Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.4.5. Bayesian methods
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.4. Statistical methods
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.4.1. Deterministic control
Other Research Topics and Application Domains
- B1. Life sciences
- B1.1.2. Molecular and cellular biology
- B1.1.4. Genetics and genomics
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B1.1.10. Systems and synthetic biology
- B2.2.4. Infectious diseases, Virology
- B4.3.1. Biofuels
1 Team members, visitors, external collaborators
Research Scientists
- Delphine Ropers [Team leader, INRIA, Senior Researcher, HDR]
- Eugenio Cinquemani [INRIA, Researcher, HDR]
- Aline Marguet [INRIA, Researcher]
- Hidde de Jong [INRIA, Senior Researcher, HDR]
Faculty Member
- Johannes Geiselmann [UGA, Professor]
Post-Doctoral Fellow
- Thibault Etienne [INRIA, until Sep 2022]
PhD Students
- Rand Asswad [INRIA, from Oct 2022]
- Ignacia Cancino Aguirre [INRIA]
- Charles Medous [UGA]
- Antrea Pavlou [INRIA, until May 2022]
- Emrys Reginato [INRIA]
- Maaike Sangster [INRIA]
Technical Staff
- Soraya Arias [Inria, Engineer]
- Eric Boucher [INRIA, Engineer, from Aug 2022 until Nov 2022]
- Ludovic Leau-Mercier [INRIA, Engineer, from Dec 2022]
Administrative Assistant
- Diane Courtiol [INRIA]
External Collaborator
- Muriel Cocaign-Bousquet [INRAE, Senior Researcher, Toulouse, HDR]
2 Overall objectives
MICROCOSME combines computational and experimental approaches for the analysis, engineering, and control of the growth of microorganisms. Understanding and controlling the dynamics of bacterial growth is vitally important in health, medicine, biotechnology, and food industries, for instance to halt the growth of pathogens or stimulate the growth of probiotics or industrial microorganisms.
We develop multiscale models of growth, where the macroscopic observable, growth of a microbial population or community, depends on various metabolic pathways and regulatory mechanisms operating at microscopic scales within the cells. We use our (deterministic or stochastic) models to interpret experimental data or to infer the underlying growth processes from the data. This requires developing a platform for the automation of experiments, as well as methods and software for model estimation and data analysis. The analysis of microbial growth calls for new methodologies at the interface of microbiology, control theory, applied mathematics, computer science, biophysics, and molecular biology, which also leads to contributions in all of these fields. Our workhorse for the realization of this research program is the bacterium Escherichia coli pictured in Figure 1. Part of the microbiota of the human gut, E. coli is the model organism par excellence in microbiology and a popular platform for bio-based chemical production. We intend to extend approaches developed in-house for this specific microbe to other microorganisms including pathogens.
MICROCOSME has been created on October 1st, 2021. A recomposition and follow-up of former IBIS project-team, MICROCOSME unites researchers from Inria Grenoble – Rhône-Alpes and the Laboratoire Interdisciplinaire de Physique at Université Grenoble Alpes (CNRS UMR 5588).
Figure 1: Microscopy image of Escherichia coli bacteria growing on a solid nutrient medium.
3 Research program
The research program of MICROCOSME is articulated around four research axes combining theory and experiments, which are illustrated in Figure 2 and detailed below.
3.1 Genome-scale analysis of microbial physiology
The molecular foundations of bacterial growth remain little understood today, because they involve large biochemical networks with physical and regulatory interactions across different levels of cellular organization. We investigate at the genome scale how the dynamics of gene expression and metabolism leads to microbial growth, using a combination of mathematical models and high-throughput data. The challenge is to integrate, in models of thousands of equations, multiple and heterogeneous datasets on the metabolic, transcriptomic, and proteomic level. We typically use constraint-based models to investigate the relations between microbial growth and metabolism, while the effect of growth on mRNA stability is analysed by means of non-linear mixed-effect models.
3.2 Natural and engineered resource allocation strategies in microorganisms
Microorganisms have evolved strategies to allocate their resources to different cellular functions and thus adjust their growth rate to fluctuating environments. We study these natural resource allocation strategies, by viewing cells as self-replicators that can be described using coarse-grained models and analysed by means of optimal and feedback control theory. The models take the form of systems of 5-10 nonlinear ordinary differential equations, with parameters estimated from published data or data obtained from dedicated experiments. Experimental work in the lab allows to validate model predictions on the single-cell and population level and to engineer new strategies for the reallocation of cell resources from growth to bioproduction.
3.3 Variability and robustness of microbial adaptation
The developments of experimental techniques and the use of video-microscopy have led to a growing number of high-quality data showing the heterogeneity among cells in a population. We combine these single-cell data with models describing the stochastic dynamics of individual cells, such as birth-death processes, branching processes, and mixed-effect models. The models allow to investigate the origins of heterogeneity and its role in the adaptation of microorganisms to environmental changes, and to leverage population heterogeneity for biotechnological applications. In practice, this requires the extension of modelling approaches by taking into account the specificities of heterogeneity, as well as the development of appropriate methods and software for the inference of models and of biological quantities from quantitative time-course profiles of the microbial response to environmental changes.
3.4 Analysis and control of microbial communities
Heterogeneity also arises within communities consisting of different microbial species. Understanding microbial interactions is a challenging task that goes well beyond the characterization of single species, and offers great opportunities for applications, such as the control of the community for bioproduction. Indeed, suitably constructed microbial consortia carry the potential to outperform single species in the accomplishment of processes of societal interest, such as biofuel synthesis. On the theoretical side, we develop (deterministic or stochastic) models of microbial dynamics similar to those in the three other research axes, which can be used to investigate new control approaches for microbial communities. On the experimental side, the application of control strategies for biotechnological applications requires the engineering of microbial strains and the automation of experiments. To that aim we have been developing a platform for feedback control experiments allowing the real-time monitoring, data processing, evaluation, and application of control laws.
