Keywords
Computer Science and Digital Science
- A6. Modeling, simulation and control
- A6.1. Methods in mathematical modeling
- A6.2. Scientific computing, Numerical Analysis & Optimization
- A6.3. Computation-data interaction
Other Research Topics and Application Domains
- B1. Life sciences
- B1.1. Biology
- B2. Health
- B2.2. Physiology and diseases
- B2.2.2. Nervous system and endocrinology
- B2.2.3. Cancer
- B2.2.4. Infectious diseases, Virology
- B2.4.1. Pharmaco kinetics and dynamics
- B2.4.2. Drug resistance
- B2.6.1. Brain imaging
1 Team members, visitors, external collaborators
Research Scientist
- Helene Leman [Inria, Researcher]
Faculty Members
- Emmanuel Grenier [Team leader, École Normale Supérieure de Lyon, Professor, HDR]
- Arthur Marly [École Normale Supérieure de Lyon, Certified Teacher, until Aug 2021]
- Paul Vigneaux [École Normale Supérieure de Lyon, Associate Professor, HDR]
Post-Doctoral Fellow
- Celine Bonnet [École Normale Supérieure de Lyon]
PhD Student
- Vianney Brouard [École Normale Supérieure de Lyon, from Sep 2021]
Technical Staff
- David Coulette [CNRS, Engineer]
Administrative Assistant
- Sylvie Boyer [Inria]
2 Overall objectives
The purpose of Numed is to develop new numerical methods and tools to simulate and parametrize complex systems arising in biology and medicine. Numed focuses on two axes:
-
Thema 1: Modeling using complex models: how to deal with multiple spatial or temporal scales (theoretical study, numerical simulations)?
This covers several aims: design of models of propagation taking into account the microscopic phenomena and starting from small scale description, importance of mechanics in the growth of tissues, peculiarities of tumor tissues, nonlinear rheology, evolutionary perspectives.
- Thema 2: Parametrization of complex models: how to find parameters for complex models, with particular emphasis on population approaches and on computationally expensive models.
Numed has also one main axe of applications, namely cancer modeling in close link with clinical data.
3 Research program
3.1 Design of complex models
Project team positioning
The originality of our work is the quantitative description of phenomena accounting for several time and spatial scales. Here, propagation has to be understood in a broad sense. This includes propagation of invasive species, chemotactic waves of bacteria, evolution of age structures populations ... Our main objectives are the quantitative calculation of macroscopic quantities as the rate of propagation, and microscopic distributions at the edge and the back of the front. These are essential features of propagation which are intimately linked in the long time dynamics.
Recent results
-
Population models.
H. Leman works at the interface between mathematics and biology, thanks to probabilist and determinist studies of models of populations. More precisely, she studies and develops probabilistic models, called agent-based models (or individual-based models) that described the population at an individual level. Each individual is characterized by one or more phenotypic traits and by its position, which may influence at the same time its ecological behavior and its motion. From a biological point of view these models are particularly interesting since they allow to include a large variety of interactions between individuals. These processes may also be studied in details to obtain theoretical results which may be simulated thanks to exact algorithms. To get quantitative results H. Leman uses changes of scales in space and time (large population, rare mutations, long time), following various biological assumptions.
In a first study , H. Leman and some collaborators studied a stochastic individual-based model of interacting plant and pollinator species through a bipartite graph: each species is a node of the graph, an edge representing interactions between a pair of species. Each species is characterized by a trait corresponding to its degree of generalism. This trait determines the structure of the interactions graph. Deterministic approximations of the stochastic measure-valued process by systems of ordinary differential equations or integro-differential equations were established and studied, when the population is large or when the graph is dense and can be replaced with a graphon. Studying the continuous limits of the interaction network and the resulting PDEs, H. Leman and her collaborators showed that nested plant-pollinator communities are expected to collapse towards a coexistence between a single pair of species of plants and pollinators.
