2023Activity reportProject-TeamSAIRPICO
RNSR: 202324398Z- Research center Inria Centre at Rennes University
- In partnership with:INSERM, Institut Curie
- Team name: Space-time imaging, artificial intelligence and computing for cellular and chemical biology
- In collaboration with:Chimie et Biologie de la Cellule
- Domain:Digital Health, Biology and Earth
- Theme:Computational Biology
Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.3. Data and knowledge analysis
- A3.3.3. Big data analysis
- A3.4. Machine learning and statistics
- A3.4.1. Supervised learning
- A3.4.5. Bayesian methods
- A3.4.6. Neural networks
- A3.4.7. Kernel methods
- A3.4.8. Deep learning
- A5.3. Image processing and analysis
- A5.3.2. Sparse modeling and image representation
- A5.3.3. Pattern recognition
- A5.3.4. Registration
- A5.4.1. Object recognition
- A5.4.4. 3D and spatio-temporal reconstruction
- A5.4.5. Object tracking and motion analysis
- A5.4.6. Object localization
- A5.9.1. Sampling, acquisition
- A5.9.2. Estimation, modeling
- A5.9.3. Reconstruction, enhancement
- A5.9.5. Sparsity-aware processing
- A5.9.6. Optimization tools
- A6.1.2. Stochastic Modeling
- A6.1.3. Discrete Modeling (multi-agent, people centered)
- A6.1.4. Multiscale modeling
- A6.1.5. Multiphysics modeling
- A6.2.3. Probabilistic methods
- A6.2.4. Statistical methods
- A6.2.6. Optimization
- A6.3. Computation-data interaction
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.3.4. Model reduction
- A6.3.5. Uncertainty Quantification
- A9.2. Machine learning
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1.1.1. Structural biology
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B2.2.3. Cancer
- B2.6. Biological and medical imaging
1 Team members, visitors, external collaborators
Research Scientists
- Charles Kervrann [Team leader, INRIA, Senior Researcher, from Apr 2023, HDR]
- Anais Badoual [INRIA, Researcher, from Apr 2023, 0.8 ETP from November 2023]
- Patrick Bouthemy [INRIA, Senior Researcher, from Apr 2023, HDR]
- Estelle Dransart [Institut Curie, Researcher, from Apr 2023, 0.25 ETP]
- Ludger Johannes [INSERM, Senior Researcher, from Apr 2023, 0.25 ETP]
- Massiullah Shafaq-Zadah [INSERM, Researcher, from Apr 2023, 0.25 ETP]
- Christian Wunder [INSERM, Researcher, from Apr 2023, 0.25 ETP]
Post-Doctoral Fellow
- Ilyes Hamitouche [Institut Curie, Post-Doctoral Fellow, from Oct 2023, 0.8 ETP]
PhD Students
- Lisa Balsollier [CNRS, from Apr 2023]
- Antonin Deschemps [INRIA, from Apr 2023 until Sep 2023]
- Sebastien Herbreteau [INRIA, from Apr 2023]
- Etienne Meunier [INRIA, from Apr 2023]
- Quentin Rapilly [INRIA, from Apr 2023]
- Quentin Tallon [IRSN, from Apr 2023]
Technical Staff
- Vincent Briane [INRIA, Engineer, from Apr 2023]
- Kevin Fournier [INRIA, Engineer, from Apr 2023 until Nov 2023]
- Arthur Masson [Inria, Engineer, from Oct 2023, SED]
- Emmanuel Moebel [INRIA, Engineer, from Apr 2023 until Nov 2023]
Interns and Apprentices
- Yasmine Hachani [INRAE, Intern, from Apr 2023 until Sep 2023]
- Sarra Khairi [INRIA, Intern, from Apr 2023 until Aug 2023]
- Maelys Risset [INRIA, Intern, from Apr 2023 until Sep 2023]
- Manuel Sanchez Laguardia [INRIA, Intern, from Apr 2023 until Sep 2023]
Administrative Assistant
- Caroline Tanguy [INRIA]
2 Overall objectives
During the past two decades many ground-breaking technologies emerged and allowed the visualization of tissues, cells, proteins, viruses, and macromolecular structures at all levels of spatial resolution (from 10 nm to 150 nm). The discovery of fluorescent labeling probes (Green fluorescence Protein, Nobel Prize in chemistry 2008) and recent advances in optics and digital sensors (e.g., PALM, STED and SIM) have been key developments which have served to overcome the theoretical optical diffraction limit (200 nm) established in the 19th century. Because of these technological breakthroughs and their impacts in life sciences, contemporary microscopy has been praised through prestigious awards, such as the Nobel Prizes awarded to inventors of the concepts of super-resolution microscopy (2014) and cryo-electron microscopy (2017). Fluorescent microscopy imaging has become the spearhead of modern biology as it is able to generate videos comprising dozens of Gigabytes of data within an hour, and can depict long-term 4D nanoscale cell behaviors with low photo-toxicity. The ability to follow nanoscale cellular events is also proving to be of immense clinical relevance, especially for the study of cancer progression and viral infections. All these technological advances in microscopy have created new challenges for researchers in signal-image processing, and have even modified conventional paradigms once digital processing became a key component in the surmounting of the diffraction barrier (e.g., PALM and SIM).
All fluorescence microscopy systems record fluorescent signals emitted by molecules tagged with genetically engineered or chemically coupled proteins within cells. In a conventional setup photons are collected and registered at a given pixel (or voxel in 3D imaging). The measured fluorescence intensity is a scalar value, generally proportional to the density of tagged-molecules representing a few dozens of nanometers within a pixel/voxel. However, fluorescence necessarily includes intensity (biomolecule density), wavelength (absorption and emission spectrum), time (fluorescence decay lifetime) and polarization (which arises from the dipole orientation). Nevertheless, it is worth noting that the orientation of dipoles cannot be measured by conventional fluorescence microscopy setups. The next generation technology will be able to provide the missing directional information which is required to better reveal the structure and function of biomolecules and organelles in cells. Among the recent progress, let us mention polarized microscopy that has the potential to probe the dipole orientation of fluorophores linked to proteins or lipids of interest and thereby, to report valuable information about the orientation and diffusive behavior of the molecule. Light polarization technology is also very flexible since it can be advantageously combined with super-resolution microscopy to characterize the nanometric structural organization of filamentous assemblies (actin filaments, microtubules), of membrane lipid orientations or the global architecture of local assembly of both proteins and lipids. Given their promising potential in terms of flexibility and production of information at high spatial resolution in vivo, polarized microscopy vector-valued images are likely to be in the future as common as confocal scalar-valued images.
As the resulting image data are 3D+time multi-valued signals, potentially depicting several fluorescently tagged molecular species, the analysis and the interpretation of these signals represents a new challenge in signal image processing and statistical machine learning, and one for which several scientific barriers must be overcome. A first barrier is to reduce the high level of noise and blur observed in 3D+time vector-valued data, which encompass information about density and orientation of biomolecules. As the processing of very large temporal series of images considerably slows down the analysis, special attention must be paid to the feasibility and scalability of the developed algorithms. A second barrier is the interpretation of dynamic and structural information content of such vector-valued images, for which no general method currently exists. A third barrier relates to the possibility of producing 3D spatial high-resolution maps of molecular motions from data generated by conventional polarized microscopy instruments. These barriers translate into unsolved digital challenges which need to be surmounted in order for this technology to be adopted in large-scale biological studies.
As the current methods are limited in handling polarized images, SAIRPICO aims to create the next generation of information processing techniques required to overcome the aforementioned barriers, and to solve challenging image processing problems induced by the acquisition of 3D+time vector-valued images. The resulting algorithms will serve to characterize the dynamics of biomolecules (e.g., proteins, lipids, …) and to decipher the molecular transport pathways or the motion (e.g., migration) and deformation of cells, which is of considerable of interest in fundamental cell biology and for precision medicine.
3 Research program
Four complementary Research Axes will be investigated with scientists who develop chemical methods (e.g. advanced imaging probes such as non-natural clickable amino acids, linker chemistry) to improve the rigidity of linkers and the photo-stability of fluorophores required for robust estimation of orientation of single molecules and components of cytosolic machinery, as well as single-molecule FRET techniques to infer and quantify interactions between membrane proteins.
Methodological Research Axis 1 - Modeling and reconstruction of multi-valued images.
Development of cutting-edge computational strategies and mathematical frameworks for reconstructing multi-valued images. Structure-based sparse representations of multi-value images will be established from the analysis of the spatiotemporal correlations and the inherent redundancy of data in multiple images. We will investigate statistical nonparametric methods and aggregation techniques, variational Bayesian methods, including shape-based models, as well as machine learning strategies to solve the underlying inverse problems.
Methodological Research Axis 2 - Methods for high-resolution spatial quantification of molecular mobility and interactions.
Characterization of molecular mobility at the nanoscale from multi-valued images. We intend to fully exploit the rich contents of microscopy images in order to build single-molecule (e.g., endocytic ligands) and biomolecule (e.g., cytosolic machinery, metabolic sensors) tracking algorithms, derive robust estimators of molecular mobility, and quantify spatially-variable interactions between molecular species and cytoskeleton. The resulting algorithms will be used to produce high-resolution spatial maps of molecular mobility given stochastic motion models and sparse representations.
Methodological Research Axis 3 - Spatiotemporal modeling of 3D shapes, motions and deformations.
Development of shape models and descriptors to capture 3D motion and deformation of macromolecular complexes (cryo-electron tomography (cryo-ET), single particle analysis (SPA)) on one hand, and on the other hand, intracellular components and tumor cells, at the scale of a single cell and tissues. We intend to represent 3D shapes by parametric surfaces controlled by key points and to segment and track structures in 3D microscopy. The main originality will be to exploit annotations and/or high-level priors to derive features for classifying molecular conformations in cryo-ET, and phenotypes induced by drugs (single cell), or controlled hypoxia conditions (tissue scale) in 3D+time fluorescence microscopy.
Transversal Research Axis 4 - Analysis of case-studies in cell biology and cancer research.
Demonstration that the methods and algorithms related to the three previous methodological axes allow one to perform image reconstruction for several 3D instruments (TIRFM, Lattice Light Sheet Microscopy, Multi-Focus Microscopy, cryo-ET), and accurately quantify the shape and motion of cell components and biomolecules that interact with membranes and the cytoskeleton. The resulting images and features will be helpful to better decipher the intracellular dynamics of trafficking and signaling events in living cells, especially membrane mechanics at the cell surface, endocytosis, as well as signal transduction to the nucleus. The methods will be developed for investigation in cellular and chemical biology, and extended further to perform analysis at the tissue scale.
4 Application domains
The advances in SAIRPICO will result in a new generation of algorithms for multi-valued microscopy instruments, which will be widely used in the future in fundamental and applied cellular and chemical biology. The team gathers researchers developing new imaging modality and computational methods, biophysicists to develop and provide adapted experimental and theoretical models, chemist to design adapted probes and cell biologists. In collaboration with other teams of U1143 and the help of dedicated engineers (to be recruited) who will stimulate the interface between experiment and data sciences, we expect to build a general approach based on theories and tools in optics, chemistry, cell biology, biophysics, statistics, and machine learning.
