Keywords
Computer Science and Digital Science
- A6.1. Methods in mathematical modeling
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.4. Multiscale modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.4. Statistical methods
- A6.2.6. Optimization
- A6.2.7. High performance computing
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.3.4. Model reduction
Other Research Topics and Application Domains
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B1.1.10. Systems and synthetic biology
- B2.2.3. Cancer
- B2.4.2. Drug resistance
- B2.6.1. Brain imaging
- B2.6.3. Biological Imaging
1 Team members, visitors, external collaborators
Research Scientists
- Olivier Saut [Team leader, CNRS, Senior Researcher, HDR]
- Sebastien Benzekry [Inria, Researcher, until Apr 2021, HDR]
- Baudouin Denis De Senneville [CNRS, Researcher]
- Christele Etchegaray [Inria, Researcher]
- Clair Poignard [Inria, Senior Researcher, HDR]
Faculty Members
- Annabelle Collin [Bordeaux INP, Associate Professor]
- David Dean [Univ de Bordeaux, Professor, from Sep 2021]
Post-Doctoral Fellows
- Kokou Atsou [Inria, until Apr 2021]
- Anne Sophie Giacobbi [Univ de Bordeaux, until Nov 2021]
- Van Linh Le [Inria]
- Oceane Saincir [Inria, until Mar 2021]
PhD Students
- Celestin Bigarre [Inria, until Apr 2021]
- Khaoula Chahdi [Inria, from Oct 2021]
- Eloise Inacio [Inria, from Oct 2021]
- Pedro Jaramillo Aguayo [Univ de Bordeaux]
- Guillaume Martinroche [Univ de Bordeaux, from Oct 2021]
- Virginie Montalibet [Univ de Bordeaux, from Oct 2021]
- Simone Nati Poltri [Institut National Polytechnique de Bordeaux, from Oct 2021]
Technical Staff
- Cedrick Copol [Inria, Engineer, until Feb 2021]
- Jerome Faure [Inria, Engineer, Dec 2021]
- Luc Lafitte [Inria, Engineer, from Nov 2021]
- Hripsime Snkhchyan [Inria, Engineer, from Apr 2021]
Interns and Apprentices
- Isabella Bicalho Frazeto [Inria, from Apr 2021 until Jun 2021]
- Pierre Charitat [École Normale Supérieure de Lyon, from May 2021 until Jun 2021]
- Manon Charmasson [Inria, from May 2021 until Jun 2021]
- Jerome Faure [Inria, from Feb 2021 until Jul 2021]
- Remy Jelin [Inria, from Feb 2021 until Aug 2021]
- Clara Mazzocco [Ministère de l'Education, Jun 2021]
- Simone Nati Poltri [Inria, from May 2021 until Aug 2021]
- Francois Sorre [Bordeaux INP, from Jun 2021 until Jul 2021]
- Ruben Taieb [Université Claude Bernard Lyon 1, from Jun 2021 until Aug 2021]
- Cedric Trabado [Inria, from Feb 2021 until Jul 2021]
- Victor Virfollet [Inria, from Apr 2021 until Aug 2021]
Administrative Assistant
- Sylvie Embolla [Inria]
External Collaborators
- Thierry Colin [Sophia Genetics, HDR]
- Amandine Crombe [Institut Bergonié]
- Charles Mesguich [Centre hospitalier universitaire de Bordeaux]
- Benjamin Taton [Centre hospitalier universitaire de Bordeaux]
- Damien Voyer [Ecole d'ingénieurs en génie des systèmes industriels]
2 Overall objectives
2.1 Objectives
The MONC project-team aims at developing new mathematical models from partial differential equations and statistical methods and based on biological and medical knowledge. Our goal is ultimately to be able to help clinicians and/or biologists to better understand, predict or control the evolution of the disease and possibly evaluate the therapeutic response, in a clinical context or for pre-clinical studies. We develop patient-specific approaches (mainly based on medical images) as well as population-type approaches in order to take advantage of large databases.
In vivo modeling of tumors is limited by the amount of information available. However, recently, there have been dramatic increases in the scope and quality of patient-specific data from non-invasive imaging methods, so that several potentially valuable measurements are now available to quantitatively measure tumor evolution, assess tumor status as well as anatomical or functional details. Using different techniques from biology or imaging - such as CT scan, magnetic resonance imaging (MRI), or positron emission tomography (PET) - it is now possible to evaluate and define tumor status at different levels or scales: physiological, molecular and cellular.
In the meantime, the understanding of the biological mechanisms of tumor growth, including the influence of the micro-environment, has greatly increased. Medical doctors now have access to a wide spectrum of therapies (surgery, mini-invasive techniques, radiotherapies, chemotherapies, targeted therapies, immunotherapies...).
Our project aims at helping oncologists in their followup of patients via the development of novel quantitative methods for evaluation cancer progression. The idea is to build phenomenological mathematical models based on data obtained in the clinical imaging routine like CT scans, MRIs and PET scans. We therefore want to offer medical doctors patient-specific tumor evolution models, which are able to evaluate – on the basis of previously collected data and within the limits of phenomenological models – the time evolution of the pathology at subsequent times and the response to therapies. More precisely, our goal is to help clinicians answer the following questions thanks to our numerical tools:
- When is it necessary to start a treatment?
- What is the best time to change a treatment?
- When to stop a treatment?
We also intend to incorporate real-time model information for improving the accuracy and efficacy of non invasive or micro-invasive tumor ablation techniques like acoustic hyperthermia, electroporation, radio-frequency, cryo-ablation and of course radiotherapies.
There is therefore a dire need of integrating biological knowledge into mathematical models based on clinical or experimental data. A major purpose of our project is also to create new mathematical models and new paradigms for data assimilation that are adapted to the biological nature of the disease and to the amount of multi-modal data available.
2.2 General strategy
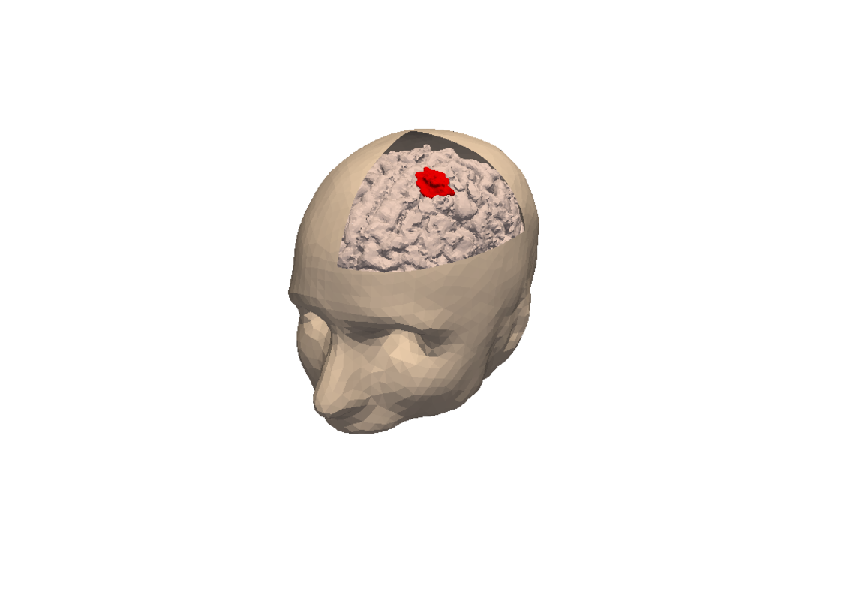
3D numerical simulation of a meningioma.