As a second example, H. Leman studied the effect of disassortative mating on the level of polymorphism. To this aim, a model of population with a single locus , where disassortative crosses between individuals with different -alleles are more successful than assortative ones, were studied. The fitness benefits associated with disassortative crosses allowing to maintain allelic polymorphism was determined using studies on dynamical systems and matrices. Then H.Leman and her collaborators investigated the successive introductions of mutants. Therefore they gave general predictions on the diversity expected in loci targeted by disassortative mate choice.
-
Landau damping.
The question of the stability of equilibria of the Vlasov Poisson equations which arise in Plasma phycis has been pioneered in the periodic case for analytic data by C. Mouhot and C. Villani in their much acclaimed "Acta Mathematica" paper (part of the Fields medal for C. Villani).
However their proof is highly technical and long (about 200 pages). It turns out that the use of ideas previously developped by T. Nguyen and E. Grenier for the study of the inviscid limit of Navier Stokes equations greatly simplify the proof. A combination of an elementary study of the Green function and of a new analytic tool leads to an elementary, 15 pages long proof of Mouhot Villani's theorem. This new approach opens the way to the study of the whole space case, which is physically more relevant.
-
Numerical analysis of complex fluids: the example of avalanches.
This deals with the development of numerical schemes for viscoplastic materials (namely with Bingham or Herschel-Bulkley laws). Recently, with other colleagues, Paul Vigneaux finished the design of the first 2D well-balanced finite volume scheme for a shallow viscoplastic model. It is illustrated on the famous Taconnaz avalanche path in the Mont-Blanc (see figure 1), Chamonix, in the case of dense snow avalanches. The scheme deals with general Digital Elevation Model (DEM) topographies, wet/dry fronts and is designed to compute precisely the stopping state of avalanches, a crucial point of viscoplastic flows which are able to rigidify [cf joint Figure and Fernandez-Nieto et al. JCP 2018]. Currently, through a collaboration with IRSTEA Grenoble, we also revisit the theory of viscoplastic boundary layers (see figure (2) by extending the Oldroyd's asymptotic scaling (1947) to the cases of moderate Bingham numbers (or Herschel-Bulkley numbers). Also with IRSTEA, we are developping a joint study (numerical and experimental) of viscoplastic avalanches in the lab, to challenge various yield stress models.

Figure 1: An example of avalanche simulation 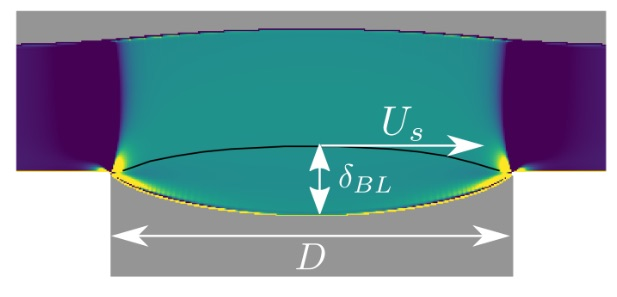
Figure 2: An example of boundary layer for complex flows
Collaborations
- Ecology: Lille (T. Rey and S. Billiard), Univ. Paris Est (C. Tran), Univ. Paris Sud (C. Coron), Toulouse (IMT, M. Costa), MNHM Paris (V. Llaurens), LISC Paris (C. Smadi).
- Landau Damping::Penn State University (T. Nguyen), Princeton University (I. Rodniansky).
- Numerical analysis of complex fluids: Enrique D. Fernandez - Nieto (Univ. de Sevilla, Spain), Jose Maria Gallardo (Univ. de Malaga, Spain).
- Comparison between numerical simulations and physical experiments for the dam-break of viscoplastic materials: collaboration with IRSTEA (now INRAE, since Jan. 2020).