Our case studies in cellular and chemical biology will be related to the analysis of intracellular transport and signaling pathways, and the migration of tumor cells in organoids, as they represent a major contributory factor to a number of diseases such as cancer and viral infection. For instance, we wish to study in detail the causal link between lectin-driven glycolipid reorganization in biological membranes and the formation of endocytic sites from which clathrin-independent endocytic carriers are generated. Since a series of pathogens (e.g., polyoma and noroviruses), pathogenic factors (e.g., Shiga and cholera toxins) and cellular proteins (integrins, CD44...) are concerned by this mechanism we expect that this study will have a general impact in the life science and membrane biophysics communities. Understanding and exploring diverse and alternative cellular entry mechanisms, by gathering as many as possible molecular information in fundamental membrane biology research, paves the way for the development of innovative cancer therapy or vaccine strategies. We expect that our results will be helpful in the design of therapeutic compounds delivered to precise intracellular locations within specialized cells for immunotherapy, or to tumors for targeted therapy.
Meanwhile, the ambition of SAIRPICO is to become the reference team in computational polarized bioimaging, with a focus on the development of advanced signal-image processing techniques for cell imaging. To that end, we will create a centralized polarized image database and disseminate the results through dedicated workshops, summer schools, mini-symposia, on-line tutorials, and publications in high-visibility journals. It is worth noting that the interdisciplinary team will be be-localized in Rennes and Paris and therefore will benefit from the scientific environment of both Inria (Applied mathematics, artificial intelligence) and Institut Curie (chemical biology, optics).
5 Social and environmental responsibility
Application of general recommendations related to transport (train, visioconferences). Participation in presential mode to conferences, workshops, and meetings, were limited to events organized in France in 2023. The team essentially published its results via the submission to international journals. This has led to a few travels by train, and then a low carbon footprint for the team in 2023.
6 Highlights of the year
6.1 Creation of the SAIRPICO Project-Team
Based on the 5-year shared experience and the foreseen benefice of grouping people from cell biology, microscopy and image processing, the SAIRPICO team involving Inria, INSERM, CNRS, and Institut Curie has been created on April 1st, 2023 and is integrated both within the INSERM Unit 1143 / UMR 3666 - Institut Curie (Paris) and the Centre Inria de l'Université de Rennes. The process of creation was jointly evaluated in 2022 by representatives of stakeholders, including INSERM (Institut Thématique “Technologies pour la Santé”), CNRS (Institut de Chimie), and Institut Curie. C. Kervrann and L. Johannes have already participated to common projects (ANR PRC DALLISH 2017-2021, Labex Cell(n) Scale PoMIMO (2022-2023), ANR PRME POLARISCOPIA (2023-2026) and have supervised one postdoc fellow (C.A. Valades-Cruz, 2017-2022). In practice, the permanent Inria and INSERM researchers are located in Rennes and Paris, respectively. M. Shafaq-Zadah (INSERM) is located in Paris and Rennes. The non-permanent researchers (PhD, post-docs) and engineers will be located in both sites. Regular meetings and reciprocal visits will be organized on a tight periodicity for team members working on the same topic, especially when it involves PhD and postdoc co-supervision. To strengthen our common work, we have developed an original computational strategy (BioImageIT middleware (S. Prigent, et al, Nature Methods 2022)) to efficiently share data and software related to our projects.
The scientific goal of the team is to analyze the geometry and the spatiotemporal organization of fluorescent probes and orientated dipoles in live-cell imaging. We plan to develop computational methods for solving image reconstruction problems, quantifying molecular mobility and interactions at a very high spatial resolution, and dynamics of cells at the tissue-scale. As such, SAIRPICO holds much promise for nanomedicine. The ambition of SAIRPICO is to become the reference team in computational bioimaging focused on the development of advanced image processing and machine learning techniques to be applied in and to receive input from the field of cellular/chemical biology.
6.2 Highlighted publications
The two following papers were published in very compoetitive and high-impact conferences:
- S. Herbreteau, E. Moebel, C. Kervrann. Normalization-equivariant neural networks with application to image denoising, Proc. of NeurIPS, New-Orleans, USA, December 2023.
- E. Meunier and P. Bouthemy. Unsupervised space-time network for temporally-consistent segmentation of multiple motions, Proc. Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, Canada, June 2023.
7 New software, platforms, open data
7.1 Plateforms
Participants: Charles Kervrann, Arthur Masson, Kevin Fournier.
7.1.1 BioImageIT for bioimage management and processing
New image acquisition systems generate large number of images and large volume images. Such data sets are hard to store, to process and to analyze for in a workstation. Many solutions exist for data management (e.g. Omero, OpenImadis), image analysis (e.g. Fiji, Icy, CellProfiler) and statistics (e.g R software). Each of them has its specificities and several bridges have been developed between pieces of software. Nevertheless, in many use-cases, we need to perform analysis using tools that are available in different pieces of software and different languages. It is then tedious to create a workflow that brings the data from one tool to another. This process requires programming skills and most of the time, custom scripts are developed to handle data processing management. To overcome these difficulties, we have already developed a framework – BioImage-IT (bioimageit.github.io) – to create a middleware application that allow any scientist to annotate, process, and analyze data using only one single high level application. This BioImage-IT application is based on 3 components:
- an image annotation method interoperable with existing databases;
- an image processing and analysis tools integration method based on packaging and wrapping techniques;
- an application with a graphical interface to easily annotate data, run processing tools, and visualize data and results.
This software architecture has three main goals. First, data are annotated with open formats and experiment can then be stored in different architectures or servers. Second, the processing tools are used as binary packages managed by the Conda or Docker technologies. These technologies enable to gently handle dependencies and several versions of the same tool. Any existing tool can then be integrated in its native programming language. Third, using a single middleware application allows to automatically generate metadata for any processed data, improving the traceability and the repeatability of any experimental result (FAIR principles).
We envision to continue to promote BioImageIT in the forthcoming years, initiated in the frame of the France-BioImaging research infrastructure (france-bioimaging.org) in order to provide a standardized image processing tool set and data management for the imaging facilities.
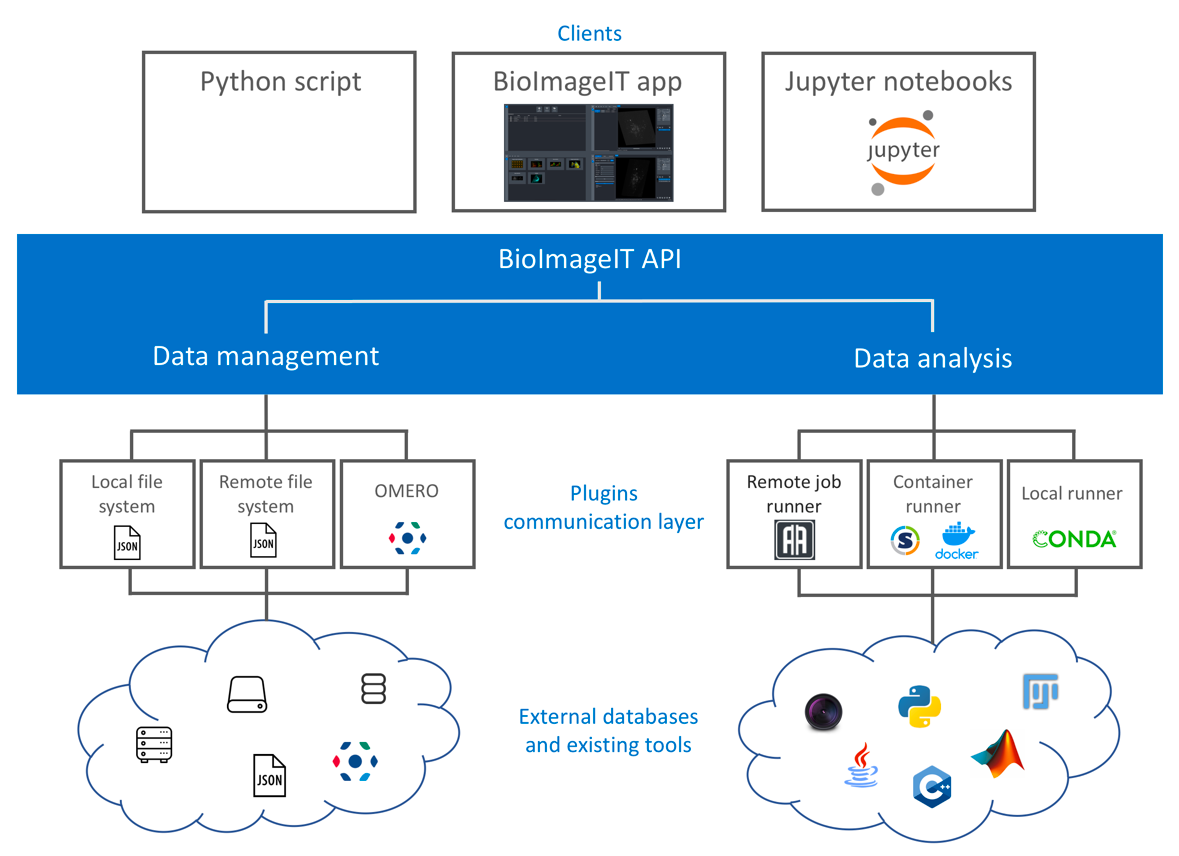
Scheme of the BioImageIT components interactions
7.2 New software
7.2.1 ST-Space-Time-Flow-segmentation
-
Keywords:
Neural networks, Unsupervised learning, Optic-flow, Movement segmentation, Image sequence
-
Functional Description:
Unsupervised segmentation of the optical flow by convolutional neural networks, taking into account the temporal dimension. The software takes an optic flow volume as input and provides an optic flow segmentation volume as output. Inference is performed in a single iteration. The software includes the trained weights of the network.
-
Authors:
Etienne Meunier, Patrick Bouthemy
-
Contact:
Etienne Meunier
7.2.2 DCT2Net
-
Name:
Trained shallow CNN (convolution neural network)-based DCT (Discrete Cosine Transform) denoiser
-
Keywords:
Deep learning, Denoising
-
Functional Description:
DCT2net software, based on the well-known DCT (Discrete Cosine Transform) image denoising algorithm, is dedicated to noise removal from images. The traditional DCT denoiser can be seen as a shallow CNN and thereby its original linear transform can be tuned through gradient descent in a supervised manner, improving considerably its performance. Consequently, DCT2net is a shallow and interpretable convolution network, whose parameters optimization allows to improve very significantly the performances of the traditional DCT denoiser. To deal with the remaining artifacts induced by DCT2net, an original hybrid solution between DCT and DCT2net is proposed, combining the best of what these two methods can offer. Experiments on artificially noisy images show that the two-layer DCT2net method provides results comparable to the BM3D method and is as fast as the DnCNN algorithm composed of more than a dozen of layers.