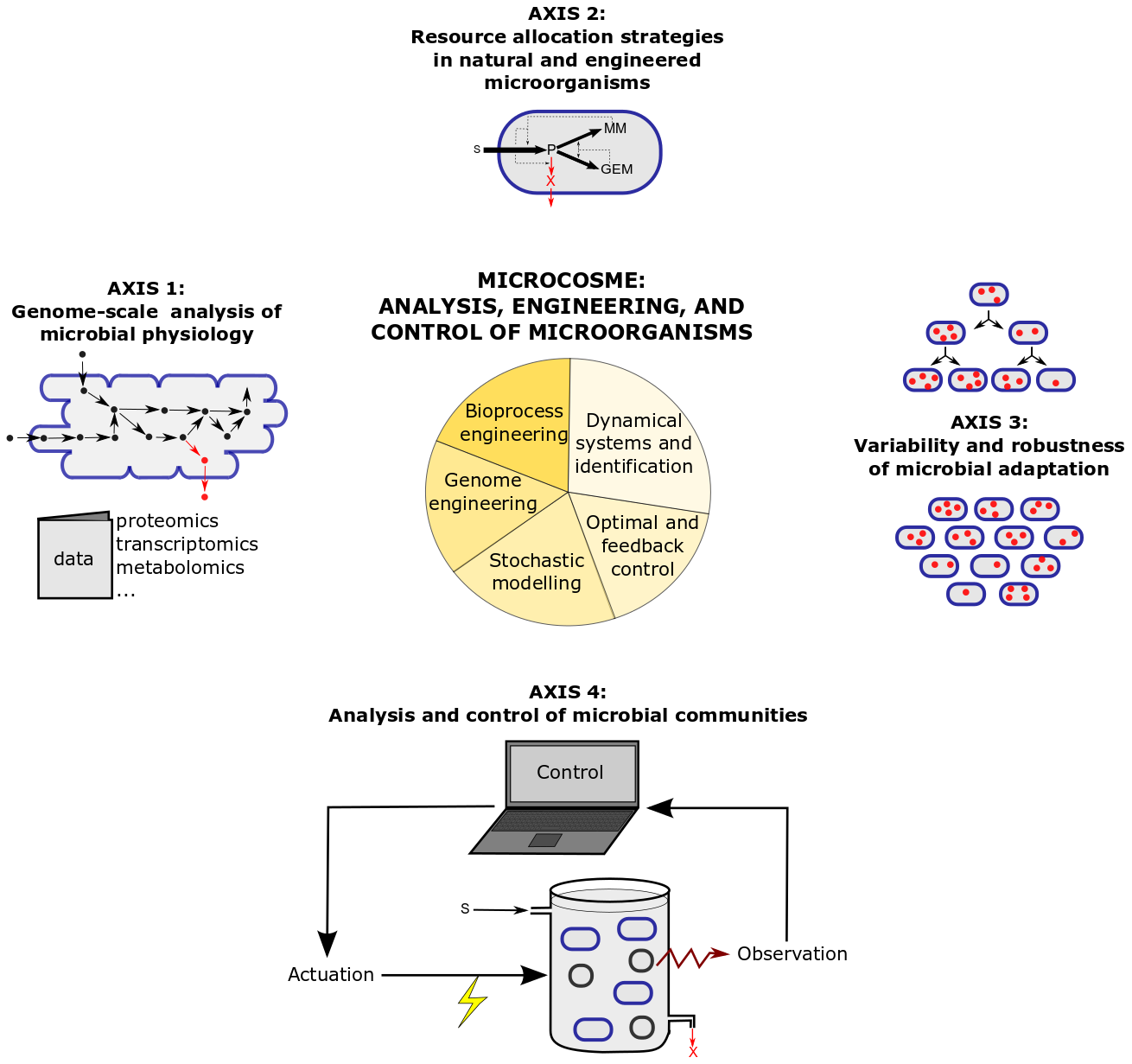
Figure 2: Research axes and methods in the MICROCOSME project-team.
4 Application domains
The research agenda of MICROCOSME is interdisciplinary in nature, driven by fundamental questions in biology, which we address by a combination of mathematical, computational, and experimental tools. This enables us to develop and share with partners a know-how useful to address challenging problems in health, bioeconomy, and biotechnology.
4.1 Biotechnology and bioeconomy
Bioproduction imposes a strong metabolic burden on microorganisms, detrimental to their growth and the production yield. Our studies of natural resource allocation strategies lead us to explore and engineer various reallocation strategies to improve bioproduction through growth control. For instance, in the past, we have successfully implemented a growth switch in E. coli bacteria, aiming at shuttling resources, away from protein synthesis (key for bacterial growth) to the high-yield production of a metabolite of interest (glycerol) 6, 24. We also develop and test control strategies for synthetic microbial communities, composed of populations of different E. coli strains or in consortia with other species. In the wake of our studies on the relation between growth and metabolism, we develop bioeconomy strategies for the valorisation of vegetal waste into value-added product. In one project, notably, we aim at designing E. coli strains able to efficiently degrade carbon sources derived from pre-treatment of agricultural and forestry residues (Section 9).
4.2 Health
Numerous Mycobacteria species pose serious threats to human and animal health. Mycobacteria tuberculosis strains are also known to withstand several of the antibiotics used to treat the infection. We have started to extend our microbial physiology analyses by means of constraint-based models to understand the molecular control of mycobacterial growth and characterize the relations between metabolism, pathogenicity, and growth phenotype of mycobacterial species. This may lead, in the long term, to the development of new treatments for curing tuberculosis and other mycobacterial infections.
5 Social and environmental responsibility
Several of our research activities have a direct societal impact: our work on Mycobacteria addresses important questions of public health, while the project on the degradation and valorisation of vegetal waste meet European efforts in Circular Bioeconomy to replace fossil feedstock with renewable resources.
6 Highlights of the year
A new line of research started in 2022, through the newly accepted associate-team GERM involving members of MICROCOSME and of the Mycobacterial Metabolism and Antibiotic Research Laboratory at the Francis Crick Institute in London. By means of computational and experimental tools, GERM aims at explaining the role of cellular metabolism in the growth-rate variability of mycobacterial species, a bacterium genus that includes dangerous human pathogens such as Mycobacterium tuberculosis and M. leprae. Part of the work within GERM is carried out in the framework of the PhD thesis of Ignacia Cancino Aguirre.
7 New software and platforms
7.1 New software
7.1.1 ODIN+
-
Name:
Platform for advanced monitoring, control and optimisation of bioprocesses
-
Keywords:
Systems Biology, Biotechnology, Automatic control, Monitoring
-
Functional Description:
This application proposes a framework for on-line supervision of bioreactors. It gathers the data sampled from different on-line and off-line sensors. ODIN+ is a distributed platform, enabling remote monitoring as well as remote data acquisition.More originally, it enables researchers and industrials to easily develop and deploy advanced control algorithms, optimisation strategies, together with estimates of state variables or process state. It also contains a process simulator which can be harnessed for experimentation and training purposes. It is modular in order to adapt to any plant and to run most of the algorithms, and it can handle the high level of uncertainties that characterises the biological processes. The architecture is based on Erlang, and communication between modules through a MQTT Broker with Python for running the algorithms. ODIN+ is developed in collaboration with the INRIA Ibis research team.