3.2 Parametrization of complex systems
Project-team positioning
Clinical data are often sparse: we have few data per patient. The number of data is of the order of the number of parameters. In this context, a natural way to parametrize complex models with real world clinical data is to use a Bayesian approach, namely to try to find the distribution of the model parameters in the population, rather than to try to identify the parameters of every single patient. This approach has been pioneered in the 90's by the Nonmem software, and has been much improved thanks to Marc Lavielle in the 2000's. Refined statistical methods, called SAEM, have been tuned and implemented in commercial softwares like Monolix.
Recent results
The main problem when we try to parametrize clinical data using complex systems is the computational time. One single evaluation of the model can be costly, in particular if this model involves partial differential equations, and SAEM algorithm requires hundreds of thousands of single evaluations. The time cost is then too large, in particular because SAEM may not be parallelized.
To speed up the evaluation of the complex model, we replace it by an approximate one, or so called metamodel, constructed by interpolation of a small number of its values. We therefore combine the classical SAEM algorithm with an interpolation step, leading to a strong acceleration. Interpolation can be done through a precomputation step on a fixed grid, or through a more efficient kriging step. The interpolation grid or the kriging step may be improved during SAEM algorithm in an iterative way in order to get accurate evaluations of the complex system only in the domain of interest, namely near the clinical values [14],[15].
We applied these new algorithms to synthetic data and are currently using them on glioma data. We are also currently trying to prove the convergence of the corresponding algorithms. We will develop glioma applications in the next section.
Moreover E. Ollier in his phD developed new strategies to distinguish various populations within a SAEM algorithm [23].
We have two long standing collaborations with Sanofi and Servier on parametrization issues:
- Servier: during a four years contract, we modelled the pkpd of new drugs and also study the combination and optimization of chimiotherapies.
- Sanofi: during a eight years contract, Emmanuel Grenier wrote a complete software devoted to the study of the degradation of vaccine. This software is used worldwide by Sanofi R&D teams in order to investigate the degradation of existing or new vaccines and to study their behavior when they are heated. This software has been used on flu, dengue and various other diseases.
Collaborations
- Academic collaborations: A. Leclerc Samson (Grenoble University)
- Medical collaborations: Dr Ducray (Centre Léon Bérard, Lyon) and Dr Sujobert (Lyon Sud Hospital)
- Industrial contracts: we used parametrization and treatment improvement techniques for Servier (four years contract, on cancer drug modeling and optimization) and Sanofi (long standing collaboration)
4 Application domains
4.1 Multiscale models in oncology
Project-team positioning
Cancer modeling is the major topic of several teams in France and Europe, including Mamba, Monc and Asclepios to quote only a few Inria teams. These teams try to model metastasis, tumoral growth, vascularisation through angiogenesis, or to improve medical images quality. Their approaches are based on dynamical systems, partial differential equations, or on special imagery techniques.
Numed focuses on the link between very simple partial differential equations models, like reaction diffusion models, and clinical data.
Results
During 2021 we developed new collaborations and strengthened previous ones with the Centre Léon Bérard (Lyon), in particular on the following topics.
- Apoptosis: the question is to investigate whether the fate of neighboring cells influence the evolution of a given cell towards apoptosis, starting from videos of in vitro drug induced apoptosis.
- Breast cancer: In vitro study of the role of metabolic adaptation in drug resistance.
Collaborations
- Centre Léon Bérard (in particular: G. Ichim, R. Mounier, P. Martinez, A. Vigneron).
4.1.1 Live-cell imaging and mathematical analysis of apoptosis
This project is in collaboration with G. Ichim (Inserm, CRCL, Lyon).
The aim of the project is to accurately estimate the variability of apoptosis induction in a cell population. Revealing the response to a given apoptotic stimulus provide a better knowledge for misunderstood phenomena such as fractional killing or apoptosis-induced proliferation, which are problematic in the context of apoptosis-based cancer therapies.
To this aim, G. Ichim and his collaborators developed a medium-throughput imaging pipeline using apoptotic cell markers, that we coupled with mathematical analysis. This allowed our team to unravel a neighboring effect for apoptosis induction, irrespective of cell death stimuli or cell density. Finally, we gave evidences that the probability of undergoing apoptosis is increased in the proximity of apoptotic cells.