Inter Deposit Digital Number: IDDN.FR.001.460033.000.S.P.2021.000.21000 21
- URL:
- Publication:
-
Contact:
Charles Kervrann
-
Participants:
Charles Kervrann, Sebastien Herbreteau
7.2.3 DeepFinder
-
Name:
Deep learning for macromolecule identification within 3D cellular cryo-electron tomograms
-
Keywords:
Image analysis, Deep learning, Cryo-electron microscopy, Object detection
-
Functional Description:
DeepFinder is a computational approach that uses artificial neural networks to accurately and jointly localize multiple types and/or states of macromolecules in 3D cellular cryo-electron tomograms. DeepFinder leverages deep learning and outperforms the commonly-used template matching method on ideal data. On synthetic image data (SHREC 2019, 2020, and 2021 challenges), DeepFinder is very fast and produces superior detection results when compared to other competitive deep learning methods, especially on small macromolecules. On experimental cryo-ET data depicting ribosomes, the detection results obtained by DeepFinder are consistent with expert annotations. We have got a high overlap of detection (86%) and a similar structure resolution that those determined by subtomogram averaging.
Inter Deposit Digital Number: IDDN.FR.001.460030.000.S.P.2021.000.21000
- URL:
- Publication:
-
Contact:
Emmanuel Moebel
-
Participants:
Emmanuel Moebel, Charles Kervrann
-
Partners:
Max Planck Institute Martinsried, Fondation Fourmentin-Guilbert, Helmholtz Pioneer Campus
7.2.4 DenseMapping
-
Name:
Dense mapping of diffusion and drift of moving particules
-
Keywords:
Microscopy, Cellular imaging
-
Functional Description:
It is of primary interest for biologists to be able to visualize the dynamics of proteins within the cell. DenseMapping software allows to estimate the diffusion and drift parameters attached to moving biomolecules within cells from 2D/3D individual trajectories. In a first step, each particle track is labeled into three motion categories: confined motion (subdiffusion), Brownian motion (free diffusion), and directed motion (superdiffusion). The long trajectories are also segmented into sub-trajectories according to these three categories. In a second step, two 2D/3D maps (diffusion, drift) are computed from a local analysis of the trajectories. The local spatio-temporal kernel estimators correspond to weighted averages of the trajectory elements. The weights allow to select and aggregate the information in an optimal way.
Inter Deposit Digital Number: IDDN.FR.001.290028.000.S.P.2021.000.21000
- URL:
- Publications:
-
Contact:
Charles Kervrann
-
Participants:
Antoine Salomon, Vincent Briane, Cesar Augusto Valades Cruz, Charles Kervrann
-
Partner:
UMR 144 CNRS - Institut Curie
7.2.5 GcoPS
-
Name:
Geo-Co-Positioning System for co-localization of image pairs of fluorescent molecules
-
Keywords:
Photonic imaging, Fluorescence microscopy, Image processing, Statistic analysis
-
Functional Description:
The GcoPS (Geo-Co-Positioning System) software is dedicated to the co-localization of fluorescence image pairs for both conventional and super-resolution microscopy. The procedure is only controlled by a p-value (type I error-rate) and tests whether the Pearson correlation between two binary images is significantly positive. Colocalization amounts here to quantifying the interaction strength by the area/volume of the intersection between the two binary images viewed as random distributions of geometrical objects. Under mild assumptions, it turns out that the appropriately normalized Pearson correlation follows a standard normal distribution under the null hypothesis if the number of image pixels is large. Unlike previous methods, GcoPS handles 2D and 3D images, variable SNRs and any kind of cell shapes. It is able to co-localize large regions with small dots, as it is the case in TIRF-PALM experiments and to detect negative co-localization. The typical processing time is two milliseconds per image pair in 2D and a few seconds in 3D, with no dependence on the number of objects per image. In addition, the method provides maps to geo-co-localize molecule interactions in specific image regions.
- URL:
- Publication:
-
Contact:
Charles Kervrann
-
Participants:
Thierry Pécot, Frédéric Lavancier, Charles Kervrann, Liu Zengzhen
-
Partners:
Université de Nantes, UMR 144 CNRS - Institut Curie, Hollings Cancer Center at the Medical University of South Carolina, Institut de Génétique & Développement de Rennes
7.2.6 SPITFIRE
-
Name:
SParse fIT for Fluorescence Image Restoration
-
Keywords:
Denoising, Fluorescence microscopy
-
Functional Description:
SPITFIR(e) (SParse fIT for Fluorescence Image Restoration) is a very flexible software designed to restore 2D-3D+Time fluorescent images and subtract undesirable out-of-focus background. We assume that the images are sparse and piece-wise smooth, and are corrupted by mixed Poisson-Gaussian noise. The principle resides in the minimization of a convex energy functional that includes a fidelity-to-data term and a Sparse-Hessian Variation regularization term. A fast primal-dual optimization algorithm allows to restore very large images in a few seconds. SPITFIR(e) is nearly parameter-free as the practitioner needs only to specify the amount of desired sparsity (weak, moderate, high). Experimental results in lattice light sheet, stimulated emission depletion, multifocus microscopy, spinning disk confocal, and wide-field microscopy demonstrate the generic ability of the SPITFIR(e) algorithm to efficiently reduce noise and blur, and to subtract undesirable fluorescent background, while avoiding the emergence of deconvolution artifacts.
Inter Deposit Digital Number: IDDN.FR.001.460029.000.S.P.2021.000.21000
- URL:
- Publication:
-
Contact:
Charles Kervrann
-
Participants:
Sylvain Prigent, Hoai Nam Nguyen, Ludovic Leconte, Cesar Augusto Valades Cruz, Jean Salamero, Charles Kervrann
-
Partner:
UMR 144 CNRS - Institut Curie
8 New results
Note: In this section, we provide details of the "scientific production" of the team created in April 2023. Each paragraph summarizes a published or submitted paper.
8.1 Supervised/unsupervised methods for image restoration and object detection in imaging and microscopy
The main stream regarding image restoration and segmentation methods advocates appropriate information processing and statistical and machine learning frameworks to make images intelligible. Over recent years, the team has promoted sparse and redundant representations, as well as deep-learning to solve the aforementioned problems in bioimaging.
DeepCristae, a CNN for the restoration of mitochondria cristae in live microscopy images
Participants: Anaïs Badoual, Charles Kervrann.
Mitochondria play an essential role in the life cycle of eukaryotic cells. However, we still don't know how their ultrastructure, like the cristae of the inner membrane, dynamically evolves to regulate these fundamental functions, in response to external conditions or during interaction with other cell components. Although high-resolution fluorescent microscopy coupled with recently developed innovative probes can reveal this structural organization, their long-term, fast and live 3D imaging remains challenging. To address this problem, we have developed a convolutional neural network (CNN), called DeepCristae, to restore mitochondrial cristae in low spatial resolution microscopy images. Our CNN is trained from 2D STED images using a novel loss specifically designed for cristae restoration. Random sampling centered on mitochondrial areas was also developed to improve training efficiency. Quantitative assessments were carried out using metrics we derived to give a meaningful measure of cristae restoration. Depending on the conditions of use indicated, DeepCristae works well on broad microscopy modalities (STED, Live-SR, AiryScan and LLSM). It is ultimately applied in the context of mitochondrial network dynamics during interaction with endo/lysosomes membranes. (in collaboration with J. Salamero, L. Leconte, C.A. Valades-Cruz, CNRS-UMR144, Institut Curie; T. Liu, Z. Chen, PKU University, Institute of Molecular Medicine, Beijing, People Republic of China)
S. Papereux, L. Leconte, C.A. Valades-Cruz, T. Liu, J. Dumont, Z. Chen, J. Salamero, C. Kervrann, A. Badoual. DeepCristae, a CNN for the restoration of mitochondria cristae in live microscopy images, bioRxiv-doi:10.1101/2023.07.05.547594, 2023. (submitted paper)
Unsupervised linear and iterative combinations of patches for image denoising
Participants: Sébastien Herbreteau, Charles Kervrann.
In the past decade, deep neural networks have revolutionized image denoising in achieving significant accuracy improvements by learning on datasets composed of noisy/clean image pairs. However, this strategy is extremely dependent on training data quality, which is a well-established weakness. To alleviate the requirement to learn image priors externally, single image (a.k.a., self-supervised or zero-shot) methods perform denoising solely based the analysis of the input noisy image without external dictionary or training dataset. This work investigates the effectiveness of linear combinations of patches for denoising under this constraint. Although conceptually very simple, we show that linear combinations of patches are enough to achieve state-of-the-art performances. The proposed parametric approach relies on quadratic risk approximation via multiple pilot images to guide the estimation of the combination weights. Experiments on images corrupted artificially with Gaussian noise but also on real-world noisy images demonstrate that our method is on par with the very best single-image denoisers, outperforming the recent neural network-based techniques, while being much faster and fully interpretable. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
S. Herbreteau, C. Kervrann. Unsupervised linear and iterative combinations of patches for image denoising, HAL-INRIA-03894346, arXiv-doi:10.48550/arXiv.2212.00422, 2023. (LIChI software) (submitted paper)
A unified framework of non-local parametric methods for image denoising
Participants: Sébastien Herbreteau, Charles Kervrann.
We propose a unified view of non-local methods for single-image denoising, for which BM3D is the most popular representative, that operate by gathering noisy patches together according to their similarities in order to process them collaboratively. Our general estimation framework is based on the minimization of the quadratic risk, which is approximated in two steps, and adapts to photon and electronic noises. Relying on unbiased risk estimation (URE) for the first step and on “internal adaptation”, a concept borrowed from deep learning theory, for the second, we show that our approach enables to reinterpret and reconcile previous state-of-the-art non-local methods. Within this framework, we propose a novel denoiser called NL-Ridge that exploits linear combinations of patches. While conceptually simpler, we show that NL-Ridge can outperform well-established state-of-the-art single-image denoisers. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
S. Herbreteau, C. Kervrann. A unified framework of non-local parametric methods for image denoising, HAL-INRIA-04472406, arXiv-doi:10.48550/arXiv.2402.13816, 2023. (NL-Ridge software) (submitted paper)
Normalization-equivariant neural networks with application to image denoising
Participants: Sébastien Herbreteau, Emmanuel Moebel, Charles Kervrann.
In many information processing systems, it may be desirable to ensure that any change of the input, whether by shifting or scaling, results in a corresponding change in the system response. While deep neural networks are gradually replacing all traditional automatic processing methods, they surprisingly do not guarantee such normalization-equivariance (scale + shift) property, which can be detrimental in many applications. To address this issue, we propose a methodology for adapting existing neural networks so that normalization-equivariance holds by design. Our main claim is that not only ordinary convolutional layers, but also all activation functions, including the ReLU (rectified linear unit), which are applied element-wise to the pre-activated neurons, should be completely removed from neural networks and replaced by better conditioned alternatives. To this end, we introduce affine-constrained convolutions and channel-wise sort pooling layers as surrogates and show that these two architectural modifications do preserve normalization-equivariance without loss of performance. Experimental results in image denoising show that normalization-equivariant neural networks, in addition to their better conditioning, also provide a much better generalization across noise levels. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
S. Herbreteau, E. Moebel, C. Kervrann. Normalization-equivariant neural networks with application to image denoising, Proc. Conference on Neural Information Processing Systems (NeurIPS), New-Orleans, USA, December 2023. (arXiv-doi:10.48550/arXiv.2306.05037) (normalization-equivariant-nn software)
8.1.1 Supervised deep-learning for detection, segmentation, and classification in microscopy
Ensembling Unets, sparse representation, and low-dimensional visualization for chromosomal aberration detection in metaphase images
Participants: Antonin Deschemps, Emmanuel Moebel, Charles Kervrann.
