Keywords
Computer Science and Digital Science
- A2.1.1. Semantics of programming languages
- A2.1.5. Constraint programming
- A2.1.10. Domain-specific languages
- A2.2.1. Static analysis
- A2.3.2. Cyber-physical systems
- A2.4. Formal method for verification, reliability, certification
- A2.4.1. Analysis
- A2.4.2. Model-checking
- A2.4.3. Proofs
- A3.4.2. Unsupervised learning
- A3.4.4. Optimization and learning
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.1.3. Discrete Modeling (multi-agent, people centered)
- A6.1.4. Multiscale modeling
- A6.2.4. Statistical methods
- A6.2.6. Optimization
- A6.3.1. Inverse problems
- A6.3.4. Model reduction
- A7.2. Logic in Computer Science
- A8.1. Discrete mathematics, combinatorics
- A8.2. Optimization
- A8.7. Graph theory
- A9.7. AI algorithmics
Other Research Topics and Application Domains
- B1. Life sciences
- B1.1.2. Molecular and cellular biology
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B1.1.10. Systems and synthetic biology
- B2.2.3. Cancer
- B2.2.6. Neurodegenerative diseases
- B2.4.1. Pharmaco kinetics and dynamics
- B9. Society and Knowledge
1 Team members, visitors, external collaborators
Research Scientists
- François Fages [Team leader, Inria, Senior Researcher, HDR]
- Aurelien Naldi [Inria, Starting Research Position, from Nov 2020]
- Sylvain Soliman [Inria, Researcher, HDR]
Faculty Member
- Anna Niarakis [Univ d'Evry Val d'Essonne, Associate Professor, from Sep 2020]
Post-Doctoral Fellows
- Mathieu Hemery [Inria, until Feb 2020]
- Aurelien Naldi [Inria, from Mar 2020 until Oct 2020]
PhD Students
- Sahar Aghakhani [Université d'Evry, from Oct 2020]
- Eleonore Bellot [Inria]
- Marine Collery [IBM France, from Oct 2020]
- Elisabeth Degrand [Inria, until Nov 2020]
- Elea Greugny [Johnson & Johnson , CIFRE]
- Jeremy Grignard [Servier]
- Julien Martinelli [INSERM]
Technical Staff
- Mathieu Hemery [Inria, Engineer, from Mar 2020]
Interns and Apprentices
- Martin Boutroux [Inria, from Nov 2020]
- Pierre Fontaine [Ecole Polytechnique, from May 2020 until Aug 2020]
- Constance Le Gac [Ecole Polytechnique, Intern, Until March 2020]
- Quentin Petitjean [Ecole normale supérieure Paris-Saclay, from May 2020 until Aug 2020]
Administrative Assistants
- Natalia Alves [Inria, from Apr 2020]
- Emmanuelle Perrot [Inria, until Mar 2020]
External Collaborator
- Denis Thieffry [École Normale Supérieure de Paris, HDR]
2 Overall objectives
This project aims at developing formal methods and experimental settings for understanding the cell machinery and establishing computational paradigms in cell biology. It is based on the vision of cells as machines, biochemical reaction networks (CRN) as programs, and on the use of concepts and tools from computer science to master the complexity of cell biochemical processes 10.
This project addresses fundamental research issues in computer science on the interplay between structure and dynamics in large interaction networks, and on the fundamental concepts of analog computability and complexity. We contribute to CRN theory and develop since 2002 a CRN modelling, analysis and synthesis software, the Biochemical Abstract Machine, BIOCHAM. The reaction rule-based language of this system allows us to reason about biochemical reaction networks at different levels of abstraction with their stochastic, differential, discrete, Boolean or hybrid semantics. We develop a variety of static analysis methods before going to simulations and dynamical analyses. We use quantitative temporal logics as a mean to formalise biological behaviours with imprecise data and to constrain model building or CRN synthesis.
A tight integration between dry lab and wet lab efforts is also essential for the success of the project. This is achieved through tight collaborations with biologists and experimentalists on concrete biological or biomedical questions.
Because of the importance of optimization techniques in our research, we keep some activity dedicated to optimization problems, in particular on constraint programming methods for computing with partial information systems and solving NP-hard static analysis problems, and on continuous optimization methods for dealing with parameters.
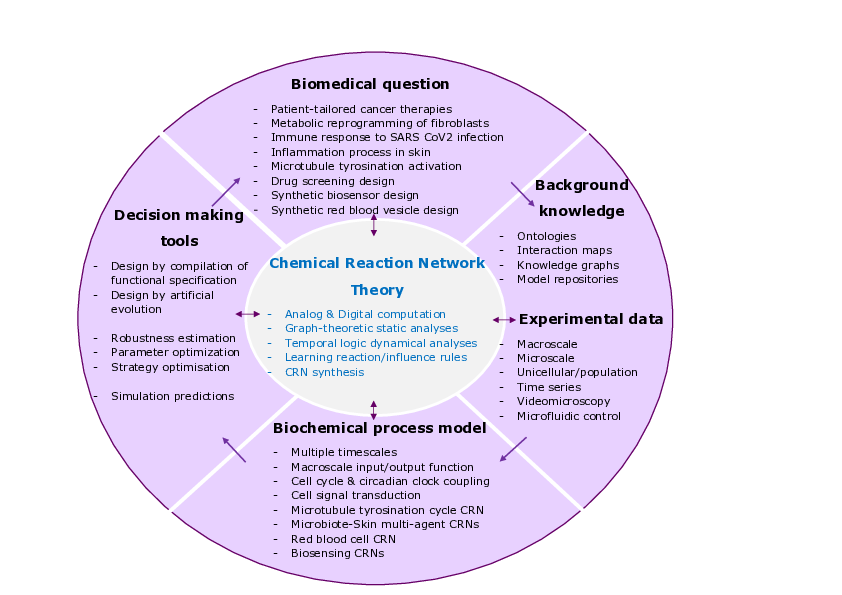
3 Research program
3.1 Computational Systems Biology
Bridging the gap between the complexity of biological systems and our capacity to model and quantitatively predict system behaviors is a central challenge in systems biology. We believe that a deeper understanding of the concept and theory of biochemical computation, and in particular of analog computation with proteins 1, is necessary to tackle that challenge. Progress in the theory is necessary for, beyond describing, understanding the structure and functions of large biochemical reaction systems, and developing static analysis, module identification and decomposition, model reductions, parameter search, and model inference methods. A measure of success on this route is the production of powerful computational modeling tools, in our case the Biochemical Abstract Machine (BIOCHAM) software, for elucidating the complex dynamics of natural biological processes, designing synthetic biochemical circuits and biosensors 2, developing novel therapy strategies, and optimizing patient-tailored therapeutics.