-
Authors:
Nicolas Niclausse, Nicolas Chleq, Jean-Luc Szpyrka, Pierre Fernique, Thibaud Kloczko, Tamas Muszbek, Amine Lahouel, Olivier Bernard, Eugenio Cinquemani
-
Contact:
Olivier Bernard
7.1.2 GNA
-
Name:
Genetic Network Analyzer
-
Keywords:
Model Checking, Bioinformatics, Gene regulatory networks, Qualitative simulation
-
Scientific Description:
Genetic Network Analyzer (GNA) is the implementation of methods for the qualitative modeling and simulation of gene regulatory networks developed in the IBIS project-team.
-
Functional Description:
The input of GNA consists of a model of the regulatory network in the form of a system of piecewise-linear differential equations (PLDEs), supplemented by inequality constraints on the parameters and initial conditions. From this information, GNA generates a state transition graph summarizing the qualitative dynamics of the system. In order to analyze large graphs, GNA allows the user to specify properties of the qualitative dynamics of a network in temporal logic, using high-level query templates, and to verify these properties on the state transition graph by means of standard model-checking tools, either locally installed or accessible through a remote web server.
-
Release Contributions:
(1) it supports the editing and visualization of regulatory networks, in an SBGN-compatible format, (2) it semi-automatically generates a prototype model from the network structure, thus accelerating the modeling process, and (3) it allows models to be exported in the SBML Qual standard.
-
News of the Year:
Tutorial on the use of the model formalism for analyzing synthetic genetic circuits, published in an edited book.
- URL:
- Publications:
-
Contact:
Hidde de Jong
-
Participants:
Hidde de Jong, Michel Page, Delphine Ropers
-
Partner:
UGA
7.1.3 WellInverter
-
Name:
WellInverter
-
Keywords:
Bioinformatics, Statistics, Data visualization, Data modeling
-
Scientific Description:
WellInverter is a web application that implements linear inversion methods for the reconstruction of gene expression profiles from fluorescent or luminescent reporter gene data. WellInverter makes the methods available to a broad audience of biologists and bioinformaticians. In particular, we have put in place a parallel computing architecture with a load balancer to distribute the analysis queries over several back-end servers, redesigned the graphical user interface, and developed a plug-in system for defining high-level routines for parsing data files produced by microplate readers from different manufacturers.
-
Functional Description:
As input, WellInverter reads the primary data file produced by a 96-well microplate reader, containing time-series measurements of the absorbance (optical density) as well as the fluorescence and luminescence intensities in each well (if available). Various modules exist to analyze the data, in particular for detecting outliers, subtracting background, estimating growth rates, promoter activities and protein concentrations, visualizing expression profiles, synchronizing replicate profiles, etc. The computational core of the web application consists of the Python library WellFARE.
-
News of the Year:
Publication in BMC Bioinformatics describing the new version of the application.
- URL:
- Publications:
-
Contact:
Hidde de Jong
-
Participants:
Delphine Ropers, Hidde de Jong, Johannes Geiselmann, Michel Page, Valentin Zulkower, Yannick Martin
-
Partner:
UGA
7.1.4 WellFARE
-
Name:
WellFARE
-
Keywords:
Bioinformatics, Statistics, Data visualization, Data modeling
-
Scientific Description:
WellFARE is a Python library implementing linear inversion methods for the reconstruction of gene expression profiles from fluorescent or luminescent reporter gene data. WellFARE form the computational core of the WellInverter web application.
-
Functional Description:
As input, WellFARE reads the primary data file produced by a 96-well microplate reader, containing time-series measurements of the absorbance (optical density) as well as the fluorescence and luminescence intensities in each well (if available). Various functions exist to analyze the data, in particular for detecting outliers, subtracting background, estimating growth rates, promoter activities and protein concentrations, visualizing expression profiles, synchronizing replicate profiles, etc. WellFARE is the computational core of the web application WellInverter.
-
News of the Year:
Publication in BMC Bioinformatics describing the new version of WellFARE
- URL:
- Publication:
-
Contact:
Hidde de Jong
-
Participants:
Delphine Ropers, Johannes Geiselmann, Hidde de Jong, Michel Page, Valentin Zulkower, Yannick Martin
-
Partner:
UGA
8 New results
8.1 Analysis of fluorescent reporter genes
Participants: E. Cinquemani, H. de Jong, J. Geiselmann, A. Pavlou.
The use of fluorescent and luminescent reporter genes allows real-time monitoring of gene expression, both at the level of individual cells and cell populations. Over the years, many useful resources have appeared, such as libraries of reporter strains for model organisms and computer tools for designing reporter plasmids. Moreover, the widespread adoption of thermostated microplate readers in experimental laboratories has made it possible to automate and multiplex reporter gene assays on the population level. This has resulted in large time-series data sets, typically comprising 100,000 - 1,000,000 measurements of absorbance, fluorescence, and luminescence for 96 wells on the microplate. In order to fully exploit these data sets, we need sound mathematical methods to infer biologically relevant quantities from the primary data and computer tools to apply the methods in an efficient and user-friendly manner.In the past few years we developed methods for the analysis of reporter gene data obtained in microplate experiments. These methods solve a range of estimation problems, notably the inference of growth rate, promoter activity, and protein concentration profiles. In the framework of her PhD thesis, Antrea Pavlou has continued this work by addressing a major issue in the interpretation of fluorescent data, namely that the inference of promoter activities is strongly dependent on the maturation properties of fluorescent proteins. We have therefore proposed an approach for incorporating maturation dynamics in the reconstruction of promoter activities. Our approach consists in developing and calibrating mechanistic maturation models for distinct fluorescent proteins. These models are then used alongside a Bayesian approach to estimate promoter activities from fluorescence data. We demonstrated by means of targeted experiments in Escherichia coli that our approach provides robust estimates, and that accounting for maturation is, in many cases, essential for the interpretation of gene expression data. This work was published in the Biophysical Journal this year 15.
8.2 Resource allocation models of bacterial growth
Participants: E. Cinquemani, H. de Jong, J. Geiselmann, A. Pavlou, D. Ropers.