In biological dosimetry a radiation dose is estimated using the average number of chromosomal aberrations per peripheral blood lymphocytes. This analysis is manually performed on 2D metaphase images depicting the 23 pairs of chromosomes as the false discovery rate of current automated detection systems is high and variable, depending on small variations in image quality (chromosome spread, illumination variations, etc). Therefore, the current systems are only assisting human experts but better automating chromosomal aberration detection has become of paramount of importance to improve diagnosis reliability and reduce human expertise time. To address this issue, we propose to build a novel object detection approach to automate chromosomal aberration detection using deep convolutional neural networks. We formulate the problem of rare aberration detection in metaphase images as a heatmap regression problem requiring the minimization of a sparsity-promoting loss to reduce the false alarm rate. Meanwhile, we describe a novel visualization approach to explore the model building during training. Finally, we demonstrate significant performance improvements on real data and provide statistical confidence intervals by using an ensemble of checkpoints that are selected at the end of each epoch during training and combined to generate a consensus model. (in collaboration, M. Benadjaoud, IRSN/SERAMED)
A. Deschemps, E. Grégoire, J.S. Martinez, A. Vaurijoux, P. Fernandez, D. Dugue, L. Bobyk, M. Valente, G. Gruel, E. Moebel, M.A. Benadjaoud, C. Kervrann. Ensembling Unets, sparse representation, and low-dimensional visualization for rare chromosomal aberration detection and identification in light microscopy images, bioRxiv-doi:10.1101/2023.09.11.557124, 2023. (submitted paper)
8.2 Motion computation, dynamics analysis, and visualization in imaging and microscopy
In this research axis, we have focused on unsupervised deep-learning-based image motion segmentation and motion saliency, while developing original concepts for optical flow estimation and investigating 3D+time data visualization techniques in fluorescence microscopy.
MorphoNet 2.0 : Efficient exploration and bio-curation of large 3D and 3D+t imaging datasets
Participants: Kevin Fournier, Charles Kervrann.
We present MorphoNet 2.0, featuring a standalone application running on all major operating systems, with bio-curation capability on large 3D+t segmented datasets. It allows multiple 3D channels of raw intensity and cell lineage images to be displayed on a local dataset. A wide range of image processing plugins is accessible to non-programming scientists through user-friendly graphical interfaces that require. Finally, MorphoNet 2.0 natively provides a Virtual Reality environment for data exploration in 3D. (in collaboration with E. Faure, LIRMM, Montpellier)
B. Gallean, T. Laurent, K. Biasuz, K. Fournier, G. Fouché, T. Cabel, G. Gay, A. Clement, J. Maizi, N. Faraj, F. Argelaguet, C. Kervrann, P. Lemaire, E. Faure. MorphoNet 2.0 : Efficient exploration and bio-curation of large 3D and 3D+t imaging datasets, 2023. (submitted paper)
EM-driven unsupervised learning for efficient motion segmentation
Participants: Etienne Meunier, Anaïs Badoual, Patrick Bouthemy.
In this paper, we present a CNN-based fully unsupervised method for motion segmentation from optical flow. We assume that the input optical flow can be represented as a piecewise set of parametric motion models, typically, affine or quadratic motion models. The core idea of our work is to leverage the Expectation-Maximization (EM) framework in order to design in a well-founded manner a loss function and a training procedure of our motion segmentation neural network that does not require either ground-truth or manual annotation. However, in contrast to the classical iterative EM, once the network is trained, we can provide a segmentation for any unseen optical flow field in a single inference step and without estimating any motion models. We investigate different loss functions including robust ones and propose a novel efficient data augmentation technique on the optical flow field, applicable to any network taking optical flow as input. In addition, our method is able by design to segment multiple motions. Our motion segmentation network was tested on four benchmarks, DAVIS2016, SegTrackV2, FBMS59, and MoCA, and performed very well, while being fast at test time. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
E. Meunier, A. Badoual and P. Bouthemy. EM-driven unsupervised learning for efficient motion segmentation, IEEE Transactions on Pattern Analysis and Machine Intelligence, 45(4):4462-4473, doi:10.1109/TPAMI. 2022.3198480, HAL-INRIA-03516617, 2023. (EM-Flow-Segmentation software)
Unsupervised space-time network for temporally-consistent segmentation of multiple motions
Participants: Etienne Meunier, Patrick Bouthemy.
Motion segmentation is one of the main tasks in computer vision and is relevant for many applications. The optical flow (OF) is the input generally used to segment every frame of a video sequence into regions of coherent motion. Temporal consistency is a key feature of motion segmentation, but it is often neglected. In this paper, we propose an original unsupervised spatio-temporal framework for motion segmentation from optical flow that fully investigates the temporal dimension of the problem. More specifically, we have defined a 3D network for multiple motion segmentation that takes as input a sub-volume of successive optical flows and delivers accordingly a sub-volume of coherent segmentation maps. Our network is trained in a fully unsupervised way, and the loss function combines a flow reconstruction term involving spatio-temporal parametric motion models, and a regularization term enforcing temporal consistency on the masks. We have specified an easy temporal linkage of the predicted segments. Besides, we have proposed a flexible and efficient way of coding U-nets. We report experiments on several VOS benchmarks with convincing quantitative results, while not using appearance and not training with any ground-truth data. We also highlight through visual results the distinctive contribution of the short- and long-term temporal consistency brought by our OF segmentation method. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
E. Meunier and P. Bouthemy. Unsupervised space-time network for temporally-consistent segmentation of multiple motions, Proc. Conf. Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, pp. 22139-22148, doi:10.1109/CVPR52729.2023.02120, 2023. (GeneralUnet software, Space-Time-Flow-Segmentation software)
Efficient local correlation volume for unsupervised optical flow estimation on small moving objects in large images
Participants: Etienne Meunier, Sarra Khairi, Patrick Bouthemy.
With the advent of deep learning methods, performance and efficiency of optical flow estimation has significantly increased, especially for supervised models. However, they do not generalize well to more specific data involving small moving objects in large images, such as high-resolution aerial, satellite or cell microscopy sequences. In addition, annotation and realistic simulation are difficult for these contents, which calls for unsupervised alternatives. Yet, the latter are still less accurate than their supervised counterparts. In this paper, we introduce an unsupervised local optical flow estimation method adapted to small moving objects in large-size images by involving no downsampling of the feature maps. We adopt a local correlation search and implement it in an original way with a per-shift computation, which minimizes memory consumption and speed up inference computation for large-scale images. We also design a loss function combining similarity, smoothness and sparsity constraints. We demonstrate the performance of our method on real aerial images, and favorably compare it to other methods. Our SMOFlow method is able to accurately capture the motion of small objects in large images, while efficiently reducing memory consumption. A paper has been submitted to a top-tier computer vision conference. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
S Khairi, E. Meunier, R. Fraisse, P. Bouthemy Efficient local correlation volume for unsupervised optical flow estimation on small moving objects in large images, 2023. (submitted paper)
Unsupervised motion segmentation in one go: smooth long-term model over a video
Participants: Etienne Meunier, Patrick Bouthemy.
Human beings have the ability to continuously analyze a video and immediately extract the main motion components. Motion segmentation methods often proceed frame by frame. We want to go beyond this classical paradigm, and perform the motion segmentation over a video sequence in one go. It will be a prominent added value for downstream computer vision tasks, and could provide a pretext criterion for unsupervised video representation learning. In this perspective, we propose a novel long-term spatiotemporal model operating in a totally unsupervised way. It takes as input the volume of consecutive optical flow (OF) fields, and delivers a volume of segments of coherent motion over the video. More specifically, we have designed a transformer-based network, where we leverage a mathematically well-founded framework, the Evidence Lower Bound (ELBO), to infer the loss function. The loss function combines a flow reconstruction term involving spatiotemporal parametric motion models combining, in a novel way, polynomial (quadratic) motion models for the (x, y)-spatial dimensions and B-splines for the time dimension of the video sequence, and a regularization term enforcing temporal consistency on the masks. We have carried out experiments on four benchmarks with convincing quantitative results. We have also highlighted through visual results the key contributions on temporal consistency brought by our method. A paper is in preparation for submission to a top-tier journal. (in collaboration with R. Fraisse, AIRBUS Defence and Space)
E. Meunier, P. Bouthemy Unsupervised motion segmentation in one go: smooth long-term model over a video, arXiv-doi:10.48550/arXiv.2310.01040, 2023.
8.3 Modeling, simulation and estimation of spatiotemporal biological mechanisms and processes
In this research axis, we have considered non-conventional spatiotemporal modeling, simulation and estimation frameworks and focused our activity on diffusion estimation and particle track analysis to address questions in cell biology that are related to cancer research.
A generative model to simulate spatiotemporal dynamics of biomolecules in cells
Participants: Lisa Balsollier, Charles Kervrann.
Generators of space-time dynamics in bioimaging have become essential to build ground truth datasets for image processing algorithm evaluation such as biomolecule detectors and trackers, as well as to generate training datasets for deep learning algorithms. In this contribution, we leverage a stochastic model, called birth-death-move (BDM) point process, in order to generate joint dynamics of biomolecules in cells. This particle-based stochastic simulation method is very flexible and can be seen as a generalization of well-established standard particle-based generators. In comparison, our approach allows us: (1) to model a system of particles in motion, possibly in interaction, that can each possibly switch from a motion regime (e.g., Brownian) to another (e.g., a directed motion); (2) to take into account finely the appearance over time of new trajectories and their disappearance, these events possibly depending on the cell regions but also on the current spatial configuration of all existing particles. This flexibility enables to generate more realistic dynamics than standard particle-based simulation procedures, by for example accounting for the colocalization phenomena often observed between intracellular vesicles. We explain how to specify all characteristics of a BDM model, with many practical examples that are relevant for bioimaging applications. As an illustration, based on real fluorescence microscopy datasets, we finally calibrate our model to mimic the joint dynamics of Langerin and Rab11 proteins near the plasma membrane, including the well-known colocalization occurrence between these two types of vesicles. We show that the resulting synthetic sequences exhibit comparable features as those observed in real microscopy image sequences. (in collaboration with F. Lavancier, ENSAI, Bruz)
L. Balsollier, F. Lavancier, J. Salamero, C. Kervrann. A generative model to synthetize spatio-temporal dynamics of biomolecules in cells, Biological Imaging, 3:e22, doi:10.1017/S2633903X2300020X (arXiv-doi:10.48550/arXiv.2303.06951), 2023.
Early prediction of the transferability of bovine embryos from video-microscopy
Participants: Yasmine Hachani, Patrick Bouthemy.