Progress on the coupling of models to data is crucial to the computational modeling of biochemical processes. Our approach based on quantitative temporal logics provides a powerful framework for formalizing experimental observations and using them as formal specification in model building or inference. Key to success is a tight integration between in vivo, in vitro and in silico work, and on the mixing of dry and wet experiments, enabled by novel biotechnologies. In particular, the use of videomicroscopy and micro-fluidic devices makes it possible to measure behaviors at both single-cell and cell population levels in vivo, provided innovative modeling, analysis and control methods are deployed in silico. For this we have constituted since nearly two decades a network of biologist partners and contributions to concrete biological questions 3456.
In synthetic biology, while the construction of simple intracellular DNA-based circuits has shown feasible and is mainstream, we develop an original approach based on analog computation with proteins, and on their implementation in artificial vesicles in tight collaboration with the CNRS Lab. Sys2Diag of Franck Molina in Montpellier 2.
3.2 Chemical Reaction Network (CRN) Theory
CRN theory has developed in several directions. Originally, Feinberg's CRN theory and Thomas's influence network analyses were created to provide sufficient and/or necessary structural conditions for the existence of multiple steady states and oscillations in complex regulatory networks. Those conditions can be verified by static analyzers without knowing kinetic parameter values nor making any simulation. In this approach, most of our work consists in analyzing the interplay between the structure (Petri net properties, influence graph, subgraph epimorphisms) and the dynamics (Boolean, CTMC, ODE, time scale separations) of biochemical reaction systems. In particular, our study of the influence graphs of reaction systems lead to the generalization of Thomas' conditions of multi-stationarity and Soulé's proof to reaction systems 78.
Our original method to infer CRNs from ODEs 9 widened the scope of this structural approach based on CRNs to the analysis of dynamical systems, and also led to our noticed Turing completeness result for finite analog CRNs 1.
We have shown that the computation of various structural invariants and the detection of model reduction relationships by subgraph epimorphisms10, made possible by using constraint programming techniques, provide solid ground for developing static analyzers, using them on a large scale in systems biology, and elucidating functional modules.
3.3 Logical Paradigm for Systems Biology
Our group was among the first ones in 2002 to apply model-checking methods to systems biology in order to reason on large molecular interaction networks, such as Kohn's map of the mammalian cell cycle (800 reactions over 500 molecules) 11. The logical paradigm for systems biology that we have subsequently developed for quantitative models can be summarized by the following identifications :
biological model = transition system
dynamical behavior specification = temporal logic formula
model validation = model-checking
model reduction = sub-model-checking, s.t.
model prediction = formula enumeration, s.t.
static experiment design = symbolic model-checking, state s.t.
model synthesis = constraint solving
dynamic experiment design = constraint solving
In particular, the definition of a continuous satisfaction degree for first-order temporal logic formulae with constraints over the reals, was the key to generalize this approach to quantitative models, opening up the field of model-checking to model optimization 1213 This line of research continues with the development of temporal logic patterns with efficient constraint solvers and their generalization to handle stochastic effects.
3.4 Computer-Aided Design of CRNs for Synthetic Biology
The continuous nature of many protein interactions leads us to consider models of analog computation, and in particular, the recent results on the theory of analog computability and complexity obtained by Amaury Pouly in his Thesis 14 and Olivier Bournez, establish fundamental links with digital computation. In 1 we have derived from those results the Turing completeness of elementary CRNs (without polymerization) under the differential semantics, closing a long-standing open problem in CRN theory. The proof of this result shows how computable function over the reals, described by Ordinary Differential Equations, namely by Polynomial Initial Value Problems (PIVP), can be compiled into elementary biochemical reactions, furthermore with a notion of analog computation complexity defined as the length of the trajectory to reach a given precision on the result. These results, implemented in BIOCHAM, open a whole research avenue to analyze biochemical circuits in Systems Biology, transform behavioural specifications into biochemical reactions for Synthetic Biology, and compare artificial circuits with natural circuits acquired through evolution, from the novel point of view of analog computation and analog computational complexity.
3.5 Constraint Solving and Optimization
Constraint solving and optimization methods are important in our research. On the one hand, static analysis of biochemical reaction networks involves solving hard combinatorial optimization problems, for which constraint programming techniques have shown particularly successful, often beating dedicated algorithms and allowing to solve large instances from model repositories. On the other hand, parameter search and model calibration problems involve similarly solving hard continuous optimization problems, for which evolutionary algorithms, and especially the covariance matrix evolution strategy (CMA-ES) 15 have been shown to provide best results in our context, for up to 100 parameters. This has been instrumental in building challenging quantitative models, gaining model-based insights, revisiting admitted assumptions, and contributing to biological knowledge 3456.
4 Application domains
4.1 Preamble
Our collaborative work on biological applications is expected to serve as a basis for groundbreaking advances in cell functioning understanding, cell monitoring and control, and novel therapy design and optimization. Our collaborations with biologists are focused on concrete biological questions, and on the building of predictive computational models of biological systems to answer them. Furthermore, one important application of our research is the development of a modeling software for computational systems biology.
4.2 Modeling software for systems biology and synthetic biology
Since 2002, we develop an open-source software environment for modeling and analyzing biochemical reaction systems. This software, called the Biochemical Abstract Machine (BIOCHAM), is compatible with SBML for importing and exporting models from repositories such as BioModels. It can perform a variety of static analyses, specify behaviors in Boolean or quantitative temporal logics, search parameter values satisfying temporal constraints, and make various simulations. While the primary reason of this development effort is to be able to implement our ideas and experiment them quickly on a large scale, BIOCHAM is used by other groups either for building models, for comparing techniques, or for teaching. A Jupyter BIOCHAM kernel has been created to use is interfaced is a web application which makes it possible to use BIOCHAM on our server without any installation. We plan to continue developing BIOCHAM for these different purposes and improve the software quality.
