in 2020. Directed by Maxime Sermesant.
10.2.4 Juries
- Xavier Pennec was member of the jury for the PhD of Oumaina El Mansouri (Univ. Toulouse, FR, Dec. 2020) and HdR of Marco Lorenzi (Univ. Côte d'Azur, FR, January 2020. ). He was president of the PhD jury of Jean Feydy (Univ. Paris Saclay, FR, July 2020).
- Marco Lorenzi was examinator of the PhD of Gia-Lac Tran (EURECOM, Sophia Antipolis, November 2020), and member of the PhD jury of Radia Zeghari (UCA, Nice, December 2020).
- Hervé Delingette was a member of the jury of Julian Krebs (Univ. Côte d'Azur, June 15th 2020) and Wen Wei (Univ. Côte d'Azur, June 5th 2020). He was a reviewer and member of the jury of Habilitation à Diriger les recherches of Florence Zara (Univ. Lyon , Oct 19th 2020) and Michaël Sdika (Univ. de Lyon, May 26th 2020).
- Maxime Sermesant was a reviewer for the PhD of Carlo Biffi (Imperial College London) and Luca Barbarotta(Einthoven, Pays Bas).
10.3 Popularization
N.Ayache co-authored the book 50 "Nouvelle enquête sur l’intelligence artificielle: Médecine, santé, technologies : ce qui va changer dans nos vies".
10.3.1 Interventions
N. Ayache participated to the round table organized in Bordeaux on "Imaging the future" as Invited Speaker.
11 Scientific production
11.1 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Scientific books
Scientific book chapters
Doctoral dissertations and habilitation theses
Reports & preprints
11.2 Other
Scientific popularization
Patents