4.1.2 Evolutionary Mechanisms of Metabolic Adaptation
This project is in collaboration with P. Martinez (Inserm, CRCL, Lyon), A. Vigneron (Univ. Lyon 1, CRCL, Lyon). C. Bonnet, post-doc at UMPA in Numed team, has been recruted on EMMA-STON (Plan Cancer, MIC, 2019) to work on the modelling part of this project.
The EMMA-STON project aims to determine how intrinsic evolutionary characteristics of cells, such as genetic instability and plasticity, can influence the dynamics of therapeutic resistance. The ultimate goal is to define whether and how combinatorial therapeutic regimens can be better tailored to these characteristics. The project focuses in particular on metabolically targeted therapies in triple negative breast cancer models.
On the theoretical side, advances focused on the development of an adequate model and the methodology to parameterize it in an efficient way, while taking into account the noise inherent to the experimentally produced biological data.
In order to verify the parameterization of the mathematical model and its identifiability, we set up a first test on human mammary epithelial cells in 3 different culture conditions. Our first results show a loss of proliferative dye over time which can disturb the estimation of death and birth rates. This loss appears to be non-linear over time and probably results from an experimental technical bias inherent to the use of the dye. We are currently working on the estimation of this loss by empirical measurements, in order to integrate it in the mathematical models.
4.2 Myogenesis
This project is a collaboration with Bénédicte Chazaud (DR INSERM), Rémi Mounier (DR CNRS), Julien Gondin (CR CNRS) from the Chazaud "MuscleStem" Team (https://www.musclestem.com) at Institut Neuromyogène (CNRS UMR 5310 - INSERM U1217, https://www.inmg.fr).
The MuscleStem project research's focus is myogenesis (muscle tissue regeneration), with an emphasis on the role of muscular stem cells in myogenesis dynamics in skeletal muscles and the impact of stem cells environment on stem cells activation. This work is of particular interest not only for a better understanding of healthy muscle behavior, but also with respect to the understanding and treatment of pathological behavior such as encountered in myopathy or cachexia.
A first step in this collaborative effort is the development of an automated data acquisition pipeline to characterize stem cell environment in skeletal muscle cells. Raw experimental data are images of transverse cuts of skeletal muscle with channels corresponding to fluorescent staining responding respectively to cellular membranes, nuclei and stems cells
We designed and developed the MYOSEG analysis pipeline, a workflow and software collection using the standard Python scientific stack. Its purpose is to accurately identify biological objects (cells, nuclei , stem cells) from raw image data and output various metrics (cross section area, colocalization ...). The workflow uses both classical parametric segmentation techniques and statistical learning techniques for objects identification. It has been validated against expert ground-truth data and exhibits accuracy on par with human experts.