Various mathematical approaches have been used in the literature to describe the networks of biochemical reactions involved in microbial growth. With different levels of detail, the resulting models provide an integrated view of these reaction networks, including the transport of nutrients from the environment and their conversion into biomass. Recent work has shown that coarse-grained models of resource allocation can account for a number of empirical regularities relating the macromolecular composition of the cell to the growth rate. A well-known example of such a growth law is the linear relation between the growth rate and the ribosomal protein concentration. While these studies focus on steady-state or balanced growth, such conditions are rarely found in natural habitats, where microorganisms are continually challenged by environmental fluctuations.In recent years, in the framework of the PhD thesis of Nils Giordano, we extended the study of microbial growth strategies to dynamical environments, using a coarse-grained resource allocation model of microbial self-replication. In collaboration with the BIOCORE project-team, we formulated dynamical growth maximization as an optimal control problem that can be solved using Pontryagin’s Maximum Principle and we compared the theoretical results thus obtained with different possible implementations of growth control in bacterial cells 4. The experimental validation of some of these results were carried out by Antrea Pavlou in the framework of her PhD project, funded by the ANR project Maximic (Section 9). We quantified resource allocation strategies in single cells of the model bacterium Escherichia coli during balanced growth and during transitions from one growth environment to another, by means of a combination of time-lapse fluorescence microscopy and statistical methods. We found a large heterogeneity of ribosome concentrations and growth rates for individual cells during balanced growth, with an unexpected absence of correlation between the two variables. A similar large heterogeneity was found during growth transitions. These results are surprising if bacteria had evolved to optimize growth, in stable or in changing environments, as is often assumed. A preliminary version of this work has been presented in the PhD thesis of Antrea Pavlou 17 and an article summarizing the results is being prepared for publication in a biology journal.
Also within the context of Maximic, we have worked on several extensions of the original self-replicator model. In collaboration with the BIOCORE project-team and Tomas Gedeon, invited professor from Montana State University in our team in 2019, we developed a coarse-grained resource allocation model to account for the observation that different strains of a microorganism growing in the same environment display a wide variety of growth rates and growth yields. We used our model to test the hypothesis that different resource allocation strategies, corresponding to different compositions of the proteome, can account for this rate-yield variability. The model predictions were verified by means of a database of hundreds of published rate-yield and uptake-secretion phenotypes of Escherichia coli strains grown in standard laboratory conditions. We found a very good quantitative agreement between the range of predicted and observed growth rates, growth yields, and glucose uptake and acetate secretion rates. These results support the hypothesis that resource allocation is a major explanatory factor of the observed variability of growth rates and growth yields across different bacterial strains. The model thus provides a fundamental understanding of quantitative bounds on rate and yield in E. coli and other microorganisms. It may also be useful for the rapid screening of strains in metabolic engineering and synthetic biology. An article based on this work is currently under revision for a biology journal 18.
8.3 Biotechnological applications of bacterial growth control
Participants: H. de Jong, J. Geiselmann, T. Clavier.
The ability to experimentally control the growth rate is crucial for studying bacterial physiology. It is also of central importance for applications in biotechnology, where often the goal is to limit or even arrest growth. Growth-arrested cells with a functional metabolism open the possibility to channel resources into the production of a desired metabolite, instead of wasting nutrients on biomass production. In recent years we obtained a foundational result for growth control in bacteria 6, in that we engineered an E. coli strain where the transcription of a key component of the gene expression machinery, RNA polymerase, is under the control of an inducible promoter. By changing the inducer concentration in the medium, we can adjust the RNA polymerase concentration and thereby switch bacterial growth between zero and the maximal growth rate supported by the medium. The publication also presented a biotechnological application of the synthetic growth switch in which both the wild-type E. coli strain and our modified strain were endowed with the capacity to produce glycerol when growing on glucose. Cells in which growth has been switched off continue to be metabolically active and harness the energy gain to produce glycerol at a twofold higher yield than in cells with natural control of RNA polymerase expression, putting the yield very close to the theoretical maximum. In a follow-up work published last year, we showed that the growth switch scales up to culture conditions that are closer to standard process conditions in biotechnology 24.
In the framework of the PhD thesis of Thibault Clavier, we addressed one of the limits of the growth switch that is inherent to the construction of synthetic networks in living microorganisms, namely that the latter evolve over time under the pressure of natural selection. In the case of the growth switch, the selection pressure is particularly high, since any spontaneously arising mutations disabling the inhibition of the expression of the RNA polymerase subunits will cause the population of growth-arrested cells to be taken over by cells that have resumed growth (at the expense of metabolite production). We improved the genetic stability of the growth switch by means of a redundant control mechanism of RNA polymerase expression. We deposited a patent of this invention. An article describing the results obtained with the extended growth switch is in preparation.
8.4 Analysis of synthetic microbial communities for bioproduction processes: real-time monitoring and optimality
Participants: S. Arias, R. Asswad, E. Boucher, E. Cinquemani, T. Clavier, H. de Jong, J. Geiselmann, L. Léau-Mercier, M. Sangster.
Modelling, analysis and control of microbial community dynamics is a fast-developing subject with great potential implications in the understanding of natural processes and the enhancement of biotechnological processes. Within the now-ended project IPL COSY, we picked up the challenge to design and investigate the dynamics of synthetically engineered microbial communities with a consortium of Inria partners, and to test control strategies in vivo.
In MICROCOSME, in particular, we have addressed the design of a bacterial community of two E. coli strains, mimicking mutualistic relationships found in nature, and with the potential to outperform a producer strain working in isolation in the production of a heterologous protein. We developed an ODE model of the key growth phenotypes of the community and their interactions, calibrated the model on literature data, and analysed the model for an in-depth understanding of the conditions supporting coexistence and of the tradeoffs encountered in this production process 10. In the context of the ongoing PhD thesis of Maaike Sangster, we have now bioengineered one version of the consortium, characterized individual species in batch as well as explored the consortium growth in continuous-flow experiments on an automated platform. Interesting results have been observed in terms of conditions for co-existence, which prompted us to further investigate widespread assumptions on E. coli overflow metabolism. Feedback control of the community for stabilization of biomasses to desirable bioproduction regimes remains in the aims of this research direction.