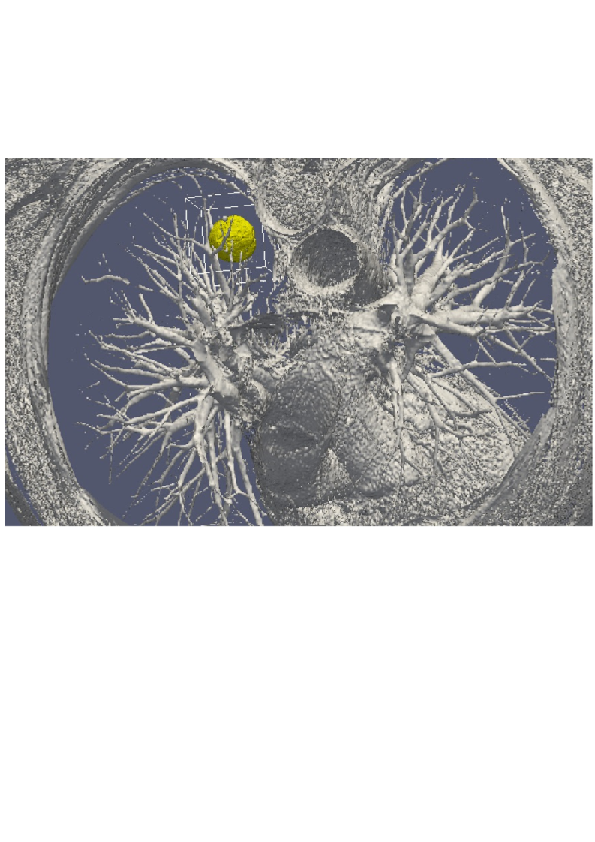
3D numerical simulation of a lung tumor.
Our general strategy may be described with the following sequence:
- Stage 1: Derivation of mechanistic models based on the biological knowledge and the available observations. The construction of such models relies on the up-to-date biological knowledge at the cellular level including description of the cell-cycle, interaction with the microenvironement (angiogenesis, interaction with the stroma). Such models also include a "macroscopic" description of specific molecular pathways that are known to have a critical role in carcinogenesis or that are targeted by new drugs. We emphasize that for this purpose, close interactions with biologists are crucial. Lots of works devoted to modeling at the cellular level are available in the literature. However, in order to be able to use these models in a clinical context, the tumor is also to be described at the tissue level. The in vitro mechanical characterization of tumor tissues has been widely studied. Yet no description that could be patient specific or even tumor specific is available. It is therefore necessary to build adapted phenomenological models, according to the biological and clinical reality.
-
Stage 2: Data collection. In the clinical context, data may come from medical imaging (MRI, CT-Scan, PET scan) at different time points. We need longitudinal data in time in order to be able to understand or describe the evolution of the disease. Data may also be obtained from analyses of blood samples, biopsies or other quantitative biomarkers. A close collaboration with clinicians is required for selecting the specific cases to focus on, the understanding of the key points and data, the classification of the grades of the tumors, the understanding of the treatment,...In the preclinical context, data may for instance be macroscopic measurements of the tumor volume for subcutaneous cases, green fluorescence protein (GFP) quantifications for total number of living cells, non-invasive bioluminescence signals or even imaging obtained with devices adapted to small animals.
- Data processing: Besides selection of representative cases by our collaborators, most of the time, data has to be processed before being used in our models. We develop novel methods for semi-automatic (implemented in SegmentIt) as well as supervized approaches (machine learning or deep learning) for segmentation, non-rigid registration and extraction of image texture information (radiomics, deep learning).
- Stage 3: Adaptation of the model to data. The model has to be adapted to data: it is useless to have a model considering many biological features of the disease if it cannot be reliably parameterized with available data. For example, very detailed descriptions of the angiogenesis process found in the literature cannot be used, as they have too much parameters to determine for the information available. A pragmatic approach has to be developed for this purpose. On the other hand, one has to try to model any element that can be useful to exploit the image. Parameterizing must be performed carefully in order to achieve an optimal trade-off between the accuracy of the model, its complexity, identifiability and predictive power. Parameter estimation is a critical issue in mathematical biology: if there are too many parameters, it will be impossible to estimate them but if the model is too simple, it will be too far from reality.
-
Stage 4: Data assimilation. Because of data complexity and scarcity - for example multimodal, longitudinal medical imaging - data assimilation is a major challenge. Such a process is a combination of methods for solving inverse problems and statistical methods including machine learning strategies.
- Personalized models: Currently, most of the inverse problems developed in the team are solved using a gradient method coupled with some MCMC type algorithm. We are now trying to use more efficient methods as Kalman type filters or so-called Luenberger filter (nudging). Using sequential methods could also simplify Stage 3 because they can be used even with complex models. Of course, the strategy used by the team depends on the quantity and the quality of data. It is not the same if we have an homogeneous population of cases or if it is a very specific isolated case.
- Statistical learning: In some clinical cases, there is no longitudinal data available to build a mathematical model describing the evolution of the disease. In these cases (e.g. in our collaboration with Humanitas Research Hospital on low grade gliomas or Institut Bergonié on soft-tissue sarcoma), we use machine learning techniques to correlate clinical and imaging features with clinical outcome of patients (radiomics). When longitudinal data and a sufficient number of patients are available, we combine this approach and mathematical modeling by adding the personalized model parameters for each patient as features in the statistical algorithm. Our goal is then to have a better description of the evolution of the disease over time (as compared to only taking temporal variations of features into account as in delta-radiomics approaches). We also plan to use statistical algorithms to build reduced-order models, more efficient to run or calibrate than the original models.
- Data assimilation of gene expression. "Omics" data become more and more important in oncology and we aim at developing our models using this information as well. For example, in our work on GIST, we have taken the effect of a Ckit mutation on resistance to treatment into account. However, it is still not clear how to use in general gene expression data in our macroscopic models, and particularly how to connect the genotype to the phenotype and the macroscopic growth. We expect to use statistical learning techniques on populations of patients in order to move towards this direction, but we emphasize that this task is very prospective and is a scientific challenge in itself.
- Stage 5: Patient-specific Simulation and prediction, Stratification. Once the mechanistic models have been parametrized, they can be used to run patient-specific simulations and predictions. The statistical models offer new stratifications of patients (i.t. an algorithm that tells from images and clinical information wheter a patient with soft-tissue sarcoma is more likely to be a good or bad responder to neoadjuvant chemotherapy). Building robust algorithms (e.g. that can be deployed over multiple clinical centers) also requires working on quantifying uncertainties.