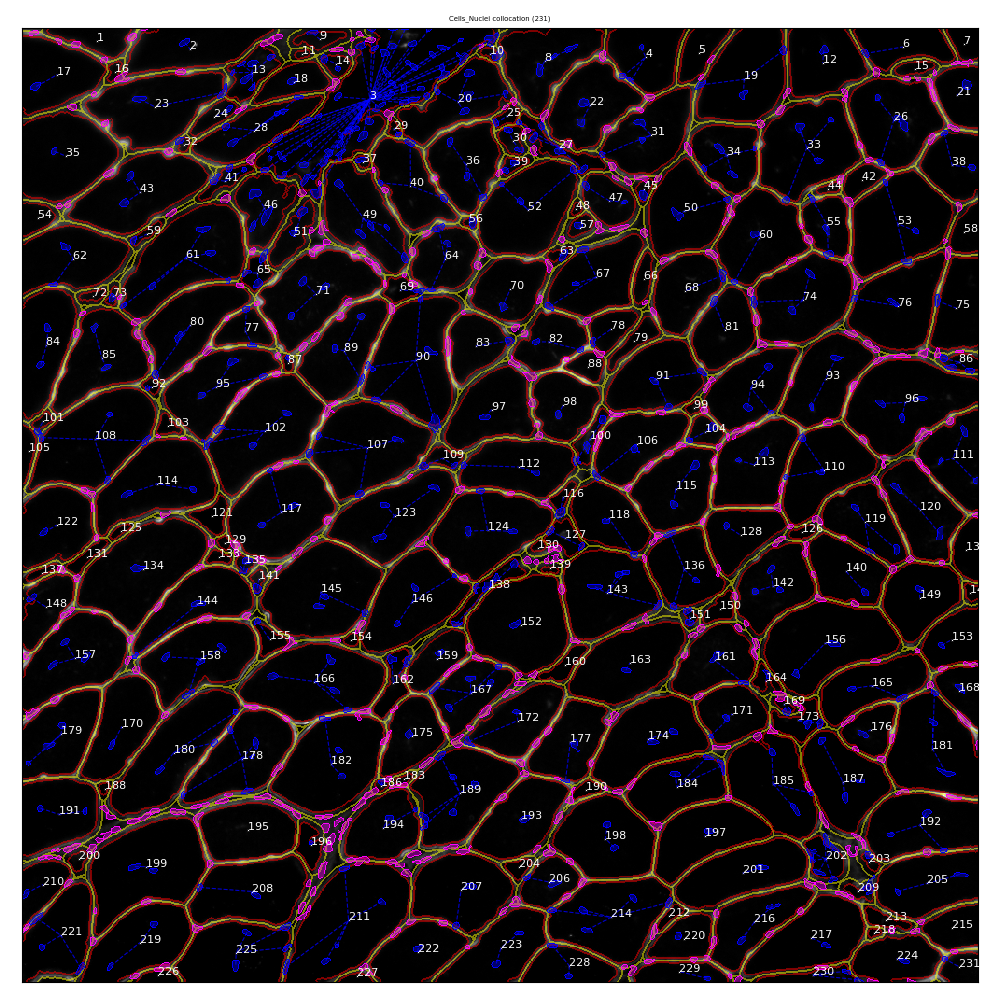
Segmentation of muscular tissue cross section and nuclei
4.3 Soil microbiote modeling using the DEMENT model
The DEMENT model depicts dynamics of litter decomposition by the soil microbiote (bacteria, fungi). It is essentially a spatial biochemical model : both nutrients, enzymes and microbes are described by their biomass concentration on a two-dimensional grid. Microbial entities act both as consumers (products of litter degradation) and producers (substrate degradation enzymes, dead matter, metabolic outputs...). Biochemical reactions and microbial metabolism are described by first order chemical reaction kinetics, completed by ad-hoc stochastic processes for microbial death, reproduction and spatial dispersion. From a genetic standpoint, the DEMENT model is an ecological one : it operates with a fixed set of microbial types defined by constant genetic parameters. In order to extend the model to allow for mutations with as little modification as possible, ie without going to a full agent stochastic model, we chose to keep the maximal number of microbial types fixed but allow for dynamical transition to one type from another during reproduction events.
Previous implementations of the DEMENT model are the seminal R implementation by S. Allison DEMENT and an updated Python port by B. Wang DEMENTpy. Though fit for their initial purpose, those implementations do not lend themselves well to extension, both in terms of problem scale and addition of new processes. We developed DEMENTmutant, a new Python implementation with a more extensible "toolbox" design in which heavy-duty computational kernels are separated as much as possible from high-level ecosystem modeling constructs. This separation allows to strike a balance between computational efficiency, design flexibility and accessibility to non-experts. Without any particular optimization the current implementation outperforms the previous ones of at least one order of magnitude both in terms of computation time and memory footprint. Parallel extensions (multithreaded and/or mpi based) are under consideration to tackle larger problems.
5 Social and environmental responsibility
We are aware of the problem of climate change and we are trying to lower our footprint. H. Leman is also responsible "sustainable development" of UMPA laboratory.