Video-microscopy is an attractive tool combined with machine learning for studying the early development of in vitro fertilized bovine embryos and assessing its transferability as soon as possible, i.e., its capacity to reach the blastocyst stage suitable for transfer to a cow uterus. We take 2D time-lapse microscopy videos as input and aim to predict transferability within four days at most. We formulate this problem as a supervised classification (transferable vs not transferable). The challenges are three-fold: 1) poorly discriminating appearance and motion, 2) class ambiguity, 3) small amount of annotated data. Indeed, the intra-class variability is high (trajectories of the embryo development may substantially vary within a given class). Conversely, the inter-class distance is short (observed development of two embryos from the two different classes may be fairly similar for the first days). Images are noisy, poorly contrasted, and subject to transparency effects. We propose a 3D convolutional neural network involving three pathways, which makes it multi-scale in time and able to handle appearance and motion in different ways, separately and in combination. For training, the focal loss was the best choice. Our model, named 3D-Fusion, compares favorably to other methods. First experiments demonstrate its efficiency and reliability for this challenging biological task, with an overall accuracy of 77% on a test set of about 200 embryo videos. (in collaboration with E. Fromont, Lacodam Team, IRISA, Rennes; A. De Paula Reis, UMR BREED, Ecole Nationale Vétérinaire d’Alfort)
Y. Hachani, P. Bouthemy, S. Ruffini, L. Laffont, I. Fromont, A. De Paula Reis. Early prediction of the transferability of bovine embryos from video-microscopy, 2023. (submitted paper)
N-BAR and F-BAR proteins - Endophilin-A3 and PSTPIP1 - control clathrin-independent endocytosis of L1CAM
Participants: Christian Wunder, Ludger Johannes.
Recent advances in the field demonstrate the high diversity and complexity of endocytic pathways. In the current study, we focus on the endocytosis of L1CAM. This glycoprotein plays a major role in the development of the nervous system, and is involved in cancer development and is associated with metastases and poor prognosis. Two L1CAM isoforms are subject to endocytosis: isoform 1, described as a clathrin-mediated cargo; isoform 2, whose endocytosis has never been studied. Deciphering the molecular machinery of isoform 2 internalisation should contribute to a better understanding of its pathophysiological role. First, we demonstrated in our cellular context that both isoforms of L1CAM are mainly a clathrin-independent cargo, which was not expected for isoform 1. Second, the mechanism of L1CAM endocytosis is specifically mediated by the N-BAR domain protein endophilin-A3. Third, we discovered PSTPIP1, an F-BAR domain protein, as a novel actor in this endocytic process. Finally, we identified galectins as endocytic partners and negative regulators of L1CAM endocytosis. In summary, the interplay of the BAR proteins endophilin-A3 and PSTPIP1, and galectins fine tune the clathrin-independent endocytosis of L1CAM. (in collaboration with H.-F. Renard, University of Namur, Department of Biology-Faculty of Sciences, Belgium)
C. Lemaigre, A. Ceuppens, C. A. Valades-Cruz, B. Ledoux, B. Vanbeneden, M. Hassan, F. R. Zetterberg, U. J. Nilsson, L. Johannes, C. Wunder, H-F. Renard, P. Morsomme. N-BAR and F-BAR proteins - Endophilin-A3 and PSTPIP1 - control clathrin-independent endocytosis of L1CAM, Traffic, doi:10.1111/tra.12883, 2023.
SLC3A2 N-glycosylation and Golgi remodeling regulate SLC7A amino acid exchangers and stress mitigation
Participants: Massiullah Shafaq-Zadah, Estelle Dransart, Ludger Johannes.
Proteostasis requires oxidative metabolism (ATP) and mitigation of the associated damage by glutathione, in an increasingly dysfunctional relationship with aging. SLC3A2 (4F2hc, CD98) plays a role as a disulfide-linked adaptor to the SLC7A5 and SLC7A11 exchangers which import essential amino acids and cystine while exporting Gln and Glu, respectively. The positions of N-glycosylation sites on SLC3A2 have evolved with the emergence of primates, presumably in synchrony with metabolism. Herein, we report that each of the four sites in SLC3A2 has distinct profiles of Golgi-modified N-glycans. N-glycans at the primate-derived site N381 stabilized SLC3A2 in the galectin-3 lattice against coated-pit endocytosis, while N365, the site nearest the membrane promoted glycolipid-galectin-3 (GL-Lect)-driven endocytosis. Our results indicate that surface retention and endocytosis are precisely balanced by the number, position, and remodeling of N-glycans on SLC3A2. Furthermore, proteomics and functional assays revealed an N-glycan-dependent clustering of the SLC3A2*SLC7A5 heterodimer with amino-acid/Na symporters (SLC1A4, SLC1A5) that balances branched-chain amino acids and Gln levels, at the expense of ATP to maintain the Na/K gradient. In replete conditions, SLC3A2 interactions require Golgi-modified N-glycans at N365D and N381D, whereas reducing N-glycosylation in the endoplasmic reticulum by fluvastatin treatment promoted the recruitment of CD44 and transporters needed to mitigate stress. Thus, SLC3A2 N-glycosylation and Golgi remodeling of the N-glycans have distinct roles in amino acids import for growth, maintenance, and metabolic stresses. (in colloration with J.W. Dennis, Samuel Lunenfeld Research Institute, Toronto, Canada; H. Clausen, University of Copenhagen, Department of Cellular and Molecular Medicine, Denmark)
C. Zhang, M. Shafaq-Zadah, J. Pawling, G.G. Hesketh, E. Dransart, K. Pacholczyk, J. Longo, A.-C. Gingras, L.Z. Penn, L. Johannes, J.W. Dennis. SLC3A2 N-glycosylation and Golgi remodeling regulate SLC7A amino acid exchangers and stress mitigation, J. Biological Chemistry 299(12), 105416, doi:10.1016/j.jbc.2023.105416, 2023.
A synthetic delivery vector for mucosal vaccination
Participants: Ludger Johannes.
The success of mRNA-based vaccines during the Covid-19 pandemic has highlighted the value of this new platform for vaccine development against infectious disease. However, the CD8 T cell response remains modest with mRNA vaccines, and these do not induce mucosal immunity, which would be needed to prevent viral spread in the healthy population. To address this drawback, we developed a dendritic cell targeting mucosal vaccination vector, the homopentameric STxB. Here, we describe the highly efficient chemical synthesis of the protein, and its in vitro folding. This straightforward preparation led to a synthetic delivery tool whose biophysical and intracellular trafficking characteristics were largely indistinguishable from recombinant STxB. The chemical approach allowed for the generation of new variants with bioorthogonal handles. Selected variants were chemically coupled to several types of antigens derived from the mucosal viruses SARS-CoV-2 and type 16 human papillomavirus. Upon intranasal administration in mice, mucosal immunity, including resident memory CD8 T cells and IgA antibodies was induced against these antigens. Our study thereby identifies a novel synthetic antigen delivery tool for mucosal vaccination with an unmatched potential to respond to an urgent medical need. (in colloration with E. Tartour, PARCC, INSERM, Université Paris Cité, Department of Immunology, Hôpital Européen Georges-Pompidou, AP-HP)
A. Billet, J. Hadjerci, T. Tran, P. Kessler, J. Ulmer, G. Mourier, M. Ghazarian, A. Gonzalez, R. Thai, P. Urquia, A.-C. Van Baelen, A. Meola, I. Fernandez, S. Deville-Foillard, E. MacDonald, L. Paolini, F. Schmidt, F.A. Rey, M.S. Kay, E. Tartour, D. Servent, L. Johannes. A synthetic delivery vector for mucosal vaccination, Biomaterials 302, 122298, doi:10.1016/j.biomaterials.2023.122298, 2023.
Engineered synthetic STxB for enhanced cytosolic delivery
Participants: Christian Wunder, Ludger Johannes.
Many molecular targets for cancer therapy are located in the cytosol. Therapeutic macromolecules are generally not able to spontaneously translocate across membranes to reach these cytosolic targets. Therefore a strong need exists for tools that enhance cytosolic delivery. Shiga toxin B-subunit (STxB) is used to deliver therapeutic principles to disease-relevant cells that express its receptor, the glycolipid Gb3. Based on its naturally existing membrane translocation capacity, STxB delivers antigens to the cytosol of Gb3-positive dendritic cells, leading to the induction of CD8 T cells. Here, we have explored the possibility of further increasing the membrane translocation of STxB to enable other therapeutic applications. For this, our capacity to synthesize STxB chemically was exploited to introduce unnatural amino acids at different positions of the protein. These were then functionalized with hydrophobic entities to locally destabilize endosomal membranes. Intracellular trafficking of these functionalized STxB was measured by confocal microscopy and their cytosolic arrival with a recently developed highly robust, sensitive, and quantitative translocation assay. From different types of hydrophobic moieties that were linked to STxB, the most efficient configuration was determined. STxB translocation was increased by a factor of 2.5, paving the path for new biomedical opportunities. (in colloration with E. Tartour, PARCC, INSERM, Université Paris Cité, Department of Immunology, Hôpital Européen Georges-Pompidou, AP-HP)
J. Hadjerci, A. Billet, P. Kessler, G. Mourier, M. Ghazarian, A. Gonzalez, C. Wunder, N. Mabrouk, E. Tartour, D. Servent, L. Johannes. Engineered synthetic STxB for enhanced cytosolic delivery, Cells, 12(9), 1291, doi:10.3390/cells12091291, 2023.
Spatial N-glycan rearrangement on α5β1integrin nucleates galectin-3 oligomers to determine endocytic fate
Participants: Massiullah Shafaq-Zadah, Estelle Dransart, Christian Wunder, Ludger Johannes.
Membrane glycoproteins frequently adopt different conformations when altering between active and inactive states. Here, we discover a molecular switch that exploits dynamic spatial rearrangements of N-glycans during such conformational transitions to control protein function. For the conformationally switchable cell adhesion glycoprotein α 5 β 1 integrin, we find that only the bent-closed state arranges N-glycans to nucleate the formation of up to tetrameric oligomers of the glycan-binding protein galectin-3. We propose a structural model of how these galectin-3 oligomers are assembled and how they clamp the bent-closed state to prime it for endocytic uptake and subsequent retrograde trafficking to the Golgi for polarized distribution in cells. Our findings highlight an unexpectedly dynamic regulation of the glycan landscape at the cell surface to achieve oligomerization of galectin-3. Galectin-3 oligomers are thereby identified as decoders of defined spatial patterns of N-glycans and as functional extracellular interactors of specifically the bent-closed conformational state of α 5 β 1 integrin and possibly other family members. (in collaboration with H. Leffler, Lund University, Division of Microbiology, Immunology and Glycobiology, Sweden; D. Roderer and S. Raunser, Leibniz-Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany)
M. Shafaq-Zadah, E. Dransart, C. Wunder, V. Chambon, C.A. Valades-Cruz, L. Leconte, N.K. Sarangi, J. Robinson, S. Bai, R. Regmi, A.D. Cicco, A. Hovasse, R. Bartels, U.J. Nilsson, S. Cianférani-Sanglier, H. Leffler, T.E. Keyes, D. Lévy, S. Raunser, D. Roderer, L. Johannes. N-glycan rearrangement on α5β1integrin nucleates galectin-3 oligomers to determine endocytic fate, bioRxiv-doi:10.1101/2023.10.27.564026 2023.
Growth factor-induced desialylation for the fast control of endocytosis
Participants: Massiullah Shafaq-Zadah, Estelle Dransart, Christian Wunder, Ludger Johannes.