Since 2018, the Casq software complements this effort by providing an interface to import large interaction maps written in SBML-qual and interpret them as Boolean influence models with various tools.
Since 2020, we participate in the CoLoMoTo notebook platform, which provides an integrated collection of software tools for the analysis of qualitative models, including CaSQ. This platform encourages the reproducibility of analysis by combining Docker images (reproducible software environment) and Jupyter notebooks (reproducible and shareable workflows).
4.3 Coupled models of the cell cycle and the circadian clock
Recent advances in cancer chronotherapy techniques support the evidence that there exist important links between the cell cycle and the circadian clock genes. One purpose for modeling these links is to better understand how to efficiently target malignant cells depending on the phase of the day and patient characterictics. These questions are at the heart of our collaboration with Franck Delaunay (CNRS Nice) and Francis Lévi (Univ. Warwick, GB, formerly INSERM Hopital Paul Brousse, Villejuif) and of our participation in the ANR Hyclock project and in the submitted EU H2020 C2SyM proposal, following the former EU EraNet Sysbio C5Sys and FP6 TEMPO projects. In the past, we developed a coupled model of the Cell Cycle, Circadian Clock, DNA Repair System, Irinotecan Metabolism and Exposure Control under Temporal Logic Constraints 16. We now focus on the bidirectional coupling between the cell cycle and the circadian clock and expect to gain fundamental insights on this complex coupling from computational modeling and single-cell experiments.
4.4 Biosensor design and implementation in non-living protocells
In collaboration with Franck Molina (CNRS, Sys2Diag, Montpellier) and Jie-Hong Jiang (NTU, Taiwan) we ambition to apply our techniques to the design and implementation of high-level functions in non-living vesicles for medical applications, such as biosensors for medical diagnosis 17. Our approach is based on purely protein computation and on our ability to compile controllers and programs in biochemical reactions. The realization will be prototyped using a microfluidic device at CNRS Sys2Diag which will allow us to precisely control the size of the vesicles and the concentrations of the injected proteins. It is worth noting that the choice of non-living chassis, in contrast to living cells in synthetic biology, is particularly appealing for security considerations and compliance to forthcoming EU regulation.
5 Highlights of the year
5.1 COVID-19 Disease Map: building of a computational knowledge repository of SARS-CoV-2 virus-host interaction mechanisms
Participants: Aurélien Naldi, Anna Niarakis, Sylvain Soliman.
This year, Anna Niarakis has been co-directing a large-scale community effort to build an open-access, interoperable, and computable repository of COVID-19 molecular mechanisms - the COVID-19 Disease Map project, with active participation of Sylvain Soliman and Aurelien Naldi for the building of the pipelines and the analyses.
The molecular pathophysiology that links SARS-CoV-2 infection to the clinical manifestations and course of COVID-19 is complex and spans multiple biological pathways, cell types and organs (Gagliardi et al. 2020; Ziegler et al. 2020). To gain the insights into this complex network, the biomedical research community needs to approach it from a systems perspective, collecting the mechanistic knowledge scattered across the scientific literature and bioinformatic databases, and integrating it using formal systems biology standards.
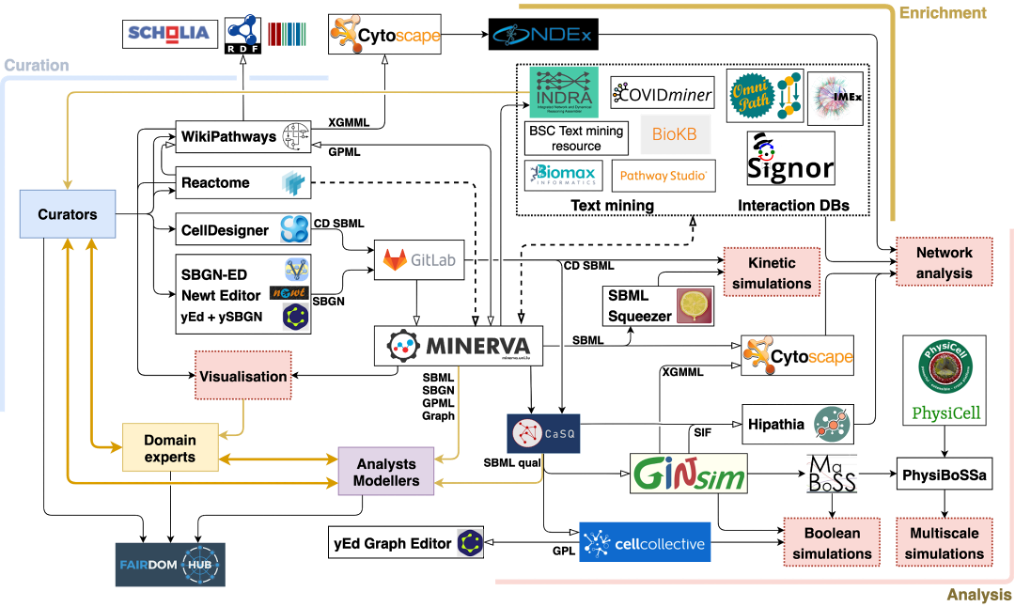
With this goal in mind, in March 2020, Anna co-initiated a collaborative effort involving over 230 biocurators, domain experts, modelers and data analysts from 120 institutions in 30 countries to develop the COVID-19 Disease Map, an open-access collection of curated computational diagrams and models of molecular mechanisms implicated in the disease (Ostaszewski et al. 2020). To this end, we aligned the biocuration efforts of the Disease Maps Community (Mazein et al. 2018; Ostaszewski et al. 2019), Reactome (Jassal et al. 2020), and WikiPathways (Slenter et al. 2018) and developed common guidelines utilising standardised encoding and annotation schemes, based on community-developed systems biology standards (Le Novere et al. 2009; Keating et al. 2020; Demir et al. 2010), and persistent identifier repositories (Wimalaratne et al. 2018). Moreover, we integrated relevant knowledge from public repositories (Turei et al. 2016; Perfetto et al. 2020; Licata et al. 2019) and text mining resources, providing a means to update and refine contents of the map. The fruit of these efforts was a series of pathway diagrams describing key events in the COVID-19 infectious cycle and host response.