The developments in 10 have spurred a number of novel questions with theoretical and practical relevance. Toward model-based feedback control of microbial communities, the problem to real-time estimate the full system state from available biomass abundance/fluorescent reporter measurements was addressed in a publication with project team VALSE in 2021 23. For a different microbial consortium, a similar question is part of the ongoing work of Ph.D. student Rand Asswad. In parallel, we have pushed further the investigation of the synthetic community design for bioproduction optimization. In collaboration with BIOCORE, we considered a generalized version of the synthetic consortium proposed in 10 where both species of the consortium can synthesize a product of interest. In addition to a theoretical analysis of equilibria and coexistence regimes, in the 2022 bioRxiv preprint 21, we showed that the generalized design can indeed outperform our original consortium design based on a single producer species, thus shifting the interest from the commonly evoked concept of division of labor (different tasks for different species) to distribution of labor (same task across different species). This work is currently under revision for possible journal publication.
The ANR Ctrl-AB puts related concepts to work for the synergistic growth of E. coli and microalgae (Section 9). As per project goals, vitamin synthesis by modified E. coli strains represents a leverage to control vitamin-dependent growth of suitable algal strains, with the potential to robustify and maximize algal lipid synthesis. On the experimental side, we advanced on the design and synthesis of the bacterial strain in the consortium. On the methodological side, in collaboration with BIOCORE, problems of modelling, model analysis and real-time state estimation are being addressed with the recently started Ph.D. thesis of Rand Asswad, which will also address consortium control analysis and design in a later phase.
8.5 Modelling and inference of mRNA degradation
Participants: E. Cinquemani, M. Cocaign-Bousquet, T. Etienne, D. Ropers.
The ability to rapidly respond to changing nutrient availability is crucial for E. coli to survive in many environments including the gut. Reorganization of gene expression is the first step for bacteria to adjust their metabolism accordingly. It involves fine-tuning of both transcription and mRNA stability by dedicated regulatory interactions. While transcriptional regulation has been largely studied, the role of mRNA stability during a metabolic switch is poorly understood. This question was addressed in previous work in collaboration with the team of Muriel Cocaign-Bousquet at the Toulouse Biotechnology Institute 22. Using combined genome-wide transcriptome and mRNA decay analyses, we investigated the role of mRNA stability in the response of E. coli to nutrient changes. We demonstrated that while transcription of most genes is down-regulated when glucose is exhausted, concomitant mRNA stabilization of many mRNAs occurs and leads to the upregulation of genes involved in responses to nutrient changes and stresses. The observation of a global stabilization of cellular mRNAs during adaptation to carbon source depletion raises questions about the regulatory mechanisms at work. Known regulators of mRNA stability such as the protein Hfq, the carbon storage regulator Csr, and several small regulatory RNAs, specifically target mRNAs. Are these regulatory mechanisms sufficient to explain the systematic adjustment of mRNA half-lives?In a follow-up study, we developed a kinetic model of mRNA degradation that allows to propose hypotheses on the regulatory mechanisms at work to adjust mRNA stability to environmental conditions 3. From a practical point of view, this amounts to infer model parameters from high-throughput biological datasets. In the framework of the ANR project RIBECO (Section 9), Thibault Etienne, Muriel Cocaign-Bousquet, Eugenio Cinquemani, and Delphine Ropers have developed an approach based on nonlinear mixed-effects modelling for the parameter estimation of large-scale mechanistic models from time-series transcriptomics data. It allows to factor out technical variability, to compensate for the limited number of conditions and time points by a population approach, and it incorporates mechanistic details to gain insight on the molecular causes of biological variability. We applied our approach for the biological interpretation of microarray and RNA-Seq gene expression profiles, but it is generalisable to numerous types of data. When integrated in a model describing the degradation kinetics of 4254 mRNAs in E. coli cells, the data allowed to identify the targets of post-transcriptional regulatory mechanisms. Our approach paves the way for the interpretation of high-throughput biological data with more comprehensive mechanistic models. An article based on this work has been submitted 19.
8.6 Inference of parameters on lineage trees
Participants: E. Cinquemani, A. Marguet, E. Reginato.
Recent technological developments have made it possible to obtain single-cell measurements of gene expression and even the associated lineage information. However, most of the existing methods for the identification of mathematical models of gene expression are not well-suited to single-cell data and make the simplifying assumptions that cells in a population are independent, thus ignoring cell lineages. The development of statistical tools taking into account the correlations between individual cells will allow in particular for the investigation of inheritance of traits in bacterial populations.Modelling and identification of gene expression models with mother-daughter inheritance have been investigated previously through a collaboration among Eugenio Cinquemani, Marc Lavielle (XPOP, Inria Saclay–Île-de-France) and Aline Marguet, which led to a publication in Bioinformatics 9. Here, a novel method generalizing the Mixed-Effects (ME) paradigm to tree-structured populations, called ARME, was developed for the inference of an Auto-Regressive inheritance model of kinetic gene expression parameters. Further exploration of the subject, including inference of inheritance dynamics from different types of data, constitutes the subject of the Ph.D. project of Emrys Reginato. The inference of inheritance parameters based on datasets where the underlying genealogy is unknown has been investigated. In this case, we explored how well the inheritance dynamics can be inferred in different cases (transient vs. stationary mean, number of observed generations, incomplete sample, etc.). Preliminary results are very promising, and suggests that even in the challenging scenario of stationary means, the inheritance dynamics can be retrieved from observation of a moderate number of generations, without any knowledge on the mother-daughter relationships in the dataset. The focus of this year has been the development of a full pipeline to infer the inheritance parameters from fluorescence measurements, with an intermediate step for the estimation of individual parameters based on mixed-effect models and associated estimation algorithms.
8.7 Mathematical analysis of structured branching populations
Participant: A. Marguet.
The investigation of cellular populations at a single-cell level has already led to the discovery of important phenomena, such as the occurrence of different phenotypes in an isogenic population. Nowadays, several experimental techniques, such as microscopy combined with the use of microfluidic devices, enable one to take investigation further by providing time-profiles of the dynamics of individual cells over entire lineage trees. The development of models that take into account the genealogy is an important step in the study of inheritance in bacterial population. And the efficient analysis of single-cell data relies on the mathematical analysis of those models.Structured branching processes allows for the study of populations, where the lifecycle of each cell is governed by a given characteristic or trait, such as the internal concentration of proteins. The dependence on this characteristic of cellular mechanisms, like division or ageing, has been explored by Aline Marguet via the mathematical analysis of those processes. In collaboration with Charline Smadi (INRAE Grenoble), Aline Marguet investigated the long-time behavior of a parasite infection in a cell population. In this work, submitted for publication 20, the dynamic of the cell population is modelled using a structured branching process where the cell cycles depends on the dynamics of the parasites contained in the cell. Obtained results focus on the asymptotic behavior of the quantity of parasites in the cells. It proved to be very sensitive to the way cell division and death rate depend on the quantity of parasites in the cell and to the law of the sharing of the parasites between the two daughter cells at division. A particular focus on the effect of various sharing strategies in the case of a constant division rate is also in progress.