It is commonly assumed that the glycan makeup of glycoproteins that reach the cell surface is final and static. Here, we challenge this notion by the discovery of a molecular switch that induces acute and reversible changes of glycans on the plasma membrane. We demonstrate that within minutes, the epidermal growth factor triggers the galectin-driven endocytosis of cell surface glycoproteins, such as integrins, that are key regulators of cell adhesion and migration. The onset of this process, mediated by the Na+/H antiporter NHE-1 and the neuraminidases Neu1/3, requires the pH-triggered enzymatic removal of sialic acids whose presence otherwise prevents galectin binding. Desialylated glycoproteins are then retrogradely transported to the Golgi apparatus where their glycan makeup is reset, and their function is repurposed to regulate EGF-dependent invasive cell migration. Glycosylation at the cell surface thereby emerges as a dynamic and reversible regulatory post-translational modification that controls a highly adaptable trafficking pathway. (in collaboration with R. Weigert, CI-NIH Bethesda, USA; H. Clausen, University of Copenhagen, Department of Cellular and Molecular Medicine, Denmark; S. Mayor, National Centre for Biological Sciences, Bangalore, India; H. Leffler, Lund University, Division of Microbiology, Immunology and Glycobiology, Sweden)
E. MacDonald, A. Forrester, C.A. Valades-Cruz, T.D. Madsen, J. Hetmanski, E. Dransart, Y. Ng, R. Godbole, A. Akhil Shp, L. Leconte, V. Chambon, D. Ghosh, A. Pinet, D.D. Bhatia, B. Lombard, D. Loew, M.R. Larsen, H. Leffler, D. Lefeber, H. Clausen, P.T. Caswell, M. Shafaq-Zadah, S. Mayor, R. Weigert, C. Wunder, L. Johannes. Growth factor-induced desialylation for the fast control of endocytosis, bioRxiv-doi:10.1101/2023.09.12.557183, 2023.
Endocytic roles of glycans on proteins and lipids
Participants: Massiullah Shafaq-Zadah, Estelle Dransart, Christian Wunder, Ludger Johannes.
Most cell surface proteins are decorated by glycans, and the plasma membrane is rich in glycosylated lipids. The mechanisms by which the enormous complexity of these glycan structures on proteins and lipids is exploited to control glycoprotein activity by setting their cell surface residence time and the ways by which they are taken up into cells are still under active investigation. Here, two mechanisms are presented, termed galectin lattices and glycolipid-lectin (GL-Lect)-driven endocytosis, which are among the most prominent to establish a link between glycan information and endocytosis. Types of glycans on glycoproteins and glycolipids are reviewed from the angle of their interaction with glycan-binding proteins that are at the heart of galectin lattices and GL-Lect-driven endocytosis. Examples are given to show how these mechanisms affect cellular functions ranging from cell migration and signaling to vascularization and immune modulation. Finally, outstanding challenges on the link between glycosylation and endocytosis are discussed.(in collaboration with H. Leffler, Lund University, Division of Microbiology, Immunology and Glycobiology, Sweden)
L. Johannes, M. Shafaq-Zadah, E. Dransart, C. Wunder, H. Leffler. Endocytic roles of glycans on proteins and lipids, Cold Spring Harb Perspect Biol., 16(1):a041398, doi:10.1101/cshperspect.a041398, 2024.
9 Bilateral contracts and grants with industry
9.1 Bilateral Grants with Industry
9.1.1 Contract with IRSN: DeepSuN – Localization of chromosomal aberrations induced by nuclear radiation dose excess
Participants: Antonin Deschemps, Charles Kervrann.
Funding: IRSN (Institut de Radioprotection et de Sureté Nucléaire) and Région-Bretagne
Duration: 36 months (Oct 2020 – Sep 2023)
Collaborator: M. Benadjaoud (IRSN/SERAMED, Fontenay-aux-Roses)
The goal of this project is to develop statistical and deep-learning methods for localizing and classifying chromosomal aberrations observed in 2D microscopy images (blood test) and estimating radiation dose following a postulated nuclear reactor accident.
This project funded by the IRSN (Institut de Radioprotection et de Sureté Nucléaire) and Région-Bretagne concerns the PhD thesis (co-funding) carried out by Antonin Deschemps. (see also ANR INCREASED)
9.1.2 Contract with AID: Localization and classification of gene translocation in FISH microscopy images
Participants: Quentin Tallon, Charles Kervrann.
Funding: AID (Agence de l'Innovation de Defense)
Duration: 36 months (Oct 2021 – Sep 2024)
Collaborator: M. Benadjaoud (IRSN/SERAMED, Fontenay-aux-Roses)
The goal of this project is to develop supervised and unsupervised machine learning methods and algorithms for detection and classification of gene translocation between chromosomes observed in FISH (Fluorescence in situ hybridization) microscopy images.
his project funded by the AID (Ministry of Defense) concerns the PhD thesis carried out by Quentin Tallon. (see also ANR INCREASED)
9.1.3 Contract with AIRBUS Defense and Space SAS: LION Chaine Image Elargie (LiChIE)
Participants: Sébastien Herbreteau, Etienne Meunier, Emmanuel Moebel, Sarra Khairi, Maelys Risset, Manuel Sanchez Laguardia, Patrick Bouthemy, Charles Kervrann.
Funding: Bpifrance / Projets Structurants pour la Compétitivité (PSPC)
Duration: 65 months (Aug 2019 – Dec 2024)
Collaborators: M. Ortner and R. Fraisse (AIRBUS Defense and Space SAS)
The goal of this large-scale project is to develop statistical and deep-learning methods for image restoration in night conditions and motion saliency detection in image sequences, respectively. The resulting algorithms will be embedded in hardware platforms for a next generation of observation satellites.
This project funded by Bpifrance concerns the PhD theses carried out by Sébastien Herbreteau and Etienne Meunier, the engineer position of Emmanuel Moebel (2022-2023), and the internships of M. Sanchez Laguardia, S. Khairy, and M. Risset.
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Participation in other International Programs
Informal international partners
Participants: Anaïs Badoual, Charles Kervrann, Ludger Johannes, Estelle Dransart, Christian Wunder, Massiullah Shafaq-Zadah.
-
–
Collaboration with EPFL (D. Sage), Biomedical Imaging Group, Lausanne, Switzerland: Dissemination and teaching of deep-learning for bioimage analysis, preparation of a workshop for Mifobio 2023 (with D. Sage). (with A. Badoual)
-
–
Collaboration with the Cambridge Advanced Imaging Centre (L. Muresan), Cambridge, UK: Analysis of transcription factors in single molecule localization microscopy; Segmentation algorithms for quantitative analysis of 3D+time live-cell images. (with C. Kervrann and A. Badoual)
-
–
Collaboration with Kyoto University Graduate School of Medicine (M. Arizono), Kyoto, Japan: analysis of astrocytic calcium activity. (with A. Badoual)
-
–
Collaboration with the PKU University (T. Liu, Z. Chen), Institute of Molecular Medicine, Beijing, People Republic of China: Development of robust mitochondria fluorescent probes and validation in LLSM, STED and LiveSR microscopy for Live cells. (with C. Kervrann, A. Badoual)
-
–
Collaboration with NCI-NIH Bethesda (R. Weigert), USA: EGF-induced desialylation for the fast control of endocytosis. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
-
–
Collaboration with Samuel Lunenfeld Research Institute (J.W. Dennis), Toronto, Canada: SLC3A2 N-glycosylation and Golgi remodeling regulate SLC7A amino acid exchangers and stress mitigation. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
-
–
Collaboration with University of Copenhagen, Department of Cellular and Molecular Medicine (H. Clausen), Denmark: EGF-induced desialylation for the fast control of endocytosis; EGF-induced desialylation for the fast control of endocytosis.
-
–
Collaboration with National Centre for Biological Sciences (S. Mayor), Bangalore, India: EGF-induced desialylation for the fast control of endocytosis. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
-
–
Collaboration with Lund University, Division of Microbiology, Immunology and Glycobiology (H. Leffler), Sweden: Spatial N-glycan rearrangement on α5β1integrin nucleates galectin-3 oligomers to determine endocytic fate; Endocytic roles of glycans on proteins and lipids; EGF-induced desialylation for the fast control of endocytosis. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
-
–
Collaboration with Leibniz-Forschungsinstitut für Molekulare Pharmakologie (D. Roderer, S. Raunser), Berlin, Germany: Spatial N-glycan rearrangement on α5β1integrin nucleates galectin-3 oligomers to determine endocytic fate. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
-
–
Collaboration with University of Namur, Department of Biology-Faculty of Sciences (H.-F. Renard), Belgium: N-BAR and F-BAR proteins - endophilin-A3 and PSTPIP1 - control the clathrin-independent endocytosis of L1CAM. (with L. Johannes, M. Shafaq-Zadah, C. Wunder, E. Dransart)
10.2 International research visitors
10.2.1 Visits to international teams
Research stays abroad
-
–
Quentin Rapilly visited for 3 months (Sep-Nov 2023) the Cambridge Advanced Imaging Centre (in collaboration with Leila Muresan), Cambridge, UK.
-
–
Ilyes Hamitouche visited for 3 months (Jul-Oct 2023) the Leibniz-Forschungsinstitut für Molekulare Pharmakologie (in collaboration with Daniel Roderer), Berlin, Germany.
10.3 European initiatives
10.3.1 Other european programs/initiatives
ESFRI initiative program: EuroBioImaging
Participants: Charles Kervrann, Arthur Masson.
Coordinator: J. Eriksson (Turku University, Finland)
Funding: Member states of the European Union
Partners: 18 European countries in 2022 (+1 observer)
As a member of the National Research Infrastructures (RI) France BioImaging, SAIRPICO is involved in the ESFRI Euro-BioImaging project, and now in the ERIC EuroBioImaging (since November 2019), one of the landmarks of biomedical science Research Infrastructures in the roadmap of the European Strategic Forum on Research Infrastructures (ESFRI 2018). The mission of Euro-BioImaging is to provide access, service and training to state-of-the-art imaging technologies and foster the cooperation and networking at the European level including multidisciplinary scientists, industry, regional, national and European authorities.
10.4 National initiatives
10.4.1 France-BioImaging project
Participants: Charles Kervrann, Arthur Masson, Kevin Fournier.
Duration: 2011 – 2024
Funding: Investissement d'Avenir, ANR INBS-PIA1 2011 and “FBI Next Generation” (ANR program 2020-2024)
Coordinator: E. Bertrand (UMR9002 CNRS)
Partners: CNRS, Aix-Marseille Université, Collège de France, Ecole Normale Supérieure, Ecole Polytechnique, Inria, Institut Curie, Institut Pasteur, INSERM, Université de Bordeaux, Université de Montpellier, Université de Nantes, Université de Paris, Université de Rennes 1.
SERPICO-SAIRPICO is a member of the French initiative, the so-called “France-BioImaging” (FBI) National Research Infrastructure which gathers several outstanding cellular imaging centers (microscopy, spectroscopy, probe engineering and signal processing). FBI is on the French Roadmap of Research Infrastructure. The mission of FBI is to build a distributed coordinated French infrastructure for photonic and electronic cellular bioimaging, dedicated to innovation, training and technology transfer. High-computing capacities are needed to exhaustively analyze image flows.