The COVID-19 Disease Map project involves three main groups: (i) biocurators, (ii) domain experts, and (iii) analysts and modellers: i. Biocurators develop a collection of systems biology diagrams focused on the molecular mechanisms of SARS-CoV-2. ii. Domain experts refine the contents of the diagrams, supported by interactive visualisation and annotations. iii. Analysts and modellers develop computational workflows to generate hypotheses and predictions about the mechanisms encoded in the diagrams. All three groups have an important role in the process of building the map, by providing content, refining it, and defining the downstream computational use of the map.
Figure 2 illustrates the ecosystem of the COVID-19 Disease Map Community, highlighting the roles of different participants, available format conversions, interoperable tools, and downstream uses. The information about the community members and their contributions are disseminated via the FAIRDOMHub (Wolstencroft et al. 2017), so that content distributed across different collections can be uniformly referenced.
6 New software and platforms
6.1 New software
6.1.1 BIOCHAM
- Name: The Biochemical Abstract Machine
- Keywords: Systems Biology, Bioinformatics
- Functional Description: The Biochemical Abstract Machine (BIOCHAM) is a software environment for modeling, analyzing and synthesizing biochemical reaction networks (CRNs) with respect to a formal specification of the observed or desired behavior of a biochemical system. BIOCHAM is compatible with the Systems Biology Markup Language (SBML) and contains some unique features about formal specifications in quantitative temporal logic, sensitivity and robustness analyses and parameter search in high dimension w.r.t. behavioral specifications, static analyses, and synthesis of CRNs.
- Release Contributions: – notebooks of Master classes – graphical user interface on top of Jupyter – synthesis of at most binary reactions with minimisation of the variables for negative values – detection of model reduction by subgraph epimorphism – option for partial tropical equilibrations
-
URL:
http://
lifeware. inria. fr/ biocham4/ - Authors: François Fages, Thierry Martinez, Sylvain Soliman
- Contact: François Fages
- Participants: François Fages, Mathieu Hemery, Sylvain Soliman
6.1.2 CaSQ
- Name: CellDesigner as SBML-Qual
- Keywords: SBML, Logical Framework, Knowledge representation
- Functional Description: CaSQ transforms a big knowledge map encoded as an SBGN-compliant SBML file in CellDesigner into an executable Logical Model in SBML-Qual
-
URL:
https://
casq. readthedocs. io/ en/ stable/ - Authors: Sylvain Soliman, Anna Niarakis
- Contact: Sylvain Soliman
- Participants: Sylvain Soliman, Anna Niarakis, Aurélien Naldi
- Partner: Université d'Evry-Val d'Essonne
6.1.3 CoLoMoTo Notebook
- Keywords: Systems Biology, Computational biology
-
Functional Description:
Dynamical models are widely used to summarize our understanding of biological networks, to challenge its consistency, and to guide the generation of realistic hypothesis before experimental validation. Qualitative models in particular enable the study of the dynamical properties that do not depend on kinetic parameters, which can be difficult to obtain. This platform integrates a wide collection of complementary software tools, along with interoperability features and a consistent Programming interface based on the popular Python language.
The platform is distributed as a Docker image, ensuring the reproducibility of the software environment. The use of Jupyter Notebooks provides a good balance between ease of use and flexibility. These notebooks can be further used to share precise analysis workflows, ensuring the dissemination of fully reproducible research work using this platform.
-
URL:
http://
colomoto. org/ notebook/ - Publication: hal-01794294
- Authors: Aurélien Naldi, Loic Paulevé
- Contact: Aurélien Naldi
- Participants: Sylvain Soliman, Aurélien Naldi
- Partners: LaBRI, Institut Curie, ENS Paris, LRI - Laboratoire de Recherche en Informatique
7 New results
7.1 Graphical Conditions for Rate Independence in Chemical Reaction Networks
Participants: Elisabeth Degrand, François Fages, Sylvain Soliman.
In their interpretation by ordinary differential equations, we say that a CRN with distinguished input and output species computes a positive real function , if for any initial concentration x of the input species, the concentration of the output molecular species stabilizes at concentration . The Turing-completeness of that notion of chemical analog computation has been established by proving that any computable real function can be computed by a CRN over a finite set of molecular species. Rate-independent CRNs form a restricted class of CRNs of high practical value since they enjoy a form of absolute robustness in the sense that the result is completely independent of the reaction rates and depends solely on the input concentrations. The functions computed by rate-independent CRNs have been characterized mathematically as the set of piecewise linear functions from input species. However, this does not provide a mean to decide whether a given CRN is rate-independent. In 8, we provide graphical conditions on the Petri Net structure of a CRN which entail the rate-independence property either for all species or for some output species. We show that in the curated part of the Biomodels repository, among the 590 reaction models tested, 2 reaction graphs were found to satisfy our rate-independence conditions for all species, 94 for some output species, among which 29 for some non-trivial output species. Our graphical conditions are based on a non-standard use of the Petri net notions of place-invariants and siphons which are computed by constraint programming techniques for efficiency reasons.
7.2 On the Complexity of Quadratization for Polynomial Differential Equations
Participants: François Fages, Mathieu Hemery, Sylvain Soliman.
The proof of Turing completeness of finite CRNs uses an encoding of real variables by two non-negative variables for concentrations, and a transformation to an equivalent quadratic PIVP (i.e. with degrees at most 2) for restricting ourselves to at most bimolecular reactions. This year, we studied the theoretical and practical complexities of the quadratic transformation. In 9 we show that both problems of minimizing either the number of variables (i.e., molecular species) or the number of monomials (i.e. elementary reactions) in a quadratic transformation of a PIVP are NP-hard. We present an encoding of those problems in MAX-SAT and show the practical complexity of this algorithm on a benchmark of quadratization problems inspired from CRN design problems.7.3 Rule model learning from data time series
Participants: Marine Collery, François Fages, Jeremy Grignard, Mathieu Hemery, Julien Martinelli, Quentin Petitjean, Sylvain Soliman.
With the automation of biological experiments and the increase of quality of single cell data that can now be obtained by phosphoproteomic and time lapse videomicroscopy, automating the building of mechanistic models from these data time series becomes conceivable and a necessity for many new applications. While learning numerical parameters to fit a given model structure to observed data is now a quite well understood subject, learning the structure of the model is a more challenging problem that previous attempts failed to solve without relying quite heavily on prior knowledge about that structure. In previous work, we gave a greedy heuristics unsupervised statistical learning algorithm to infer reaction rules with a time complexity for inferring one reaction in where n is the number of species and t the number of observed transitions in the traces.