The study of the asymptotic behavior of general non-conservative semigroups is important for several aspects of branching processes, especially to prove the efficiency of statistical procedures. Vincent Bansaye from École Polytechnique, Bertrand Cloez from INRAE Montpellier, Pierre Gabriel from Université Versailles Saint-Quentin and Aline Marguet obtained necessary and sufficient conditions for uniform exponential contraction in weighted total variation norm of non-conservative semigroups. It ensures the existence of Perron eigenelements and provides quantitative estimates of spectral gaps, complementing Krein-Rutman theorems and generalizing recent results relying on probabilistic approaches. This work has been published in the Journal of the London Mathematical Society14.
9 Partnerships and cooperations
9.1 International initiatives
9.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
Associate-team in the framework of the Inria London Program: GERM
Participants: I. Cancino Aguirre, H. de Jong, D. Ropers.
-
Title:
Growth-rate control in mycobacteria: Computational exploration of metabolic strategies
-
Duration:
2022 -> 2024
-
Coordinator:
Delphine Ropers
-
Partners:
- Luiz de Carvalho, Francis Crick Institute (United Kingdom)
-
Summary:
Numerous Mycobacterium species pose serious threats to human and animal health. Genome-scale mathematical models of Mycobacterium metabolism are promising avenues to uncover bottlenecks explaining the growth-rate variability and pathogenicity observed across the genus. We employ these models to integrate and analyze diverse types of experimental data, including measurements of doubling times and metabolite concentrations. The results will allow us to formulate hypotheses on the molecular mechanisms responsible for growth-rate variability and pathogenicity observed across mycobacteria. The hypotheses will be tested by targeted experiments.
9.1.2 Informal international partners
H. de Jong and D. Ropers collaborate with T. Gedeon, former invited researcher in our former team IBIS, on research allocation strategies in microorganisms.
9.1.3 Visits to international teams
Research stays abroad
I. Cancino Aguirre
-
Visited institution:
Francis Crick Institute
-
Country:
United Kingdom
-
Dates:
01/11/2022 - 18/11/2022
-
Context of the visit:
visit to the Carvalho's laboratory in the context of the associate-team GERM
-
Mobility program/type of mobility:
research stay
D. Ropers
-
Visited institution:
Francis Crick Institute
-
Country:
United Kingdom
-
Dates:
01/11/2022 - 04/11/2022
-
Context of the visit:
visit to the Carvalho's laboratory in the context of the associate-team GERM
-
Mobility program/type of mobility:
research stay
9.2 National initiatives
| Project name | MAXIMIC: Optimal control of microbial cells by natural and synthetic strategies |
| Coordinator | H. de Jong |
| MICROCOSME participants | E. Cinquemani, T. Clavier, J. Geiselmann, H. de Jong, A. Pavlou, D. Ropers |
| Type | ANR project (2017-2023) |
| Web page | Link to project description |
| Project name | RIBECO (RIBonucleotide ECOnomy): Engineering RNA life cycle to optimize economy of microbial energy |
| Coordinator | M. Cocaign-Bousquet |
| MICROCOSME participants | E. Cinquemani, T. Etienne, D. Ropers |
| Type | ANR project (2018-2023) |
| Web page | Link to project description |
| Project name | Ctrl-AB : Optimization and control of the productivity of an algal-bacterial consortium |
| Coordinator | J.-L. Gouzé |
| MICROCOSME participants | R. Asswad, S. Arias, E. Boucher, E. Cinquemani, Th. Clavier, H. de Jong, J. Geiselmann, L. Léau-Mercier, A. Marguet, M. Sangster |
| Type | ANR project (2020-2024) |
| Project name | PlugNBio: A plug-and-play platform for reproducible microbial culture control experiments |
| Coordinator | E. Cinquemani |
| MICROCOSME participants | S. Arias, E. Boucher, E. Cinquemani, J. Geiselmann, L. Léau-Mercier |
| Type | Inria ADT (2022-2024) |
In addition to the above projects, A. Marguet contributes to the ANR JCJC NOLO of Bertrand Cloez (INRAE Montpellier) on non-local branching processes.
9.3 Regional initiatives
| Project name | ATLAS: Analysis of brain energy metabolism in the context of Parkinson’s disease |
| Coordinator | F. Fauvelle (Grenoble Institute of Neurosciences) |
| MICROCOSME participants | D. Ropers |
| Type | IXXI/BioSyl project (2022 – 2024) |
| Web page | Link to project description |
| Project name | MOSTIC: Stochastic modelling and inference for cell communities in interaction |
| Coordinator | A.Marguet |
| MICROCOSME participants | E. Cinquemani, J. Geiselmann, A.Marguet, C.Medous |
| Type | IXXI/BioSyl project (2020 – 2022) |
| Web page | Link to project description |
| Project name | AnaComBa: Analyse de Communautés Bactériennes : modélisation stochastique |
| Coordinator | A.Marguet & L. Coquille |
| MICROCOSME participants | E. Cinquemani, A.Marguet, C.Medous |
| Type | Equipe-Action du LABEX Persyval (2021 – 2024) |
10 Dissemination
Participants: R. Asswad, I. Cancino Aguirre, E. Cinquemani, Th. Clavier, M. Cocaign-Bousquet, H. de Jong, Th. Etienne, J. Geiselmann, A. Marguet, Ch. Medous, A. Pavlou, E. Reginato, D. Ropers, M. Sangster.