3 Research program
3.1 Introduction
We are working in the context of data-driven medicine against cancer. We aim at coupling mathematical models with data to address relevant challenges for biologists and clinicians in order for instance to improve our understanding in cancer biology and pharmacology, assist the development of novel therapeutic approaches or develop personalized decision-helping tools for monitoring the disease and evaluating therapies.
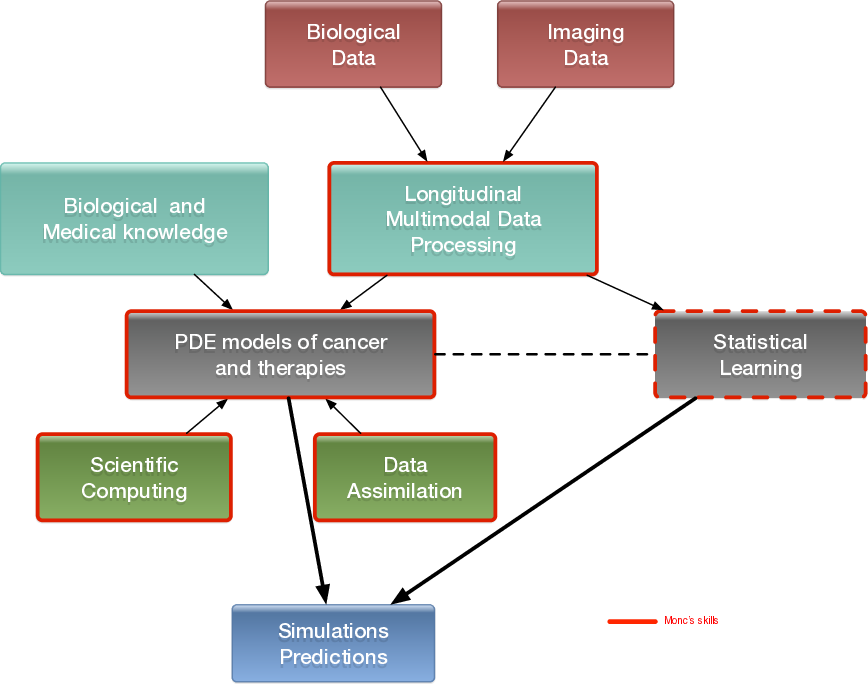
More precisely, our research on mathematical oncology is three-fold:
- Axis 1: Tumor modeling for patient-specific simulations: Clinical monitoring. Numerical markers from imaging data. Radiomics.
- Axis 2: Bio-physical modeling for personalized therapies: Electroporation from cells to tissue. Radiotherapy.
- Axis 3: Quantitative cancer modeling for biological, clinical and preclinical studies: Biological mechanisms. Metastatic dissemination. Pharmacometrics.
In the first axis, we aim at producing patient-specific simulations of the growth of a tumor or its response to treatment starting from a series of images. We hope to be able to offer a valuable insight on the disease to the clinicians in order to improve the decision process. This would be particularly useful in the cases of relapses or for metastatic diseases.
The second axis aims at modeling biophysical therapies like electroporation, but also radiotherapy, thermo-ablations, radio-frequency ablations or electroporation that play a crucial role for a local treatment of the disease if possible limiting the metastatic dissemination, which is precisely the clinical context where the techniques of axis 1 will be applied.
The third axis is essential since it is a way to better understand and model the biological reality of cancer growth and the (possibly complex) effects of therapeutic intervention. Modeling in this case also helps to interpret the experimental results and improve the accuracy of the models used in Axis 1. Technically speaking, some of the computing tools are similar to those of Axis 1.
3.2 Axis 1: Tumor modeling for patient-specific simulations
The gold standard treatment for most cancers is surgery. In the case where total resection of the tumor is possible, the patient often benefits from an adjuvant therapy (radiotherapy, chemotherapy, targeted therapy or a combination of them) in order to eliminate the potentially remaining cells that may not be visible. In this case personalized modeling of tumor growth is useless and statistical modeling will be able to quantify the risk of relapse, the mean progression-free survival time...However if total resection is not possible or if metastases emerge from distant sites, clinicians will try to control the disease for as long as possible. A wide set of tools are available. Clinicians may treat the disease by physical interventions (radiofrequency ablation, cryoablation, radiotherapy, electroporation, focalized ultrasound,...) or chemical agents (chemotherapies, targeted therapies, antiangiogenic drugs, immunotherapies, hormonotherapies). One can also decide to monitor the patient without any treatment (this is the case for slowly growing tumors like some metastases to the lung, some lymphomas or for some low grade glioma). A reliable patient-specific model of tumor evolution with or without therapy may have different uses:
- Case without treatment: the evaluation of the growth of the tumor would offer a useful indication for the time at which the tumor may reach a critical size. For example, radiofrequency ablation of pulmonary lesion is very efficient as long as the diameter of the lesion is smaller than 3 cm. Thus, the prediction can help the clinician plan the intervention. For slowly growing tumors, quantitative modeling can also help to decide at what time interval the patient has to undergo a CT-scan. CT-scans are irradiative exams and there is a challenge for decreasing their occurrence for each patient. It has also an economical impact. And if the disease evolution starts to differ from the prediction, this might mean that some events have occurred at the biological level. For instance, it could be the rise of an aggressive phenotype or cells that leave a dormancy state. This kind of events cannot be predicted, but some mismatch with respect to the prediction can be an indirect proof of their existence. It could be an indication for the clinician to start a treatment.
- Case with treatment: a model can help to understand and to quantify the final outcome of a treatment using the early response. It can help for a redefinition of the treatment planning. Modeling can also help to anticipate the relapse by analyzing some functional aspects of the tumor. Again, a deviation with respect to reference curves can mean a lack of efficiency of the therapy or a relapse. Moreover, for a long time, the response to a treatment has been quantified by the RECIST criteria which consists in (roughly speaking) measuring the diameters of the largest tumor of the patient, as it is seen on a CT-scan. This criteria is still widely used and was quite efficient for chemotherapies and radiotherapies that induce a decrease of the size of the lesion. However, with the systematic use of targeted therapies and anti-angiogenic drugs that modify the physiology of the tumor, the size may remain unchanged even if the drug is efficient and deeply modifies the tumor behavior. One better way to estimate this effect could be to use functional imaging (Pet-scan, perfusion or diffusion MRI, ...), a model can then be used to exploit the data and to understand in what extent the therapy is efficient.
- Optimization: currently, we do not believe that we can optimize a particular treatment in terms of distribution of doses, number, planning with the model that we will develop in a medium term perspective.