We investigate the same kind of rule inference algorithm in the thesis of Marine Collery at IBM France in the domain fraud detection from bank transaction traces.
7.4 Model learning to identify systemic regulators of the peripheral circadian clock
Participants: François Fages, Sylvain Soliman, Julien Martinelli.
In a submitted paper, we employ a model learning approach to identify systemic regulators of the peripheral circadian clock
Personalized medicine aims at providing patient-tailored therapeutics based on multi-type data towards improved treatment outcomes. Chronotherapy that consists in adapting drug administration to the patient's circadian rhythms may be improved by such approach. Recent clinical studies demonstrated large variability in patients' circadian coordination and optimal drug timing. Consequently, new eHealth platforms allow the monitoring of circadian biomarkers in individual patients through wearable technologies (rest-activity, body temperature), blood or salivary samples (melatonin, cortisol), and daily questionnaires (food intake, symptoms). A current clinical challenge involves designing a methodology predicting from circadian biomarkers the patient peripheral circadian clocks and associated optimal drug timing. The mammalian circadian timing system being largely conserved between mouse and humans yet with phase opposition, the study was developed using available mouse datasets.
We investigated at the molecular scale the influence of systemic regulators (e.g. temperature, hormones) on peripheral clocks, through a model learning approach involving systems biology models based on ordinary differential equations. Using as prior knowledge our existing circadian clock model, we derived an approximation for the action of systemic regulators on the expression of three core-clock genes: Bmal1, Per2 and Rev-Erb. These time profiles were then fitted with a population of models, based on linear regression. Selected models involved a modulation of either Bmal1 or Per2 transcription most likely by temperature or nutrient exposure cycles. This agreed with biological knowledge on temperature-dependent control of Per2 transcription. The strengths of systemic regulations were found to be significantly different according to mouse sex and genetic background.
7.5 CRN model reductions by tropical equilibrations
Participants: Eléonore Bellot, François Fages, Sylvain Soliman.
In the framework of the ANR-DFG SYMBIONT project we investigate mathematical justification of SEPI reductions based on Tikhonov's theorem and their computation using tropical algebra methods and constraint programming techniques 18.
This year we contributed to a deliverable on performance evaluation comparing polyhedral, SAT modulo theories (Z3) and constraint programming (BIOCHAM) methods.
7.6 Buffering semantics for qualitative models
Participants: Aurélien Naldi.
In collaboration with colleagues in the discrete biomathematics group at FU Berlin, we study extended Boolean models with buffer variables representing unknown delays on the interactions. This approach eliminates implicit kinetic assumptions in the classical updating modes of these models, not only to recover relevant dynamical behaviours, but also to provide a framework for reasoning on kinetic constraints which are sufficient to enable these behaviours. An article is in preparation.
7.7 Automated inference of Boolean models from molecular interaction maps using CaSQ
Participants: Martin Boutroux, Aurélien Naldi, Anna Niarakis, Sylvain Soliman.
Molecular interaction maps have emerged as a meaningful way of representing biological mechanisms in a comprehensive and systematic manner. However, their static nature provides limited insights to the emerging behavior of the described biological system under different conditions. Computational modelling provides the means to study dynamic properties through in silico simulations and perturbations. In 2 we describe how to bridge the gap between static and dynamic representations of biological systems with CaSQ, a software tool that infers Boolean rules based on the topology and semantics of molecular interaction maps built with CellDesigner (see Section 6.1.2). We developed CaSQ by defining conversion rules and logical formulas for inferred Boolean models according to the topology and the annotations of the starting molecular interaction maps. We used CaSQ to produce executable files of existing molecular maps that differ in size, complexity and the use of SBGN standards. We also compared, where possible, the manually built logical models corresponding to a molecular map to the ones inferred by CaSQ. The tool is able to process large and complex maps built with CellDesigner (either following SBGN standards or not) and produce Boolean models in a standard output format, SBML-qual, that can be further analyzed using popular modelling tools. References, annotations and layout of the CellDesigner molecular map are retained in the obtained model, facilitating interoperability and model reusability.
7.8 Setting the basis of best practices and standards for curation and annotation of logical models in biology
Participants: Aurélien Naldi, Anna Niarakis, Sylvain Soliman.
The fast accumulation of biological data calls for their integration, analysis and exploitation through more systematic approaches. The generation of novel, relevant hypotheses from this enormous quantity of data remains challenging. Logical models have long been used to answer a variety of questions regarding the dynamical behaviours of regulatory networks. As the number of published logical models increases, there is a pressing need for systematic model annotation, referencing and curation in community-supported and standardised formats. In 5 we summarise the key topics and future directions of a meeting entitled ‘Annotation and curation of computational models in biology’, organised as part of the 2019 [BC]2 conference. The purpose of the meeting was to develop and drive forward a plan towards the standardised annotation of logical models, review and connect various ongoing projects of experts from different communities involved in the modelling and annotation of molecular biological entities, interactions, pathways and models. This article defines a roadmap towards the annotation and curation of logical models, including milestones for best practices and minimum standard requirements.
In 4, we present a practical hands-on protocol breaking down the process of mechanistic modeling of biological systems in a succession of precise steps, aimed at those who are new to computational modeling.
In 7 , we review the different types of public resources with causal interactions, the different views on biological processes that they represent, the various data formats they use for data representation and storage, and the data exchange and conversion procedures that are available to extract and download these interactions. Indeed, broad sets of causal interactions are available in a variety of biological knowledge resources. However, different visions, based on distinct biological interests, have led to the development of multiple ways to describe and annotate causal molecular interactions. It can therefore be challenging to efficiently explore various resources of causal interaction and maintain an overview of recorded contextual information that ensures valid use of the data.
7.9 SBML Level 3: an extensible format for the exchange and reuse of biological models
Participants: Aurélien Naldi, Sylvain Soliman.