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
Member of organizing committees
| MICROCOSME members | Conference, workshop, school | Date |
| Ignacia Cancino Aguirre | Inria PhD seminar, Montbonnot | 2022 |
| Hidde de Jong | Summer school on Economic Principles in Cell Physiology, Paris | Jul 2022 |
10.1.2 Scientific events: selection
Chair of conference program committees
| MICROCOSME member | Conference, workshop, school | Role |
| Eugenio Cinquemani | European Control Conference (ECC 2022) | Associate editor |
| Eugenio Cinquemani | BMC Supplements CMSB 2021 (post-proceedings) | Supplement editor (with Loïc Paulevé) |
Member of conference program committees
| MICROCOSME member | Conference, workshop, program |
| Eugenio Cinquemani | ECC 2022, ECC 2023, CMSB 2022 |
| Hidde de Jong | IFAC DYCOPS-CAB 2022 |
| Delphine Ropers | ECCB 2022 |
10.1.3 Journal
Member of editorial boards
| MICROCOSME member | Journal |
| Hidde de Jong | Journal of Mathematical Biology |
| Hidde de Jong | Biosystems (reviews editor) |
| Hidde de Jong | ACM/IEEE Transactions on Computational Biology and Bioinformatics |
| Johannes Geiselmann | Frontiers in Microbiology (review editor) |
10.1.4 Invited talks and other presentations
Ignacia Cancino Aguirre
| Title | Event and location | Date |
| Computational analysis of metabolic strategies of mycobacteria | Inria PhD Seminar, Montbonnot | May 2022 |
| Computational analysis of metabolic strategies of mycobacteria | Mycobacterial metabolism Laboratory Seminar, London | Nov 2022 |
Hidde de Jong
| Title | Event and location | Date |
| Optimal cell behavior in time | Summer school on Economic Principles in Cell Physiology, Paris (with M. Köbis) | Jul 2022 |
| Mathematical modeling of bacterial metabolism | Invited talk and blackboard session at Spring School on Bioinformatics and Computational Approaches in Microbiology, Nottwil (Switzerland) (with M. Sangster) | May 2022 |
Aline Marguet
| Title | Event and location | Date |
| Parasite infection in a cell population with deaths | Séminaire de Probabilités de l'IRMAR, Rennes | Jan 2022 |
| Parasite infection in a cell population with deaths | Séminaire de Probabilités de l'ICJ et l'UMPA, Lyon | Mar 2022 |
Delphine Ropers
| Title | Event and location | Date |
| Integrative analysis of big data with genome-scale metabolic models: Application in precision medicine | Lecture and blackboard course at Summer school of MSc Health and Medical Data Analytics: Learning from health data | Sep 2022 |
| Computational analysis of metabolic strategies of mycobacteria | Mycobacterial metabolism Laboratory Seminar, London | Nov 2022 |
Maaike Sangster
| Title | Event and location | Date |
| Analysis and control of an E. coli cross-feeding consortium on an automated experimental platform | Microbial communities: current approaches and open challenges, Cambridge (UK) | Oct 2022 |
| Mathematical modeling of bacterial metabolism | Blackboard session at Spring School on Bioinformatics and Computational Approaches in Microbiology, Nottwil (Switzerland) (with H. de Jong) | May 2022 |
10.1.5 Scientific evaluation and expertise
| MICROCOSME member | Organism | Role |
| Johannes Geiselmann | ANR | Member committee CE30 |
| Johannes Geiselmann | UMR5240 CNRS-UCBL-INSA-BayerCropScience | Member scientific council |
| Hidde de Jong | Microbiology and Food Chain Department, INRAE | Member scientific council |
| Hidde de Jong | Univ Grenoble Alpes | Member scientific council of Pôle MSTIC |
| Hidde de Jong | International Human Frontier Science Program (HFSP) | Member grant selection committee |
| Hidde de Jong | Univ Grenoble Alpes | Member of Vice-Présidence Recherche et Innovation élargie |
| Hidde de Jong | Univ Grenoble Alpes | Member of Collège des Ecoles Doctorales |
| Hidde de Jong | Univ Grenoble Alpes | Membre jury Cross-Disciplinary Projects (CDP) |
10.1.6 Research administration
| MICROCOSME member | Committee | Role |
| Ignacia Cancino Aguirre | Inria - Univ. Grenoble Alpes | Member Comité du centre |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité des Emplois Scientifiques (CES) |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité des Utilisateurs des Moyens Informatiques (CUMI) |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité Développement Technologique (CDT) |
| Hidde de Jong | Inria - Univ. Grenoble Alpes | President scientific council (COS) |
| Hidde de Jong | Inria | Member working group on International Relations of Conseil d'Orientation Scientifique et Technique (COST) |
| Hidde de Jong | Inria | Member Commission d'évaluation (CE) |
| Aline Marguet | Inria - Univ. Grenoble Alpes | Member Comité du centre |
| Aline Marguet | Inria - Univ. Grenoble Alpes | Member Comité des études doctorales |
| Delphine Ropers | Inria - Univ. Grenoble Alpes | Référente chercheurs |
10.1.7 Recruitment committees
| MICROCOSME member | Organism | Recruitment |
| Eugenio Cinquemani | Inria - Univ Grenoble Alpes | CRCN (jury d’admissibilité) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | CRCN/IFSP (jury d'admissibilité) |
| Hidde de Jong | Inria national | DR0 (jury de promotion) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | Responsable de service de recrutement |
| Johannes Geiselmann | Grenoble INP | Selection committee Assistant Professor |
| Delphine Ropers | Inria national | CRCN/IFSP (jury d'admission) |
| Delphine Ropers | Institut Pasteur, Paris | Research Engineer |
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
J. Geiselmann is full professor at Univ Grenoble Alpes. He therefore has a full teaching service (at least 192 hours per year) and administrative duties related to the organization and evaluation of the university course programs on all levels. The following people have also contributed to courses last year.