6 Highlights of the year
The project of H. Leman, in collaboration with A. Lambert and H. Morlon (ENS, Paris), entitled "Stochastic Evolution of Trait Diversity" has been selected for funding by the "Institut des mathématiques pour la planète Terre" (Institut web page). Thuy Vo has been recruted as a 2 years post-doc at UMPA on this funding.
7 New software and platforms
Work with E. Abs and DEMENT software.
7.1 New software
7.1.1 Bingham flows
-
Functional Description:
A 1D and 2D code with a new method for the computation of viscoplatic flows with free-surface. It essentially couples Optimization methods and Well-Balanced Finite-Volumes schemes for viscous shallow-water equations (induced by the viscoplastic nature of the fluid). Currently applied to avalanches of dense snow, it is a private code currently actively developed (in C++). One of the key feature is that its well-balanced property allows to obtained the stationary states which are linked to the stopping of the snow avalanche for this highly non-linear type of fluid.
-
Contact:
Emmanuel Grenier
7.1.2 OptimChemo
-
Functional Description:
OptimChemo is a userfriendly software designed to study numerically the effect of multiple chemotherapies on simple models of tumour growth and to optimize chemotherapy schedules.
-
Contact:
Emmanuel Grenier
-
Participants:
Ehouarn Maguet, Emmanuel Grenier, Paul Vigneaux, Violaine Louvet
7.1.3 SETIS
-
Keywords:
Health, DICOM, Medical imaging, Drug development
-
Functional Description:
SETIS software is a GUI allowing to treat DICOM medical images to extract pathological data. These data can then be exported and used in a SAEM software (including Monolix (Inria & Lixoft)) for the parameters' estimation of models in the context of population approaches. As an example SETIS can be used to segment and compute the tumor size of a patients from MRI scans taken at different times. The software is sufficiently general to be used in various situations by clinicians (already done by colleagues in Lyon Hospital).
-
Contact:
Paul Vigneaux
-
Participants:
Ehouarn Maguet, Paul Vigneaux
-
Partner:
ENS Lyon
7.1.4 SIMPHYT
-
Keywords:
Bioinformatics, Cancer, Drug development
-
Functional Description:
SimPHyt is an implementation in Python of the low grad glioma model. The aim is to predict the evolution of the glioma size of patients.
-
Contact:
Benjamin Ribba
-
Participant:
Benjamin Ribba
7.1.5 SITLOG
-
Contact:
Emmanuel Grenier
-
Participants:
Benjamin Ribba, Morgan Martinet
7.1.6 VAXSIMSTAB
-
Keywords:
Bioinformatics, Health, Drug development
-
Functional Description:
VAXSIMSTAB is a modeler stability prediction of vaccine software.
-
Contact:
Benjamin Ribba
-
Participants:
Benjamin Ribba, Emmanuel Grenier, Vincent Calvez
8 New results
E. Grenier, in collaboration with T. Nguyen (Penn State University) and I. Rodniansky (Princeton University) have given a simplified proof of the Landau damping for analytic initial data, a famous result by C. Mouhot and C. Villani.
9 Partnerships and cooperations
9.1 National initiatives
Paul Vigneaux is the Principal investigator of the ANR-20-CE46-0006 VPFLOWS PRC
Paul Vigneaux participates to the INRIA-INRAE Action Exploratoire (AEx) GRANIER GRAvitatioNal hazards in mountaIns in the contExt of Risks prediction — Nonsmooth modeling and simulation with data in Geomechanics. Coordinators : Vincent Acary, Franck Bourrier
Emmanuel Grenier is coordinateur of EmmaStone, an Inserm - Plan Cancer project on the modeling of bitherapies in oncology.
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: selection
Reviews for Mathematical Models and Methods in Applied Sciences.