Systems biology has experienced dramatic growth in the number, size, and complexity of computational models. To reproduce simulation results and reuse models, researchers must exchange unambiguous model descriptions. We review the latest edition of the Systems Biology Markup Language (SBML), a format designed for this purpose. A community of modelers and software authors developed SBML Level 3 over the past decade. Its modular form consists of a core suited to representing reaction-based models and packages that extend the core with features suited to other model types including constraint-based models, reaction-diffusion models, logical network models, and rule-based models. The format leverages two decades of SBML and a rich software ecosystem that transformed how systems biologists build and interact with models. More recently, the rise of multi-scale models of whole cells and organs, and new data sources such as single-cell measurements and live imaging, has precipitated new ways of integrating data with models. In 3 we provide with a large international community of colleagues our perspectives on the challenges presented by these developments and how SBML Level provides the foundation needed to support this evolution.
7.10 Dynamical modelling of type I IFN responses in SARS-CoV2 infection
Participants: Aurélien Naldi, Anna Niarakis, Sylvain Soliman.
In this work, we make use of the type I IFN graphical model developed during the COVID19 Disease Map project and available in the C19DM repository and the map-to-model translation framework 2, to obtain an executable, dynamic model of type I IFN signalling for in silico experimentation. Our goal is to understand how we can maximize the Antiviral Immune response (production of ISGs), limit Inflammation, and amplify the inhibition of viral replication through the production of the IFNs. Starting from the CellDesigner map of the type I Interferon signalling we use the tool CaSQ to obtain an SBML qual file with preliminary Boolean rules. To cope with specificities of the COVID19 Disease Map repository, the tool CaSQ was adapted (version 9.0.2) to include: a) automatic merging of identical SBML species that can appear for visual purposes on the XML CellDesigner graph, b) the active suffix in the species name when the dotted circle was used to denote activation of a certain species. The SBML qual file was processed to ensure compatibility regarding name display in GINsim, and also control nodes were added in an attempt to reduce the number of inputs (from 37 to 7) and ease the computational cost. Given the size of the model (given the number of nodes and edges), an exhaustive attractors search would not be feasible. Therefore, we searched for attractors for a given set of initial conditions that cover different biological scenarios of the type I IFN pathway with or without the infection, and in the presence or absence of drugs. For the analysis we used the CoLoMoTo notebook implementation. An article is in preparation.
7.11 Fibroblasts as therapeutic targets in rheumatoid arthritis (RA) and cancer. Computational modeling of the metabolic reprogramming (glycolytic switch) in RA synovial fibroblasts (RASFs) and cancer associated fibroblasts (CAFs)
Participants: Sahar Aghakhani, Anna Niarakis, Sylvain Soliman.
Fibroblasts are critical regulators of several physiological processes linked to extracellular matrix regulation. Under certain conditions, fibroblasts can also transform into more aggressive phenotypes and contribute to disease pathophysiology. In this work, we highlight metabolic reprogramming as a critical event toward the transition of fibroblasts from quiescent to activated and aggressive cells, in rheumatoid arthritis and cancer. We draw obvious parallels and discuss how systems biology approaches and computational modeling could be employed to highlight targets of metabolic reprogramming and support the discovery of new lines of therapy. The objectives of this project are: a) First, to study similarities in the molecular and signalling pathways of RASFs and CAFs, which are involved in the regulation of glucose metabolism. b) Second, to create dynamic models that could be used for in silico perturbations in order to study the cells’ response to different conditions such as inflammation, hypoxia, and elevated ROS levels, to name a few. Dynamical modelling is indispensable if we want to understand the emergent behaviour of the cells when complex and intertwined pathways are involved. c) Lastly, to identify and propose a possible mechanism (common or not) that could explain the fibroblasts’ metabolic reprogramming regarding glucose in disease- specific conditions. An in-depth review focusing on RASFs and CAFs was recently published in Cancers 1.
7.12 Inference of a modular, large-scale Boolean network for modelling the Rheumatoid Arthritis fibroblast-like synoviocytes
Participants: Aurélien Naldi, Anna Niarakis, Sylvain Soliman.
Rheumatoid arthritis fibroblast-like synoviocytes (RA FLS) are central players in the disease pathogenesis, as they are involved in the secretion of cytokines and proteolytic enzymes, exhibit invasive traits, high rate of self-proliferation and an apoptosis-resistant phenotype. Using the RA map and the tool CaSQ we build a large-scale dynamic model of RA FLS to study apoptosis, cell proliferation and growth, osteoclastogenesis and bone erosion, matrix degradation and cartilage destruction and inflammation outcomes. The RA FLS network was created selecting fibroblast-relevant sub-parts of the state-of-the-art RA map (Singh et al. 2020), and the Boolean rules were added using the tool CaSQ that infers logical formulae based on the topology and the semantics of the network 2. To cope with complexity, we also employ a “divide and conquer” strategy by creating separate executable sub-modules that comprise each only one phenotype. We present also reduced model versions that facilitate downstream analysis. Systematic testing of different initial conditions could further lead to predictions regarding the outcomes of specific perturbations, such as single or combined effects, simulated with the model by forcing or suppressing the activity of various factors of interest systematically. The goal is to gain a better understanding of the mechanisms that drive inflammation, resistance to apoptosis, high proliferation rate, and cartilage and bone degradation, and their coordination, to delineate and gain control of these outcomes (paper in preparation).
7.13 Computational model of the microtubule tyrosination cycle
Participants: François Fages, Jeremy Grignard, Sylvain Soliman.
Microtubules and their post-translational modifications involved in major cellular processes such as: mitosis, cardiomyocyte contraction, and neuronal differentiation. More precisely, in neurons, the post-translational modifications of detyrosination and tyrosination are crucial for neuronal plasticity, axon regeneration, recruitment and transports of proteins and correct neuronal wiring. We hypothesize that the decrease of density and length of microtubules and the loss of neuronal structures such as synapses, dendritic spine and growth cone which are correlated with the progressive cognitive decline may be the consequence of the dysregulation of the cycle detyrosination/tyrosination in neurodegenerative disorder. This hypothesis is investigated in collaboration with Servier by combining experimental approaches with mathematical modelling in a submitted paper.
7.14 Computational model of inflammation process in skin
Participants: François Fages, Eléa Greugny.
Skin protects the body against external agents, for instance pathogens, irritants, or UV radiation, that can trigger inflammation. Inflammation is a complex phenomenon that is classified in two main types, acute and chronic. They are distinguished by different parameters such as the duration, the underlying mechanisms, the components involved like the type of immune cells, and the nature and intensity of the associated clinical signs. The computational models developed in collaboration with Johnson&Johnson France, combine mathematical and multi-agent modelling using BIOCHAM and EPISIM modelling tools.