The scientific challenge is therefore as follows: given the history of the patient, the nature of the primitive tumor, its histopathology, knowing the treatments that patients have undergone, some biological facts on the tumor and having a sequence of images (CT-scan, MRI, PET or a mix of them), are we able to provide a numerical simulation of the extension of the tumor and of its metabolism that fits as best as possible with the data (CT-scans or functional data) and that is predictive in order to address the clinical cases described above?
Our approach relies on the elaboration of PDE models and their parametrization with images by coupling deterministic and stochastic methods. The PDE models rely on the description of the dynamics of cell populations. The number of populations depends on the pathology. For example, for glioblastoma, one needs to use proliferative cells, invasive cells, quiescent cells as well as necrotic tissues to be able to reproduce realistic behaviors of the disease. In order to describe the relapse for hepatic metastases of gastro-intestinal stromal tumor (gist), one needs three cell populations: proliferative cells, healthy tissue and necrotic tissue.
The law of proliferation is often coupled with a model for the angiogenesis. However such models of angiogenesis involve too many non measurable parameters to be used with real clinical data and therefore one has to use simplified or even simplistic versions. The law of proliferation often mimics the existence of an hypoxia threshold, it consists of an ODE. or a PDE that describes the evolution of the growth rate as a combination of sigmoid functions of nutrients or roughly speaking oxygen concentration. Usually, several laws are available for a given pathology since at this level, there are no quantitative argument to choose a particular one.
The velocity of the tumor growth differs depending on the nature of the tumor. For metastases, we will derive the velocity thanks to Darcy's law in order to express that the extension of the tumor is basically due to the increase of volume. This gives a sharp interface between the metastasis and the surrounding healthy tissues, as observed by anatomopathologists. For primitive tumors like glioma or lung cancer, we use reaction-diffusion equations in order to describe the invasive aspects of such primitive tumors.
The modeling of the drugs depends on the nature of the drug: for chemotherapies, a death term can be added into the equations of the population of cells, while antiangiogenic drugs have to be introduced in a angiogenic model. Resistance to treatment can be described either by several populations of cells or with non-constant growth or death rates. As said before, it is still currently difficult to model the changes of phenotype or mutations, we therefore propose to investigate this kind of phenomena by looking at deviations of the numerical simulations compared to the medical observations.
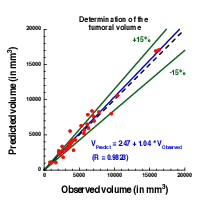
The calibration of the model is achieved by using a series (at least 2) of images of the same patient and by minimizing a cost function. The cost function contains at least the difference between the volume of the tumor that is measured on the images with the computed one. It also contains elements on the geometry, on the necrosis and any information that can be obtained through the medical images. We will pay special attention to functional imaging (PET, perfusion and diffusion MRI). The inverse problem is solved using a gradient method coupled with some Monte-Carlo type algorithm. If a large number of similar cases is available, one can imagine to use statistical algorithms like random forests to use some non quantitative data like the gender, the age, the origin of the primitive tumor...for example for choosing the model for the growth rate for a patient using this population knowledge (and then to fully adapt the model to the patient by calibrating this particular model on patient data) or for having a better initial estimation of the modeling parameters. We have obtained several preliminary results concerning lung metastases including treatments and for metastases to the liver.
3.3 Axis 2: Bio-physical modeling for personalized therapies
In this axis, we investigate locoregional therapies such as radiotherapy, irreversible electroporation. Electroporation consists in increasing the membrane permeability of cells by the delivery of high voltage pulses. This non-thermal phenomenon can be transient (reversible) or irreversible (IRE). IRE or electro-chemotherapy – which is a combination of reversible electroporation with a cytotoxic drug – are essential tools for the treatment of a metastatic disease. Numerical modeling of these therapies is a clear scientific challenge. Clinical applications of the modeling are the main target, which thus drives the scientific approach, even though theoretical studies in order to improve the knowledge of the biological phenomena, in particular for electroporation, should also be addressed. However, this subject is quite wide and we focus on two particular approaches: some aspects of radiotherapies and electro-chemotherapy. This choice is motivated partly by pragmatic reasons: we already have collaborations with physicians on these therapies. Other treatments could be probably treated with the same approach, but we do not plan to work on this subject on a medium term.
-
Radiotherapy (RT) is a common therapy for cancer. Typically, using a CT scan of the patient with the structures of interest (tumor, organs at risk) delineated, the clinicians optimize the dose delivery to treat the tumor while preserving healthy tissues. The RT is then delivered every day using low resolution scans (CBCT) to position the beams. Under treatment the patient may lose weight and the tumor shrinks. These changes may affect the propagation of the beams and subsequently change the dose that is effectively delivered. It could be harmful for the patient especially if sensitive organs are concerned. In such cases, a replanification of the RT could be done to adjust the therapeutical protocol. Unfortunately, this process takes too much time to be performed routinely. The challenges faced by clinicians are numerous, we focus on two of them:
- Detecting the need of replanification: we are using the positioning scans to evaluate the movement and deformation of the various structures of interest. Thus we can detect whether or not a structure has moved out of the safe margins (fixed by clinicians) and thus if a replanification may be necessary. In a retrospective study, our work can also be used to determine RT margins when there are no standard ones. A collaboration with the RT department of Institut Bergonié is underway on the treatment of retroperitoneal sarcoma and ENT tumors (head and neck cancers). A retrospective study was performed on 11 patients with retro-peritoneal sarcoma. The results have shown that the safety margins (on the RT) that clinicians are currently using are probably not large enough. The tool used in this study was developed by an engineer funded by INRIA (Cynthia Périer, ADT Sesar). We used well validated methods from a level-set approach and segmentation / registration methods. The originality and difficulty lie in the fact that we are dealing with real data in a clinical setup. Clinicians have currently no way to perform complex measurements with their clinical tools. This prevents them from investigating the replanification. Our work and the tools developed pave the way for easier studies on evaluation of RT plans in collaboration with Institut Bergonié. There was no modeling involved in this work that arose during discussions with our collaborators. The main purpose of the team is to have meaningful outcomes of our research for clinicians, sometimes it implies leaving a bit our area of expertise.
- Evaluating RT efficacy and finding correlation between the radiological responses and the clinical outcome: our goal is to help doctors to identify correlation between the response to RT (as seen on images) and the longer term clinical outcome of the patient. Typically, we aim at helping them to decide when to plan the next exam after the RT. For patients whose response has been linked to worse prognosis, this exam would have to be planned earlier. This is the subject of collaborations with Institut Bergonié and CHU Bordeaux on different cancers (head and neck, pancreas). The response is evaluated from image markers (e.g. using texture information) or with a mathematical model developed in Axis 1. The other challenges are either out of reach or not in the domain of expertise of the team. Yet our works may tackle some important issues for adaptive radiotherapy.