SAIRPICO is co-head of the IPDM (Image Processing and Data Management) node of the FBI network composed of 7 nodes. In this context, we address the following scientific problems: i/ exhaustive analysis of bioimaging data sets; ii/ deciphering of key steps of biological mechanisms at organ, tissular, cellular and molecular levels through the systematic use of time-lapse 3D microscopy and image processing methods; iii/ storage and indexing of extracted and associated data and metadata through an intelligent data management system. SERPICO-SAIRPICO recruited R&D engineers to disseminate image processing software, to build the Mobyle@serpico web portal and to manage the IGRIDA-SERPICO cluster opened for end-users and dedicated to large scale computing and data sets processing.
Two projects involving the SERPICO-SAIRPICO Team were selected to FBI open calls in 2021 and 2022:
-
–
"BioImageIT Dissemination" project, 18 months (2021-2023), coordinated by J. Salamero, with the participation of S. Prigent C.-A. Valades-Cruz, L. Leconte, and C.Kervrann. This project concerns the engineer position of L. Maury.
-
–
POLARIMAGING project, 18 months (2022-2024), coordinated by S. Brasselet (Institut Fresnel, Marseille), with the participation of J. Salamero, C.-A. Valades-Cruz, L. Leconte, and C.Kervrann.
This project concerned the 6-month engineer position of Kevin Fournier in 2023.
10.4.2 ANR INCREASED project: Artificial intelligence for the detection of chromosomal aberrations in dosimetry
Participants: Antonin Deschemps, Quentin Tallon, Emmanuel Moebel, Charles Kervrann.
Duration: 36 months (Oct 2020 – Sept 2023)
Funding: ANR (Agence Nationale de la Recherche) ASTRID
Coordinator: Gaetan Gruel (Institut de Radioprotection et de Sureté Nucléaire (IRSN/LRAcc), Fontenay-aux-Roses)
Partners: IRSN/LRAcc and IRSN/SERAMED, SAIRPICO Team, IRBA (Institut de Recherche Biomédicale des Armées)
During insidious scenarios, when the assessment of ionizing radiation exposure condition is difficult or impossible, the quantification, on samples taken from the victims, of radio-induced chemical or biological effects is more suitable for an individualized dose reconstruction compared to theoretical calculation or Monte-Carlo simulations in regard to their great uncertainties. The INCREASED project aims to adapt the most powerful algorithms of modern artificial intelligence to the context of automatic detection of chromosomal aberrations in dosimetry. This project proposes to revisit the semi-automatic or automatic detection methods currently used for the recognition of dicentrics in GIEMSA imagery in the light of the most recent advances in artificial intelligence and deep-learning. These modern methods, which demonstrated their indisputable superiority in other areas of computer vision, will be deployed on GIEMSA images for an exhaustive multi-class count not only of dicentric chromosomes but also of ring-centric aberrations, acentric fragments, and even tricentrics. Furthermore, the INCREASED initiative is also quite innovative in dosimetry based on FISH imaging. To date, this type of dosimetric reconstruction is based on manual counting of non-exhaustive and very simplified annotations of the different possible forms of translocations. This protocol is therefore as heavy as imprecise. The INCREASED project addresses two major improvements: i/ design of a universal, rigorous and exhaustive scoring of the different forms of observable FISH translocations (3 colors). ii/ development of modern artificial intelligence algorithms able to detect and translate co-locations/co-neighborhoods into different categories of translocations.
This project concerned the PhDs of Quentin Tallon and Antonin Deschemps in 2023.
10.4.3 ANR POLARISCOPIA project: Next generation information processing of microscopy vector-valued images : application in cell polarized imaging
Participants: Charles Kervrann, Vincent Briane.
Duration: 48 months (Oct 2022 – Sept 2026)
Funding: ANR (Agence Nationale de la Recherche) PRME
Coordinator: Charles Kervrann
Collaborators: U1143/UMR3666 (L. Johannes) and UMR168 (B. Hajj)
The objective of the project is to create the next generation of information processing techniques required to overcome the three aforementioned barriers, and to solve challenging image processing problems induced by the acquisition of 3D+Time vector-valued images. This will be achieved here by integrating concepts in statistical signal-image processing and machine learning, combined with innovative developments in fluorescence microscopy. The resulting algorithms will serve to characterize the dynamics of biomolecules and to decipher the molecular transport pathways, which are of considerable of interest in fundamental cell biology and for future precision medicine.
This project concerned the postdoc position of Vincent Briane in 2023.
10.4.4 ANR DEEPNER project: Deciphering chromatin rearrangements in response to UV irradiation using new deep learning based cryo-electron tomography data analysis tools
Participants: Charles Kervrann.
Duration: 48 months (Oct 2023 – Sept 2027)
Funding: ANR (Agence Nationale de la Recherche) PRC
Coordinator: Mikhail Eltsov (IGBMC, Strasboug)
Partners: Sorbonne University (IMPMC), Inria Rennes (SAIRPICO Team)
The goal is to reveal, with unprecedented resolution, chromatin reorganization during genotoxic stress. We will analyse the following three structural levels of the chromatin: (1) spatial organization of chromatin domains (nucleosome distribution, density, local order); (2) geometry of DNA linkers and (3) conformation and disassembly of nucleosomes. This analysis will reveal the chromatin structure-based mechanisms enabling detection and repair of UV-induced lesions in the chromatin context. To enable this analysis, we will develop appropriate DL approaches and software, in particular those for in situ cryo-ET data annotation and analysis. The added value of this research will be cryo-ET data analysis methods and algorithms embedded in software enabling not only an automated in situ annotation and analysis of nucleosomes but also other molecular complexes, in health and disease.
10.4.5 Labex Cell(n)Scale PoMIMO – Polarization Microscopy for Imaging of Membrane Organization
Participants: Charles Kervrann.
Duration: 36 months (2022 – 2024).
Funding: Labex Cell(n)Scale
Coordinator: J. Salamero.
Partners: U1143/UMR3666 (L. Johannes) and UMR168 (P. Bassereau), Institut Curie, and GATACA Systems company.
The main objective is to study the molecular organization of cargo proteins and membrane lipids by adding information on their dipole orientations. To do so, we develop innovative polarization modalities adapted to available microscopic setups and appropriate computational methods. We plan to use chemical engineering of lipid and protein-based fluorescent labels to make them rigid for polarization imaging. We aim to create a new imaging approach that will not only provide novel information about endocytosis, but will have the potential to be applied to all topics where the spatial and molecular organization of lipids/proteins defines both the structure and function of these assemblies (i.e., mitochondria cristae, MAMs....) in reconstituted systems and in living cells.
10.4.6 CNRS Project 80|Prime ModSpatioTempCell: Spatiotemporal stochastic models for the comprehension of membrane trafficking at the single cell scale
Participants: Lisa Balsolier, Frédéric Lavancier, Jean Salamero, Charles Kervrann.
Duration: 36 months (Oct 2021 – March 2023).
Funding: CNRS
Coordinator: F. Lavancier.
Partners: SAIRPICO Team, UMR144 Institut Curie, Laboratoire de Mathématiques Jean-Leray Nantes, UMR6629.
Endocytosis and exocytosis mechanisms involve multiple molecular components that interact through space and time according to a complex process. In this project, we plan to analyze the spatiotemporal activity of some of these proteins thanks to a tailored mathematical model, fitted to sequences of real data acquired by fluorescence microscopy. The mathematical model at hand characterizes the dynamics of a system of interacting particles with possible appearance and disappearance of some particles through time, in accordance with the observed dynamics of proteins in cell membrane trafficking.
This project concerns the PhD thesis carried out by Lisa Balsollier (since October 2021).
10.4.7 INRAE-Inria call - Early prediction of the transferability of bovine embryos from video-microscopy
Participant: Patrick Bouthemy.
Duration: 36 months (2023 – 2025).
Funding: INRIA
Coordinator: E. Fromont (Lacodam team, IRISA, Rennes)
Partners: UMR BREED (Ecole Nationale Vétérinaire d’Alfort), Lacodam Team
This project concerns with the development of deep learning methods for the supervised comparison and classification of videos of in vitro fertilized (IVF) bovine embryos. The biological objective is to improve our knowledge of early phenotype formation, with ultimate applications in cattle breeding, where the challenge is to make early decision on embryo transferability. The research will also encompass the recognition of specific events during embryonic development as cell division. Challenging issues are diverse. The videomicroscopy data have specific and complex characteristics, the volume of available annotated data is limited, and the targeted classification is not that obvious.
This project concerns the PhD thesis carried out by Yasmine Hachani (since October 2023).
10.4.8 INRAE-Inria call - MreB: Role of membrane microdomains in bacterial morphogenesis
Participant: Charles Kervrann.
Duration: 36 months (Oct 2020 – Sep 2023)
Funding: INRAE
Coordinator: R. Carballido-Lopez (INRAE UMR MICALIS, Jouy-en-Josas)
Partners: INRAE UMR MICALIS (Jouy-en-Josas), SAIRPICO Team
This project concerns the PhD thesis carried out by Claire-Jing Rouchet (since October 2020 in the INRAE UMR MICALIS, Jouy-en-Josas). The goal is to understand how dynamic molecular interactions are regulated in time and in space to form functional machineries that establish long-range orders and cellular functions. To this end we combine cutting-edge high-resolution fluorescence microscopy and spectroscopy techniques with powerful genetic, biophysical, biochemical and systems biology approaches available in model bacteria, in particular the rod-shaped Gram-positive bacterium Bacillus subtilis, to determine mechanistic details underlying basic cellular and developmental processes: the molecules, macromolecular assemblies, cellular events and physiological adaptations involved. Understanding how bacterial cells grow has important implications for the identification of new antimicrobial targets and for the development of synthetic biology tools for bioproduction and biotechnology purposes.
10.5 Regional initiatives
10.5.1 Deep-SuN: Localization of chromosomal aberrations induced by nuclear radiation dose excess
Participants: Antonin Deschemps, Charles Kervrann.
Funding: IRSN (Institut de Radioprotection et Sureté Nucléaire), Région-Bretagne
Duration: 36 months (Oct 2020 – Sep 2023)
This project concerns the PhD thesis carried out by Antonin Deschemps, in collaboration with IRSN/SERAMED (see DeepSun projet - Section 9.1.1).
11 Dissemination
11.1 Promoting scientific activities
11.1.1 Scientific events: selection
Participants: Charles Kervrann, Patrick Bouthemy.
Member of the conference program committees
-
–
Charles Kervrann was member of the scientific committee of IABM'2023 (Colloque Français d'Intelligence Artificielle en Imagerie Biomédicale).
-
–
Patrick Bouthemy was Associate Editor for the Intl ISBI'2023 conference and member of the scientific committee of IABM'2023 (Colloque Français d'Intelligence Artificielle en Imagerie Biomédicale).
11.1.2 Journal
Participants: Charles Kervrann, Ludger Johannes, Emmanuel Moebel, Sébastien Herbreteau, Etienne Meunier.
Member of the editorial boards
-
–
Charles Kervrann is Associate Editor for the "Transactions on Image Processing" and "Biological Imaging" journals since 2021. He was Guest Associate Editor for a Special Issue of "Journal of Selected Topics in Quantum Electronics" (published in 2023).