Rand Asswad
- Course: Fundamental mathematical tools for life sciences, L1, Life Sciences, Univ Grenoble Alpes (16 h)
Ignacia Cancino Aguirre
- Practicals: Cellular biology, L2, Life Sciences, Univ Grenoble Alpes (14 h)
Eugenio Cinquemani
- Course: Statistics for systems biology, M1, Master Approches Interdisciplinaires du Vivant, CRI/Univ Paris Descartes (20 h, also in charge of 20 h of practicals)
- Course: Modelling and identification of metabolic networks, M1, Phelma, INP Grenoble (4 h)
Hidde de Jong
- Course and practicals: Modeling and simulation of gene regulatory networks, M2, BIM, INSA de Lyon (25 h)
Aline Marguet
- Practicals: Biostatistics, M2, Univ Grenoble Alpes (27 h)
Charles Medous
- Course and practicals: Introduction to mathematical biology and population dynamics, L1, Life sciences, Univ Grenoble Alpes (32 h)
Emrys Reginato
- Tutorial: Statistics, L1, Computer sciences, Univ Grenoble Alpes (18 h)
- Course and tutorials: Fundamental mathematical tools for life sciences, L1, Life Sciences, Univ Grenoble Alpes (16 h)
Delphine Ropers
- Course and practicals: Modelling in systems biology, M1, Phelma, INP Grenoble (16 h)
- Course and practicals: Cell systems biology and modelling cell functions, M1, Master ingéniérie de la santé, Univ Grenoble Alpes (24 h)
- Course: Modeling and simulation of genetic regulatory networks, M2, INSA de Toulouse (4 h)
- Course: Metabolic modelling with omics data, M2, IA4 Health International master course, Univ Grenoble Alpes (6h)
10.2.2 Supervision
- PhD completed: Antrea Pavlou, Experimental and computational analysis of bacterial self-replicators. Supervisors: Hidde de Jong and Johannes Geiselmann. Defense: Jul 2022.
- PhD in progress: Rand Asswad, Development of control strategies for synthetic microbial consortia. Supervisors: Eugenio Cinquemani and Jean-Luc Gouzé (Inria - Univ Côte d'Azur)
- PhD in progress: Ignacia Cancino Aguirre, Computational analysis of metabolic strategies in pathogenic bacteria. Supervisors: Delphine Ropers and Hidde de Jong
- PhD in progress: Thibault Clavier Genetic growth control to maximize the bioproduction in E. coli. Supervisors: Johannes Geiselmann and Hidde de Jong
- PhD in progress: Charles Medous, Analysis of bacterial communities: stochastic modelling. Supervisors: Loren Coquille (Institut Fourier, Grenoble), Aline Marguet, Charline Smadi (Inrae Grenoble)
- PhD in progress: Emrys Reginato, Development, analysis, and inference of stochastic models of gene expression in growing cell populations. Supervisors: Eugenio Cinquemani and Aline Marguet
- PhD in progress: Maaike Sangster, Development, characterization and control of E. coli communities on an automated experimental platform. Supervisors: Eugenio Cinquemani and Johannes Geiselmann
10.2.3 Juries
PhD thesis committees
| MICROCOSME member | Role | PhD student | University | Date |
| Hidde de Jong | Directeur | Antrea Pavlou | Univ Grenoble Alpes | Jul 2022 |
| Hidde de Jong | Rapporteur | Julien Hurbain | Univ de Lille | Nov 2022 |
| Johannes Geiselmann | Examinateur | Yizhong YUAN | Univ Grenoble Alpes | Jun 2022 |
| Johannes Geiselmann | Rapporteur | Charlotte Roux | INSA Toulouse | Jul 2022 |
| Johannes Geiselmann | Examinateur | Daniele di Bari | Univ Grenoble Alpes | Jul 2022 |
| Johannes Geiselmann | Rapporteur | Mickael Dinclaux | INSA Toulouse | Oct 2022 |
| Johannes Geiselmann | Rapporteur | Paul Anziani | Univ. Claude Bernard Lyon I | Oct 2022 |
| Delphine Ropers | Examinatrice | Elias Ventre | ENS Lyon | Sep 2022 |
| Delphine Ropers | Rapportrice | Arnaud Belcour | Univ de Rennes 1 | Oct 2022 |
PhD advisory committees
| MICROCOSME member | PhD student | University |
| Eugenio Cinquemani | Marielle Peré | Univ Côte d’Azur |
| Johannes Geiselmann | Clément Caffaratti | Univ Grenoble Alpes |
| Delphine Ropers | Paul Ahavi | Univ Paris Saclay |
10.3 Popularization
10.3.1 Interventions
Delphine Ropers
| Title | Event and location | Date |
| Des mathématiques pour comprendre l'infiniment petit - La croissance bactérienne | Invited talk at the "cérémonie de remise de prix des Olympiades de Mathématiques", Académie de Grenoble | May 2022 |
11 Scientific production
11.1 Major publications
- 1 articleEstimation of time-varying growth, uptake and excretion rates from dynamic metabolomics data.Bioinformatics33142017, i301-i310
- 2 articleStochastic reaction networks with input processes: Analysis and application to gene expression inference.Automatica1012019, 150-156
- 3 articleCompetitive effects in bacterial mRNA decay.Journal of Theoretical Biology504November 2020
- 4 articleDynamical allocation of cellular resources as an optimal control problem: Novel insights into microbial growth strategies.PLoS Computational Biology123March 2016, e1004802
- 5 articleStatistical estimation in a randomly structured branching population.Stochastic Processes and their Applications129122019, 5236-5277
- 6 articleA synthetic growth switch based on controlled expression of RNA polymerase.Molecular Systems Biology1111November 2015, 840
- 7 articleWhat population reveals about individual cell identity: Single-cell parameter estimation of models of gene expression in yeast.PLoS Computational Biology122February 2016, e1004706
- 8 articleOptimal proteome allocation and the temperature dependence of microbial growth laws.npj Systems Biology and Applications7142021
- 9 articleInheritance and variability of kinetic gene expression parameters in microbial cells: modeling and inference from lineage tree data.Bioinformatics35142019, i586-i595
- 10 articleEnhanced production of heterologous proteins by a synthetic microbial community: Conditions and trade-offs.PLoS Computational Biology1642020, e1007795
- 11 articleThe Csr System Regulates Escherichia coli Fitness by Controlling Glycogen Accumulation and Energy Levels.mBio85October 2017, 1-14
- 12 articleThe post-transcriptional regulatory system CSR controls the balance of metabolic pools in upper glycolysis of Escherichia coli.Molecular Microbiology1004May 2016, 686-700
- 13 articleAcetate metabolism and the inhibition of bacterial growth by acetate.Journal of Bacteriology20113July 2019, 147 - 166
11.2 Publications of the year
International journals
Doctoral dissertations and habilitation theses
Reports & preprints
11.3 Cited publications
- 22 articleGenomewide Stabilization of mRNA during a ''Feast-to- Famine'' Growth Transition in Escherichia coli.MSphere53June 2020
- 23 articleState observation in microbial consortia: A case study on a synthetic producer-cleaner consortium.International Journal of Robust and Nonlinear ControlDecember 2021, 1-12
- 24 articleMultiomics Study of Bacterial Growth Arrest in a Synthetic Biology Application.ACS Synthetic Biology1011November 2021, 2910-2926