-
Both IRE and electrochemotherapy are anticancerous treatments based on the same phenomenon: the electroporation of cell membranes. This phenomenon is known for a few decades but it is still not well understood, therefore our interest is two fold:
- We want to use mathematical models in order to better understand the biological behavior and the effect of the treatment. We work in tight collaboration with biologists and bioeletromagneticians to derive precise models of cell and tissue electroporation, in the continuity of the research program of the Inria team-project MC2. These studies lead to complex non-linear mathematical models involving some parameters (as less as possible). Numerical methods to compute precisely such models and the calibration of the parameters with the experimental data are then addressed. Tight collaborations with the Vectorology and Anticancerous Therapies (VAT) of IGR at Villejuif, Laboratoire Ampère of Ecole Centrale Lyon and the Karlsruhe Institute of technology will continue, and we aim at developing new collaborations with Institute of Pharmacology and Structural Biology (IPBS) of Toulouse and the Laboratory of Molecular Pathology and Experimental Oncology (LMPEO) at CNR Rome, in order to understand differences of the electroporation of healthy cells and cancer cells in spheroids and tissues.
- This basic research aims at providing new understanding of electroporation, however it is necessary to address, particular questions raised by radio-oncologists that apply such treatments. One crucial question is "What pulse or what train of pulses should I apply to electroporate the tumor if the electrodes are located as given by the medical images"? Even if the real-time optimization of the placement of the electrodes for deep tumors may seem quite utopian since the clinicians face too many medical constraints that cannot be taken into account (like the position of some organs, arteries, nerves...), one can expect to produce real-time information of the validity of the placement done by the clinician. Indeed, once the placement is performed by the radiologists, medical images are usually used to visualize the localization of the electrodes. Using these medical data, a crucial goal is to provide a tool in order to compute in real-time and visualize the electric field and the electroporated region directly on theses medical images, to give the doctors a precise knowledge of the region affected by the electric field. In the long run, this research will benefit from the knowledge of the theoretical electroporation modeling, but it seems important to use the current knowledge of tissue electroporation – even quite rough –, in order to rapidly address the specific difficulty of such a goal (real-time computing of non-linear model, image segmentation and visualization). Tight collaborations with CHU Pellegrin at Bordeaux, and CHU J. Verdier at Bondy are crucial.
- Radiofrequency ablation. In a collaboration with Hopital Haut Leveque, CHU Bordeaux we are trying to determine the efficacy and risk of relapse of hepatocellular carcinoma treated by radiofrequency ablation. For this matter we are using geometrical measurements on images (margins of the RFA, distance to the boundary of the organ) as well as texture information to statistically evaluate the clinical outcome of patients.
- Intensity focused ultrasound. In collaboration with Utrecht Medical center, we aim at tackling several challenges in clinical applications of IFU: target tracking, dose delivery...
3.4 Axis 3: Quantitative cancer modeling for biological and preclinical studies
With the emergence and improvement of a plethora of experimental techniques, the molecular, cellular and tissue biology has operated a shift toward a more quantitative science, in particular in the domain of cancer biology. These quantitative assays generate a large amount of data that call for theoretical formalism in order to better understand and predict the complex phenomena involved. Indeed, due to the huge complexity underlying the development of a cancer disease that involves multiple scales (from the genetic, intra-cellular scale to the scale of the whole organism), and a large number of interacting physiological processes (see the so-called "hallmarks of cancer"), several questions are not fully understood. Among these, we want to focus on the most clinically relevant ones, such as the general laws governing tumor growth and the development of metastases (secondary tumors, responsible of 90% of the deaths from a solid cancer). In this context, it is thus challenging to exploit the diversity of the data available in experimental settings (such as in vitro tumor spheroids or in vivo mice experiments) in order to improve our understanding of the disease and its dynamics, which in turn lead to validation, refinement and better tuning of the macroscopic models used in the axes 1 and 2 for clinical applications.
In recent years, several new findings challenged the classical vision of the metastatic development biology, in particular by the discovery of organism-scale phenomena that are amenable to a dynamical description in terms of mathematical models based on differential equations. These include the angiogenesis-mediated distant inhibition of secondary tumors by a primary tumor the pre-metastatic niche or the self-seeding phenomenon Building a general, cancer type specific, comprehensive theory that would integrate these dynamical processes remains an open challenge. On the therapeutic side, recent studies demonstrated that some drugs (such as the Sunitinib), while having a positive effect on the primary tumor (reduction of the growth), could accelerate the growth of the metastases. Moreover, this effect was found to be scheduling-dependent. Designing better ways to use this drug in order to control these phenomena is another challenge. In the context of combination therapies, the question of the sequence of administration between the two drugs is also particularly relevant.
One of the technical challenge that we need to overcome when dealing with biological data is the presence of potentially very large inter-animal (or inter-individual) variability.
Starting from the available multi-modal data and relevant biological or therapeutic questions, our purpose is to develop adapted mathematical models (i.e. identifiable from the data) that recapitulate the existing knowledge and reduce it to its more fundamental components, with two main purposes:
- to generate quantitative and empirically testable predictions that allow to assess biological hypotheses or
- to investigate the therapeutic management of the disease and assist preclinical studies of anti-cancerous drug development.
We believe that the feedback loop between theoretical modeling and experimental studies can help to generate new knowledge and improve our predictive abilities for clinical diagnosis, prognosis, and therapeutic decision. Let us note that the first point is in direct link with the axes 1 and 2 of the team since it allows us to experimentally validate the models at the biological scale (in vitro and in vivo experiments) for further clinical applications.
More precisely, we first base ourselves on a thorough exploration of the biological literature of the biological phenomena we want to model: growth of tumor spheroids, in vivo tumor growth in mice, initiation and development of the metastases, effect of anti-cancerous drugs. Then we investigate, using basic statistical tools, the data we dispose, which can range from: spatial distribution of heterogeneous cell population within tumor spheroids, expression of cell markers (such as green fluorescent protein for cancer cells or specific antibodies for other cell types), bioluminescence, direct volume measurement or even intra-vital images obtained with specific imaging devices. According to the data type, we further build dedicated mathematical models that are based either on PDEs (when spatial data is available, or when time evolution of a structured density can be inferred from the data, for instance for a population of tumors) or ODEs (for scalar longitudinal data). These models are confronted to the data by two principal means:
- when possible, experimental assays can give a direct measurement of some parameters (such as the proliferation rate or the migration speed) or
- statistical tools to infer the parameters from observables of the model.
This last point is of particular relevance to tackle the problem of the large inter-animal variability and we use adapted statistical tools such as the mixed-effects modeling framework.
Once the models are shown able to describe the data and are properly calibrated, we use them to test or simulate biological hypotheses. Based on our simulations, we then aim at proposing to our biological collaborators new experiments to confirm or infirm newly generated hypotheses, or to test different administration protocols of the drugs. For instance, in a collaboration with the team of the professor Andreas Bikfalvi (Laboratoire de l'Angiogénèse et du Micro-environnement des Cancers, Inserm, Bordeaux), based on confrontation of a mathematical model to multi-modal biological data (total number of cells in the primary and distant sites and MRI), we could demonstrate that the classical view of metastatic dissemination and development (one metastasis is born from one cell) was probably inaccurate, in mice grafted with metastatic kidney tumors. We then proposed that metastatic germs could merge or attract circulating cells. Experiments involving cells tagged with two different colors are currently performed in order to confirm or infirm this hypothesis.