7.15 Computational model of red blood cells
Participants: Elisabeth Degrand, François Fages, Constance Le Gac.
In the framework of the AEx GRAM we have developed a computational model of erythrocytes, i.e. red blood cells. Their most obvious function concerns the respiratory system since erythrocytes allow gas exchanges at the level of the organism by transporting dioxygen and carbon dioxide between the lungs and the tissues. However, red blood cells also have an important buffer function in the blood, which is necessary to keep blood pH in the physiological range. Modelling the red blood cells with CRNs using BIOCHAM gives us insight as to which biological objects are necessary to allow the cell to process its functions correctly. At the level of Systems Biology, it allows us to understand the links between the different biological functions of erythrocytes and investigate their synthesis in artificial vesicles. Unfortunately, due to the Covdi-19 crisis our biologist partner and his lab was not available to make the planned experiments.
7.16 Biomachines For Medical Diagnosis
Participants: Elisabeth Degrand, François Fages, Jeremy Grignard.
Current evolutions in medical practices induce a change of paradigm with the convergence of diagnosis and therapy, going to precision medicine and “theranostics”. One can observe the new role of biomarkers in biomedical and therapeutic applications, for instance in the development of molecular multiplex biosensors (nucleic acid, proteins, and metabolites). In addition, there is an increasing interest for point-of-care (POC) and of home monitoring/testing technologies devoted to probe patient parameters in his direct environment. The obvious constraints for such a kind of new clinical practices are simplification, drastic cost reduction while keeping high performances. Within this context, synthetic biology provides new opportunities to develop a novel generation of biological biosensors able to perform multiplexed biomarkers detection, simple computation and returning simplified relevant results. In 6 we present an original methodology to design robust synthetic to design robust synthetic biochemical circuits ensuring biochemical implementation of logical tasks within nonliving artificial cells for biosensor systems reliable in a clinical context. This methodology covers in silico design, simulation, microfluidics production and clinical validation on human samples. It ends up in very simple assay like for instance the new insulin-resistance assay, which is also quick and easy to run out of a laboratory and at low cost. In 11 we present recent theoretical foundations for this approach.
7.17 Algorithmic modeling of the immune system
Participants: François Fages, Pierre Fontaine.
The immune system is involved in the defense of the organism against the in- trusion of pathogens. The various mechanisms involved in this defense against harmful agents are likely to interest computer science for topics related to the recognition and memorization of intrusions, the adaptation of a specific defense or learning according to the chronology of intrusions. Such a system is moreover autonomous, decentralized and based on numerous random mutations. Thus, multiple factors are brought together to justify a study of the Bio- logical Immune System (BIS) in order to build an Artificial Immune System (AIS). In an unpublished report, we describe a learning algorithm for intrusion detection that accounts for the properties of the biological immune system. Such an algorithm makes it possible, on the one hand, to formalize and better understand certain immunological processes, and on the other hand, to analyze a machine learning algorithm which living organisms have shown to be conclusive for the recognition of intrusions and other tasks.
8 Bilateral contracts and grants with industry
8.1 Bilateral contracts with industry
In the framework of the Cifre PhD thesis of Jeremy Grignard at Servier, we work on the coupling between computational modeling and biological experiment design, and on chemical reaction network inference methods from data time series.
In the framework of the Cifre PhD thesis of Eléa Greugny at Johnson&Johnson Santé Beauté France, we work on the computational modeling of inflammatory process in the skin, using multi-scale modeling and multi-agent simulation.
In the framework of the (non-Cifre) PhD thesis of Marine Collery at IBM France, we work on rule learning from time series data in the context of learning fraud detection rules in bank transactions.
9 Partnerships and cooperations
9.1 International research visitors
9.1.1 Visits of international scientists
The following researchers have been invited for short visits:
- Pierre-Jean Meyer, U.C. Berkeley
- Alexandru Iosif, JRC-COMBINE, Aachen, Germany
9.2 National initiatives
9.2.1 ANR Projects
- ANR IFFERENCE on "Complexity theory with discrete ODEs", coordinated by Olivier Bournez, Ecole Polytechnique, with F. Fages, F. Chyzak Inria SIF, A. Durand Univ. Paris-Diderot, F. Madelaine P. Valarché Univ. Créteil, N20 Jerôme Durand-Lose, Orléans - Moulay Barkatou Thomas Cluzeau, Limoges - Mathieu Sablik, Toulouse
- ANR-DFG Symbiont (2018-2021) on “Symbolic Methods for Biological Systems”, coordinated by T. Sturm (CNRS, LORIA, Nancy, France) and A. Weber (Univ. Bonn, Germany) with F. Fages and F. Boulier (U. Lille), O. Radulescu (U. Montpellier), A. Schuppert (RWTH Aachen), S. Walcher (RWTH Aachen), W. Seiler (U. Kassel);
- ANR-MOST Biopsy (2016-2020) on “Biochemical Programming System”, coordinated by F. Molina (CNRS, Sys2diag, Montpellier) and J.H. Jiang (National Taiwan University), with F. Fages;
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
General chair, scientific chair
François Fages is member of the Steering Committee of the conference series Computational Methods for Systems Biology (CMSB) since 2008.
Anna Niarakis was co-organizer of the following events
- Computational modelling of cellular processes: mechanistic and machine learning approaches, workshop of ECCB 2020, Sitges Barcelone, 5-6 September
- 2. 5th Disease Map Community meeting, November 2020, Virtual
- ISMB/ECCB COSI SysMod, Lyon, France, July 2021 Workshop
Member of the conference program committees
François Fages was PC member of
- IJCAI-PRICAI 2020, 29th International Joint Conference on Artificial Intelligence and 17th Pacific Rim International Conference on Artificial Intelligence. Yokohama, Japan, July 11-17 2020.
- CMSB'20 18th International Conference on Computational Methods in Systems Biology. Unifversity of Konstanz, Germany, September 2020.
- BIOINFORMATICS'20 11th International Conference on Bioinformatics Models, Methods and Algorithms” co-located with BIOSTEC'20, Valetta, Malta, 24-26 Feb 2020.
Sylvain Soliman was PC member of
- BIOINFORMATICS'20, the 11th International Conference on Bioinformatics Models, Methods and Algorithms co-located with BIOSTEC'20, Valetta, Malta, 24–26 Feb. 2020.