Reviews for Journal of Computational Physics, ESAIM : M2AN (Mathematical Modelling and Numerical Analysis), Computers and Mathematics with Applications, Journal of Non-Newtonian Fluid Mechanics,
10.1.2 Invited talks
P. Vigneaux gave a talk at the Séminaire EDP/AIRSEA/CVGI au LJK, Grenoble. 25 Mars 2021.
H. Leman has been invited to the following international conferences and seminars:
- Oct. 2021 Séminaire de probabilités, Institut Fourier (Grenoble).
- Oct. 2021 Conférence Readinet 2021, online. Juin 2021 and Conférence “Probability and Evolution”, CIRM, Marseille.
- Mars 2021 Séminaire de probabilités, IMT (Toulouse).
10.1.3 Leadership within the scientific community
Paul Vigneaux is director of the national GdR CNRS MathGeoPhy (number 3485) (2021-).
Paul Vigneaux is scientific co-head of the national CNRS research group GdR EGRIN (2019-2021)
10.1.4 Scientific expertise
H. Leman has evaluated projects for HFSP Research Grant application.
E. Grenier has been member of the Scientific Council of the CLARA (Cancéropole)
E. Grenier is member of the CSS2 Scientific Committee of INSERM (Cancerology and genetic diseases)
10.1.5 Research administration
H. Leman is part of the "conseil de laboratoire" of UMPA.
10.2 Teaching - Supervision - Juries
P. Vigneaux is the head of the 1st year of the Advanced Mathematics Master of ENS de Lyon (ENSL) (2019-2021)
P. Vigneaux is a member of the Board of Doctoral School ED 512 Informatique-Mathématiques (2021-)
H. Leman taught M1 class of Stochastic Processes at ENS Lyon and L2 class of maths for medical school students at Univ. Lyon 1.
P. Vigenaux taught in the Preparation for the Agrégation of Mathematics, national competitive examination.
P. Vigneaux taught a course "Computational Sciences: an introduction to modelling" for L3-M2 for students from (physics, computer science, biology, geology, etc) at ENS de Lyon.
E. Grenier taught in L3, M1 and M2 (PDE lectures).
H. Leman gave the following M2 seminars:
- Avr. 2021, Graduate Seminar, Universidade Federal do ABC, Sao Paulo.
- Fev. 2021, Séminaire du M2 Mathématiques pour les sciences du vivant (Univ. Paris Sud).
- Jan. 2021, Master Class du M2 Mathématiques appliquées CEPS: Mathématiques pour la biologie et l'écologie (Univ. Marseille).
10.2.1 Supervision
P. Vigneaux supervised Clément Berger. PhD Grant élève normalien. 2021- and Colin Ginot. PhD Grant ANR. 2021-. Co-advised with Guillaume Chambon (INRAE Grenoble) and Pierre Philippe (INRAE Aix)
H. Leman supervised:
- Hanane Hassainia (M2, 2021),"Bayesian estimation for metabolic reconstruction", with Arnaud Vigneron (Univ. Lyon 1, CRCL).
- Céline Bonnet (Post-doc, 2020-), "Mathematical modelling of cancer", with Pierre Martinez (Inserm, CRCL).
- Thuy Vo (Post-doc, 2021-), "Multi-scales studies of genetic diversity", with Amaury Lambert, Hélène Morlon (IBENS, ENS Paris).
- Vianney Brouard (Doc, 2021-), "Cellular dynamics of multitype populations in oncology", with Vincent Calvez (ICJ, Univ. Lyon 1, Dracula team).
10.2.2 Juries
Paul Vigneaux has been member of PhD Committee: (13 Déc.) Rapporteur. G. Dongmo. Univ. Paris-Saclay. Encadrants : Ch. Berthon and Q. H. Tran (IFPEN). (2 Déc.) F. Henri. Univ. Bordeaux. Encadrants : P. Lubin and M. Coquerelle.
10.3 Popularization
10.3.1 Education
H. Leman and D. Coulette hosted 4 pupils (3eme and 2nd) between 1 day and 1 week in 2021.
11 Scientific production
11.1 Publications of the year
International journals