Eventually, we use the large amount of temporal data generated in preclinical experiments for the effect of anti-cancerous drugs in order to design and validate mathematical formalisms translating the biological mechanisms of action of these drugs for application to clinical cases, in direct connection with the axis 1. We have a special focus on targeted therapies (designed to specifically attack the cancer cells while sparing the healthy tissue) such as the Sunitinib. This drug is indeed indicated as a first line treatment for metastatic renal cancer and we plan to conduct a translational study coupled between A. Bikfalvi's laboratory and medical doctors, F. Cornelis (radiologist) and A. Ravaud (head of the medical oncology department).
4 Application domains
4.1 Tumor growth monitoring and therapeutic evaluation
Each type of cancer is different and requires an adequate model. More specifically, we are currently working on the following diseases:
- Glioma (brain tumors) of various grades,
- Metastases to the lung, liver and brain from various organs,
- Soft-tissue sarcoma,
- Kidney cancer and its metastases,
- Non small cell lung carcinoma.
In this context our application domains are:
- Image-driven patient-specific simulations of tumor growth and treatments,
- Parameter estimation and data assimilation of medical images,
- Machine and deep learning methods for delineating the lesions and stratifying patients according to their responses to treatment or risks of relapse.
4.2 Biophysical therapies
- Modeling of electrochemotherapy on biological and clinical scales.
- Evaluation of radiotherapy and radiofrequency ablation.
4.3 In-vitro and animals experimentations in oncology
- Theoretical biology of the metastatic process: dynamics of a population of tumors in mutual interactions, dormancy, pre-metastatic and metastatic niche, quantification of metastatic potential and differential effects of anti-angiogenic therapies on primary tumor and metastases.
- Mathematical models for preclinical cancer research: description and prediction of tumor growth and metastatic development, effect of anti-cancerous therapies.
5 Social and environmental responsibility
5.1 Footprint of research activities
Numerical computations on (GPU) clusters like Plafrim and mission travels (nearly inexistent this year because of the Covid crisis).
5.2 Impact of research results
In the long run, our research could yield interesting outcomes for cancer patients. Yet we are mostly building proofs of concept that would have to be taken over by an industrial partner for any transfer towards clinics (like we did with Sophia Genetics in the past).
6 Highlights of the year
- Sébastien Benzekry has left the team for creating a new team at INRIA Sophia Antipolis: Compo
- Some new grants were awarded to the team: Plan Cancer, Plan de relance with Sophia Genetics, 2 PhD grants from Region Nouvelle Aquitaine, 1 from the Plan IA, 1 from INRIA and 1 engineer position as well.
- Research highlights/
- New direction of research: electroporation in cardiology with IHU Lyric and INRIA team Carmen,
- New direction of research around auto-immune diseases with CHU Bordeaux.
7 New results
7.1 Joint state-parameter estimation for tumor growth model
Authors: A. Collin, T. Kritter, C. Poignard, O. Saut
Abstract: We present a shape-oriented data assimilation strategy suitable for front-tracking tumor growth problems. A general hyperbolic/elliptic tumor growth model is presented as well as the available observations corresponding to the location of the tumor front over time extracted from medical imaging as MRI or CT scans. We provide sufficient conditions allowing to design a state observer by proving the convergence of the observer model to the target solution, for exact parameters. In particular, the similarity measure chosen to compare observations and simulation of tumor contour is presented. A specific joint state-parameter correction with a Luenberger observer correcting the state and a reduced-order Kalman filter correcting the parameters is introduced and studied. We then illustrate and assess our proposed observer method with synthetic problems. Our numerical trials show that state estimation is very effective with the proposed Luenberger observer, but specific strategies are needed to accurately perform parameter estimation in a clinical context. We then propose strategies to deal with the fact that data is very sparse in time and that the initial distribution of the proliferation rate is unknown. The results on synthetic data are very promising and work is ongoing to apply our strategy on clinical cases.
7.2 MRI-Based Radiomics Input for Prediction of 2-Year Disease Recurrence in Anal Squamous Cell Carcinoma
Authors: Nicolas Giraud, Olivier Saut et al.
Abstract: Purpose: Chemo-radiotherapy (CRT) is the standard treatment for non-metastatic anal squamous cell carcinomas (ASCC). Despite excellent results for T1-2 stages, relapses still occur in around 35% of locally advanced tumors. Recent strategies focus on treatment intensification, but could benefit from a better patient selection. Our goal was to assess the prognostic value of pre-therapeutic MRI radiomics on 2-year disease control (DC). Methods: We retrospectively selected patients with non-metastatic ASCC treated at the CHU Bordeaux and in the French FFCD0904 multicentric trial. Radiomic features were extracted from T2-weighted pre-therapeutic MRI delineated sequences. After random division between training and testing sets on a 2:1 ratio, univariate and multivariate analysis were performed on the training cohort to select optimal features. The correlation with 2-year DC was assessed using logistic regression models, with AUC and accuracy as performance gauges, and the prediction of disease-free survival using Cox regression and Kaplan-Meier analysis. Results: A total of 82 patients were randomized in the training (n = 54) and testing sets (n = 28). At 2 years, 24 patients (29%) presented relapse. In the training set, two clinical (tumor size and CRT length) and two radiomic features (FirstOrderEntropy and GLCMJointEnergy) were associated with disease control in univariate analysis and included in the model. The clinical model was outperformed by the mixed (clinical and radiomic) model in both the training (AUC 0.758 versus 0.825, accuracy of 75.9% versus 87%) and testing (AUC 0.714 versus 0.898, accuracy of 78.6% versus 85.7%) sets, which led to distinctive high and low risk of disease relapse groups (HR 8.60, p = 0.005). Conclusion: A mixed model with two clinical and two radiomic features was predictive of 2-year disease control after CRT and could contribute to identify high risk patients amenable to treatment intensification with view of personalized medicine.
7.3 Deep correction of breathing-related artifacts in real-time MR-thermometry
Authors: Baudouin Denis de Senneville, Pierrick Coupé, Mario Ries, Laurent Facq, Chrit Moonen
Abstract: Real-time MR-imaging has been clinically adapted for monitoring thermal therapies since it can provide on-the- fly temperature maps simultaneously with anatomical information. However, proton resonance frequency based thermometry of moving targets remains challenging since temperature artifacts are induced by the respiratory as well as physiological motion. If left uncorrected, these artifacts lead to severe errors in temperature estimates and impair therapy guidance. In this study, we evaluated deep learning for on-line correction of motion related errors in abdominal MR- thermometry. For this, a convolutional neural network (CNN) was designed to learn the apparent temperature perturbation from images acquired during a preparative learning stage prior to hyperthermia. The input of the designed CNN is the most recent magnitude image and no surrogate of motion is needed. During the subsequent hyperthermia procedure, the recent magnitude image is used as an input for the CNN-model in order to generate an on-line correction for the current temperature map. The method’s artifact suppression performance was evaluated on 12 free breathing volunteers and was found robust and artifact-free in all examined cases. Furthermore, thermometric precision and accuracy was assessed for in vivo ablation using high intensity focused ultrasound. All calculations involved at the different stages of the proposed workflow were designed to be compatible with the clinical time constraints of a therapeutic procedure.