- CSBio 2020the 11th International Conference on Computational Systems-Biology and Bioinformatics, Bangkok, Thailand, 19–21 Nov. 2020.
Reviewer
Sylvain Soliman was a (sub)reviewer for ECCB'2020 and CMSB'2020.
Julien Martinelli was a (sub)reviewer for ECCB'2020.
10.1.2 Journal
Member of the editorial boards
François Fages is member of
- the Editorial Board of the Computer Science area of the Royal Society Open Science journal, since 2014;
- the Editorial Board of the journal RAIRO OR Operations Research, since 2004.
Anna Niarakis is member of
- the Editorial Board of Scientific Reports Nature Publishing.
Reviewer - reviewing activities
François Fages made reviews for the journals Bioinformatics, Machine Learning, ACS Synthetic Biology, Machine Learning, PLOS Computational Biology and two book chapters at ISTE.
Anna Niarakis made reviews for: Database Oxford, Frontiers in Applied Mathematics and Statistic, Systems Biology category, Frontiers in Physiology, Molecular Medicine, Cancers, MDPI, Biology.
Sylvain Soliman made reviews for the journals PLoS Computational Biology, IET Systems Biology.
10.1.3 Scientific expertise
François Fages evaluated applications for
- BBSRC grant, UK,
- WWTF Vienna Research Group Leader Position, Austria
- and for one application to the Institut Universitaire de France.
10.1.4 Research administration
François Fages is member of
- the "Bureau du Comité des équipes-projets du centre Inria SIF"
- the "Bureau du département Informatique Données et Intelligence Artificielle (IDIA) de l'Institut Polytechnique de Paris"
- the Scientific committee of the Doctorate School "ED 474 - Frontière de l'Innovation en Recherche (FIRE)"
Sylvain Soliman is member of the “Commission Scientifique” of Inria Saclay-IdF.
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
- International advanced course: Anna Niarakis, Ben Hall (co-organizers), Aurélien Naldi, Sylvain Soliman (trainers) Computational Systems Biology, MRC University of Cambridge, Wellcome Trust course WGC Advanced Course, 6-11 December 2020, Hinxton, Cambridge UK.
- International advanced course: Aurélien Naldi (trainer) Logical Modeling for Experimental Design in Biotechnology and Biomedicine, August 17-28, NTNU, Trondheim, Norway.
- International advanced course: Aurélien Naldi (trainer, 3h) Qualitative models in Systems Biology, Nov 4-5, IGC PhD Program in Integrative Biology and Biomedecine, Oeiras, Portugal.
- Master 2: François Fages (coordinator module 24h and teacher 12h) C2-19 - Biochemical Programming, Master Parisien de Recherche en Informatique (MPRI), Paris.
- Master 1: François Fages, Sylvain Soliman (co-coordinators and teachers 36h) INF555 - Constraint-based Modeling and Algorithms for Decision Making ProblemsMaster Artificial Intelligence, Ecole Polytechnique.
- Master 1: François Fages, Sylvain Soliman (co-coordinators and teachers 60h) INF473L - Programming in the language of the living
- Bachelor 3: François Fages, Sylvain Soliman (co-coordinators and teachers 36h) CSE 307 - Constraint Logic Programming, Ecole Polytechnique.
- Bachelor 2: Eléonore Bellot (teacher 6h) CSE201 Object-oriented Programming in C++ TD and project supervision
- L2: Julien Martinelli (teaching assistant 42h) Real Analysis, Université de Paris / (teaching assistant 18h) Multivariate functions, Université de Paris
10.2.2 Supervision
- PhD in progress: Sahar Aghakhani, “Fibroblasts as therapeutic targets in rheumatoid arthritis (RA) and cancer. Computational modeling of the metabolic reprogramming (glycolytic switch) in RA synovial fibroblasts (RASFs) and cancer associated fibroblasts (CAFs)” , ED Paris-Saclay, Sep. 2020, Anna Niarakis & Sylvain Soliman (50%)
- PhD in progress : Eléonore Bellot, “Réduction de modèles différentiels par résolution de contraintes d’algèbre tropicale (min,+)”, ED IPP, Ecole Polytechnique, Sept. 2018, F. Fages & S. Soliman (50-50%) ges & S. Soliman (50-50%)
- PhD in progress : Marine Collery, “Apprentissage de règles à partir de données dépendantes du temps appliqué à la détection de fraudes”, ED IPP, Ecole Polytechnique, Sept. 2020, F. Fages & P. Bonnard (50-50%)
- PhD stopped : Elisabeth Degrand, “Chemical Programming of Non-living Vesicles”, ED IPP, Ecole Polytechnique, Oct. 2019- Oct. 2020, F. Fages & S. Soliman (50-50%)
- PhD in progress : Eléa Greugny, “Development and Implementation of a Mathematical Model of Inflammation in the Human Skin”, ED IPP, Ecole Polytechnique, Aug. 2019, F. Fages & J. Bensaci & G. Stamatas, Johnson&Johnson Santé Beauté France (1/3-2/3))
- PhD in progress : Jérémy Grignard, “Apprentissage de modèles à partir de données pour la conception d’expériences de criblage et la recherche de médicaments”, ED IPP, Ecole Polytechnique, Mar. 2019, F. Fages & T. Dorval, Servier (50-50%)
- PhD in progress : Julien Martinelli, “Apprentissage de modèles mécanistes à partir de données temporelles, application à la personnalisation de la chronothérapie des cancers”, ED IPP, Ecole Polytechnique, Oct. 2018, F. Fages & A. Ballesta, Inserm (50-50%)
10.2.3 Juries
François Fages participated in the jurys of
- HDR Loïc Paulevé, Rapporteur, Université Paris-Sud, 3 Sep. 2020.
- PhD Eugenia Oshurko Rapporteur, Président du jury, 6 Jul. 2020.
10.3 Popularization
10.3.1 Articles and contents
Our article 11 is a gentle introduction to our vision of cells as analog computers, and in what sense the language of chemical reactions provides a universal Turing-complete programming language.
Our article 10 on "Artificial Intelligence in Biological Modelling" is a book chapter of an encyclopedia of AI published this year.
11 Scientific production
11.1 Publications of the year
International journals
International peer-reviewed conferences
Scientific book chapters