7.4 A model-strengthened imaging biomarker for survival prediction in EGFR-mutated non-small-cell lung carcinoma patients treated with tyrosine kinase inhibitors
Authors: Annabelle Collin, Vladimir Groza, Louise Missenard, François Chomy, Thierry Colin, Jean Palussière, Olivier Saut
Abstract: Non-small-cell lung carcinoma is a frequent type of lung cancer with a bad prognosis. Depending on the stage, genomics, several therapeutical approaches are used. Tyrosine Kinase Inhibitors (TKI) may be successful for a time in the treatment of EGFR-mutated non-small cells lung carcinoma. Our objective is here to propose a survival assessment as their efficacy in the long run is challenging to evaluate. The study includes 17 patients diagnosed as of EGFR-mutated non-small cell lung cancer and exposed to an EGFR-targeting TKI with 3 computed tomography (CT) scans of the primitive tumor (one before the TKI introduction and two after). An imaging biomarker based on the texture heterogeneity evolution between the first and the third exams is derived and computed from a mathematical model and patient data. Defining the overall survival as the time between the introduction of the TKI treatment and the patient death, we obtain a statistically significant correlation between the overall survival and our imaging marker (p = 0:009). Using the ROC curve, the patients are separated into two populations and the comparison of the survival curves is statistically significant (p = 0:025). The baseline exam seems to have a significant role in the prediction of response to TKI treatment. More precisely, our imaging biomarker defined using only the CT scan before the TKI introduction allows to determine a first classification of the population which is improved over time using the imaging marker as soon as more CT scans are available. This exploratory study leads us to think that it is possible to obtain a survival assessment using only few CT scans of the primary tumor.
7.5 Improved 18-FDG PET/CT diagnosis of multiple myeloma diffuse disease by radiomics analysis
Authors: Charles Mesguich, Elif Hindie, Baudouin Denis de Senneville, Ghoufrane Tlili, Jean-Baptiste Pinaquy, Gerald Marit, Olivier Saut
Abstract: In Multiple Myeloma (MM) patients, diffuse infiltration of bone marrow can be diagnosed on MRI and is associated with poorer prognosis. On 18-FDG PET/CT, the other important imaging modality in MM, the diagnosis of diffuse disease by visual analysis can be challenging. Radiomics allows the extraction of large amount of data from images to individualize disease specific diagnostic or prognostic patterns. We aimed todevelop a radiomics-based model derived from PET and CT images, that could improve the diagnosis of multiple myeloma diffuse disease on 18-FDG PET/CT.
7.6 Deciphering Tumour Tissue Organization by 3D Electron Microscopy and machine learning
Authors: Baudouin Denis de Senneville, Fatma Zohra Khoubai, Marc Bevilacqua, Alexandre Labedade, Kathleen Flosseau, Christophe Chardot, Sophie Branchereau, Jean Ripoche, Stefano Cairo, Etienne Gontier, Christophe Grosset
Abstract: Despite recent progress in the characterization of tumour components, the tri-dimensional (3D) organization of this pathological tissue and the parameters determining its internal architecture remain elusive. Here, we analysed the spatial organization of patient-derived xenograft tissues generated from hepatoblastoma, the most frequent childhood liver tumour, by serial block-face scanning electron microscopy using an integrated workflow combining 3D imaging, manual and machine learning-based semi-automatic segmentations, mathematics and infographics. By digitally reconstituting an entire hepatoblastoma sample with a blood capillary, a bile canaliculus-like structure, hundreds of tumour cells and their main organelles (e.g. cytoplasm, nucleus, mitochondria), we report unique 3D ultrastructural data about the organization of tumoral tissue. We found that the size of hepatoblastoma cells correlates with the size of their nucleus, cytoplasm and mitochondrial mass. We also discovered that the blood capillary controls the planar alignment and size of tumour cells in their 3D milieu. Finally, a set of tumour cells polarized in the direction of a hot spot corresponding to a bile canaliculus-like structure. In conclusion, this pilot study allowed the identification of bioarchitectural parameters that shape the internal and spatial organization of tumours, thus paving the way for new investigations in an emerging field that we call onconanotomy.
7.7 On a magnetic skin effect in eddy current problems: the magnetic potential in magnetically soft materials
Authors: Victor Peron, Clair Poignard
Abstract: This work is concerned with the time-harmonic eddy current problem in a bidimen-sional setting with a high contrast of magnetic permeabilities between a conducting medium and a dielectric medium. We describe a magnetic skin effect by deriving rigorously a multiscale expansion for the magnetic potential in power series of a small parameter ε which represents the inverse of the square root of a relative permeability. We make explicit the first asymptotics up to the order ε3. As an application we obtain impedance conditions up to the fourth order of approximation for the magnetic potential. Finally we measure this skin effect with a characteristic length that depends on the scalar curvature of the boundary of the conductor.
8 Bilateral contracts and grants with industry
8.1 Bilateral contracts with industry
- Research contract between Roche and the MONC team.
- Collaboration contract with Sophia Genetics in the context of the Pimiento project.
- Collaboration contract with Sophia Genetics for the Plan de relance.
8.2 Bilateral grants with industry
Pimiento project from MSDAvenir through INRIA Foundation.
9 Partnerships and cooperations
9.1 International research visitors
9.1.1 Visits to international teams
Research stays abroad
- C. Poignard : Centre de Recherche en Mathematiques, Univ. Montreal. 15-16 Nov.2021.
9.2 National initiatives
9.2.1 Plan Cancer
Systems Biology of Renal Carcinoma
- Title: Plan Cancer Systems Biology of Renal Carcinoma using a Mouse RCC model
- Partners : LAMC, INSERM-Univ. Bordeaux.
- Duration - June 2018 to June 2021
- Team participants: O. Saut, S. Benzekry (co-PI)
- Funding: 116.64k€
QUANTIC
- Plan Cancer QUANTIC: 2020–2022. QUANTitative modeling combined to statistical learning to understand and predict resistance to Immune-checkpoint inhibition in non-small cell lung Cancer
- Funding: 338 k€
- Partners: Inria Team MONC, SMARTc (Centre de Recherche sur le Cancer de Marseille, Inserm, CNRS), Assistance Publique Hôpitaux de Marseille
- Duration: December 2019 — December 2022
- Project leader: S. Benzekry
- Co-PI: D. Barbolosi (SMARTc), F. Barlési (AP-HM)
NUMEP
- Plan Cancer NUMEP. NUMerical modeling of ElectroPorationn in non-small cell lung Cancer
- Funding: 460 k€
- Partners: Inria Team MONC, Hopital J. Verdier-APHP, Institut de Pharmacologie et Biologie Structurale
- Duration: 2016–2021
- Project leader: C. Poignard
- Co-PIs: O. Séror (AP-HP), M-P Rols (IPBS)
9.2.2 Transnation call: INCA/ARC
- Title: Minimally and non-invasive methods for early detection and/or progression of low grade glioma
- Partners: INRIA Monc, INRIA SISTM, INSERM, Humanitas Research Hostital, Univ. Bergen
- Acronym: Glioma PRD
- Team participants: A. Collin, C. Etchegaray, C. Poignard, O. Saut (local PI)
- Total funds: 1M150, Monc's share 275k€.
9.3 Regional initiatives
Two PhD grants were awarded to the team by Region Nouvelle Aquitaine.
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: selection
Chair of conference program committees
- C. Poignard : Co-chair of International Conference by Center for Mathematical Modeling and Data Science, Osaka University
10.1.2 Journal
Member of the editorial boards
- C. Poignard : DCDS-S, AIMS Bioengineering.
Reviewer - reviewing activities
- C. Etchegaray : Technology in Cancer Research and Treatment, PLOS Computational Biology.
- O. Saut : European Radiology, MICCAI
10.1.3 Invited talks
- C. Etchegaray : Seminar of the Probability and Statistics team, LAMA (Univ. Gustave Effeil).
- C. Poignard. CRM, Univ. de Montréal (Canada).
10.1.4 Leadership within the scientific community
- O. Saut : in charge of Interdisciplinarity at the math institute (INSMI) of CNRS.
10.1.5 Scientific expertise
- C. Etchegaray : reviewer for the phase 2 ANR program.
- O. Saut : Member of the panel of CES 45 of ANR.
- O. Saut : Expert for the MESRI
- O. Saut : Member of the math panel of NCN (Poland)
10.1.6 Research administration
- C. Etchegaray : local correspondent for the national MATHSAV Research Group.
- C. Etchegaray : co-organizer of the IMB CSM team seminar.
- C. Etchegaray : head of IMB’s parity committee.
- C. Etchegaray : co-head of the parity committee of MARGAUx Federation.
- C. Poignard : member of the Scientific Committee of MATHSAV Research Group.
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
- Master : S. Benzekry, Cours "Modélisation de la croissance tumorale", 3h, niveau M2, Université de Tours, France.
- Master : S. Benzekry, Cours "Mathematical tools for pharmacometrics", 20h, niveau M2, Aix-Marseille Université, France.
- Ecole doctorale : S. Benzekry, "Introduction to modeling and data science in oncology", 12h.
- Master: A. Collin, Pratical C++ programming, 96h, niveau M1, INP Bordeaux, France.
- Master: A. Collin, Mesh theory, 36h, niveau M2, INP Bordeaux, France.
- Master: A. Collin, Machine Learning, niveau M2, INP Bordeaux, France.
- PhD: A. Collin, Modeling Life Science Module, Digital Public Health, Graduate Program.
- Licence : C. Etchegaray, Undergraduate teaching in Harmonic analysis, 32h, niveau L2, Univ. Bordeaux, France.
- Licence : C. Etchegaray, Undergraduate teaching in Mathematical Representation of Physical phenomena, 13.33h, niveau L1, Univ. Bordeaux, France.
- Licence : C. Poignard, Undergraduate teaching in Numerical and Applied Mathematics, 80h, L3-M1, INP Bordeaux, ENSAM, France.
10.2.2 Supervision
- Manon Charmasson (L3 Maths La Rochelle) : Modeling the intracellular actin dynamics in quiescent cells ; 6 weeks internship, cosupervision by C. Etchegaray, C. Poignard, O. Saut.
- Pierre Charitat (L3 ENS Lyon) : Modeling the Snowflake Yeast Dynamics ; 6 weeks internship, cosupervision by C. Etchegaray and C. Poignard.
- François Sorre (2nd year Bordeaux INP) : Numerical simulations of an active particle in a Stokes fluid ; 8 weeks internship, cosupervision by C. Etchegaray and C. Poignard.
- Inaara Kassamaly, Sonia Quertinmont, Julia Rossi (M2 Cancer biology) : Pancreatic cancer: experimental and mathematical approach to evaluate DC-based vaccine therapeutic potential ; 6 weeks project, cosupervision by C. Etchegaray and Vanja Sisirak (ImmunoConcept).
- O. Saut : PhD candidates (K. Chahdi, V. Montalibet, G. Martinroche) + interns.
10.2.3 Juries
- C. Etchegaray : PhD committee of Alessandro Cucchi (Univ. Paris), as guest.
- C. Etchegaray : Jury member in the Inria BSO CRCN-ISFP campaign.
- O. Saut : HDR G. Sciume, Univ. Bordeaux.
- O. Saut : PhD committee of E. Villain (INSA Toulouse).
10.3 Popularization
10.3.1 Interventions
- C.Etchegaray : Chiche! interventions (Igon, Lormont (2), Dax (3)).
- C.Etchegaray : IREM Conference, IMB.
- O. Saut : Participation "Images et Cancer" of Asperon and Co (cancer patient association), 2021.
11 Scientific production
11.1 Major publications
- 1 articleMathematical modeling of tumor-tumor distant interactions supports a systemic control of tumor growth.Cancer Research2017
- 2 articleClassical Mathematical Models for Description and Forecast of Experimental Tumor Growth.PLoS Computational Biology108August 2014, e1003800
- 3 articleModeling Spontaneous Metastasis following Surgery: An In Vivo-In Silico Approach.Cancer Research763February 2016, 535 - 547
- 4 articleComputational Modeling Of Solid Tumor Growth: The Avascular Stage.SIAM Journal on Scientific Computing324August 2010, 2321--2344
- 5 articlePatient specific simulation of tumor growth, response to the treatment and relapse of a lung metastasis: a clinical case.Journal of Computational Surgery2015, 18
- 6 articleSuperconvergent second order Cartesian method for solving free boundary problem for invadopodia formation.Journal of Computational Physics339June 2017, 412 - 431
- 7 articleRevisiting bevacizumab + cytotoxics scheduling using mathematical modeling: proof of concept study in experimental non-small cell lung carcinoma.CPT: Pharmacometrics and Systems Pharmacology2018, 1-9
- 8 articleSpatial modelling of tumour drug resistance: the case of GIST liver metastases Mathematical Medicine and Biology Advance.Mathematical Medicine and Biology002016, 1 - 26
- 9 articleConducting and permeable states of cell membrane submitted to high voltage pulses: Mathematical and numerical studies validated by the experiments.Journal of Theoretical Biology360November 2014, 83-94
- 10 articleComputational Trials: Unraveling Motility Phenotypes, Progression Patterns, and Treatment Options for Glioblastoma Multiforme.PLoS ONE111January 2016
11.2 Publications of the year
International journals
Scientific books
Scientific book chapters
Reports & preprints