-
–
Ludger Johannes is member of the Editorial Board of the "Biology of the Cell" journal (since 2010) and Academic Editor for the "PLoS ONE" journal (since 2013).
Reviewer - reviewing activities
-
–
Charles Kervrann was reviewer for "Nature Methods" in 2023.
-
–
Sébastien Herbreteau was reviewer for "IEEE Transactions on Image Processing" in 2023.
-
–
Étienne Meunier was reviewer for "IEEE Transactions on Image Processing" in 2023, and for the international conferences ISBI'2023 and CVPR'2024.
-
–
E. Moebel was reviewer for "Nature Communications" in 2023.
11.1.3 Invited talks
Participants: Charles Kervrann, Ludger Johannes, Arthur Masson, Sébastien Herbreteau, Lisa Balsollier, Quentin Rapilly, Antonin Deschemps.
-
–
Charles Kervrann:
-
>"Statistical and artificial intelligence methods for live-cell fluorescence imaging", Anniversaire 20 ans du GdR ImaBio, Paris, France, July.
-
>"Finding is believing: AI reveals the presence of macromolecules in 3D cellular cryogenic electron tomograms", Colloque Français d’Intelligence Artificielle en Imagerie Biomédicale (IABM), Paris, France, March.
-
>"Sparse denoising and adaptive estimation enhances the resolution and contrast of fluorescence emission difference microscopy based on array detector", Journée GdR MIA-ISIS-Ondes "Co-conception : capteurs hybrides et algorithmes pour des systèmes innovants", Paris, France, November.
-
>"NAVISCOPE: image-guided navigation and visualization of large data sets in live cell imaging and microscopy", Sci-Rennes, Inria Rennes, March. (seminar)
-
>"Statistical and artificial intelligence methods for live-cell fluorescence imaging", MUSCA Team, Inria Paris-Saclay, France, April. (seminar)
-
>"Statistical and artificial intelligence methods for live-cell fluorescence imaging", INSERM-U1143,"Endocytic Trafficking and Intracellular Delivery" Team, Institut Curie, Paris, France, October. (seminar)
-
>
-
–
Ludger Johannes:
-
>"Growth factor induced desialylation for the fast control of endo"ytosis", Gordon Conf. Molecular Membrane Biology, Andover, USA, July.
-
>"Desialylation glycoswitch to acutely control endocytosis", EMBO keynote lecture at 26th Intl. Symposium on Glycoconjugates (Glyco26), Taipei, Taiwan, August.
-
>"Conformational glycoswitch to drive the formation of galectin-3 oligomer", EMBO keynote lecture at Galectin Satellite Symp. of the Glyco26, Taipei, Taiwan, August.
-
>"Mechanisms of acute regulation of lycoLipid-Lectin (GL-Lect) endocytosis", 7th EMBO Endocytosis meeting, San Servolo Island, Venice, Italy, September.
-
>"Glycoswitches for the control of endocytosis", Leibniz-Institute for Molecular Pharmacology (FMP), Berlin, Germany, October. (seminar)
-
>"Glycoswitches for the control of endocytosis", Indian Institute of Science Education and Research, Pune, India, December. (seminar)
-
>"Glycoswitches for the control of endocytosis", National Centre for Cell Science, Pune, India), December. (seminar)
-
>
-
–
Arthur Masson:
-
>"BioImageIT: a solution for data and image management and analysis", 12e Journées Scientifiques et Techniques du RµI - INRAE, Montpellier, France, November.
-
>
-
–
Sébastien Herbreteau
-
>"Towards better conditioned and interpretable neural networks: a study of the normalization-equivariance property with application to image denoising", TU Darmstadt, Germany, September. (seminar)
-
>"Towards better conditioned and interpretable neural networks: a study of the normalization-equivariance property with application to image denoising", EPFL-BIG, Lausanne, Switzerland, October. (seminar)
-
>
-
–
Antonin Deschemps:
-
>"Ensembling Unets: a deep-learning approach for automated chromosomal aberration detection in cytogenetic imaging", Intl. Congress of Radiation Research (ICRR), Montreal, QC, Canada, August.
-
>
-
–
Lisa Balsollier:
-
>"A generative model to simulate spatio-temporal dynamics of biomolecules in cells", Intl. Workshop Spatial Analysis for Biological Imaging, Imperial College London, UK, March.
-
>
-
–
Quentin Rapilly:
-
>"Hybrid methods : CNN-snakes, for cell segmentation on 3D+time bio images", University of Cambridge, UK, October. (seminar)
-
>
11.1.4 Leadership within the scientific community
Participants: Charles Kervrann, Ludger Johannes.
-
–
Charles Kervrann is co-head of the "BioImage Informatics" node (ANR France-BioImaging project, National Research Infrastructure" for Biology and Health) since 2011. He is member of the IEEE Signal Processing Society (since 2010).
-
–
Ludger Johannes is member of the American Society for Cell Biology (since 1996), Société de Biologie Cellulaire de France (since 1996), French Society for Biophysics (SFB) and Membrane Study Group (GEM) (since 2012), Société de Chimie Thérapeutique (SCT) (since 2014), Biophysical Society (since 2015), Société Française pour l'Etude des Toxines (SFET) (since 2021), Société Française du Cancer (SFC) (since 2022), Société Chimique de France (SCF) (since 2023).
11.1.5 Scientific expertise
Participants: Charles Kervrann, Ludger Johannes.
-
–
Charles Kervrann was member of the selection committee for the recruitment of a Professor (University of Caen Basse Normandie, CNU 27) et Junior Professor Chair (University of Montpellier, CNU 26)
-
–
Ludger Johannes is member of the scientific council of the Indo-French Centre for the Promotion of Advanced Research (IFCPAR/CEFIPRA (since 2021) and member of the permanent SVE3 HCERES expert panel (independent agency for the reviewing of research structures in France) (since 2021). He was member of the HCERES panel at Institut Jacques Monod, December. He is member of the Scientific Council of the "Membrane Study Group"since 2021, member of the scientific council of the Chemical Biology Interest Group (GDR Chémobiologie) since 2021, member of the scientific committee of the Cellular and Tissular Imaging Platform (PICT) at CurieCoreTech of Institut Curie since 2021, and member of the scientific council of Doctoral School BIOSIGNE (ED568) since 2014.
11.1.6 Research administration
Participants: Charles Kervrann, Patrick Bouthemy, Ludger Johannes.
-
–
Charles Kervrann:
-
>Member of the executive board of the project committee "Bureau du Comité des Projets" of the Inria Rennes - Bretagne Atlantique centre since 2010.
-
>Co-head of the "BioImage Informatics" node (ANR France-BioImaging project, National Research Infrastructure" for Biology and Health) since 2011.
-
>
-
–
Patrick Bouthemy:
-
>Member of the executive board of Excellence Lab (Labex) CominLabs.
-
>Member of the Selection and Validation Committee of the Images & Réseaux competitivity cluster.
-
>Head of the four evaluation committees of the ANR call for proposals on Specific Topics in Artificial Intelligence (Thématiques Spécifiques en Intelligence Artificielle – TSIA).
-
>Member of the steering committee of the medical imaging Neurinfo platform.
-
>
-
–
Ludger Johannes:
-
>Head of Traffic, Signaling and Delivery team at Curie Institute since 2001.
-
>Director of Cellular and Chemical Biology unit (U1143 INSERM, UMR3666 CNRS) since 2014.
-
>Scientific director of the Metabolomics and Lipidomics Platform at CurieCoreTech of Institut Curie since 2021.
-
>Representative of Doctoral School BIOSIGNE (ED568) in the Doctoral College of PSL University since 2016.
-
>Institut Curie coordinator of institutional partnership between Institut Curie, CNRS and the National Centre for Biological Sciences (NCBS) à Bangalore (India) since 2012.
-
>
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
Participants: Charles Kervrann, Anaïs Badoual, Ludger Johannes, Sébastien Herbreteau, Etienne Meunier, Lisa Balsollier, Quentin Rapilly.
-
–
Charles Kervrann:
-
>Master 2: "From Bioimage Processing to BioImage Informatics", 3 hours (4.5 hours TD), coordinator of the module (30 hours / equiv 45 hours TD), Master 2 Research IRIV, Telecom-Physique Strasbourg and University of Strasbourg.
-
>Engineer Degree (3rd year) and Master 2 Statistics and Mathematics: Statistical Models and Image Analysis", 27.5 hours (41.25 hours TD), Ecole Nationale de la Statistique et de l'Analyse de l'Information (ENSAI), Bruz.
-
>
-
–
Ludger Johannes
-
>Master 2: "Endocytic trafficking" for "Chemical Frontiers of Living Cells", 2 hours, PSL Research University.
-
>
-
–
Anaïs Badoual:
-
>Engineer Degree (3rd year): "Statistical Models and Image Analysis", 15 hours (TP / Practical courses), Ecole Nationale de la Statistique et de l'Analyse de l'Information (ENSAI), Bruz.
-
>Master degree: "Analysis of Image Sequences", 8 hours (12 hours TD) and 6 hours TD, Master 2 Research SISEA, ISTIC & University of Rennes 1.
-
>Master degree: "Object Tracking in microscopy", 1.75 hours (3.5 hours TD), Master 2 Research IRIV, Telecom-Physique Strasbourg.
-
>
-
–
Sébastien Herbreteau:
-
>Master 2 : Advanced statistical learning: reinforcement learning and model-based clustering (14 hours TD). University of Nantes.
-
>
-
–
Etienne Meunier:
-
>Master degree: "Machine Learning", 22 hours TP, EFREI, Paris.
-
>
-
–
Lisa Balsollier:
-
>Licence (L1) degree : "Mathematics for biology", 30 hours TD, University of Rennes 1.
-
>Master degree : "Probabilities and statistics (for "agrégation interne"), 3 hours (4,5 hours TD) and 28 hours TD, University of Rennes 1.
-
>
-
–
Quentin Rapilly:
-
>Licence (L2) degree: "Introduction to probabilities theory", 40 hours TD, INSA Rennes.
-
>
11.2.2 Supervision of students in team
Participants: Charles Kervrann, Patrick Bouthemy, Anaïs Badoual, Ludger Johannes, Massiullah Shafaq-Zadah, Estelle Dransart, Emmanuel Moebel, Sébastien Herbreteau, Etienne Meunier.
-
–
PhD supervision
-
>Sébastien Herbreteau (PhD thesis defended in Dec 2023): "Machine learning and interpretable convolutional networks for supervised and unsupervised image denoising with application to satellite imagery", tel-04403632 (supervised by C. Kervrann).
-
>Antonin Deschemps (PhD thesis defended in Dec 2023): "Machine learning and convolutional networks for automated biological dosimetry" (supervised by C. Kervrann and M.A. Benadjaoud (IRSN/SERAMED, Fontenay-aux-Roses)).
-
>Etienne Meunier (PhD thesis defended in Dec 2023): "Unsupervised learning for motion segmentation and motion saliency in videos" (supervised by P. Bouthemy).
-
>Quentin Tallon (PhD in progress): "Statistical learning for localization and classification of chromosomal aberrations in FISH microscopy images" (started in October 2021, supervised by C. Kervrann and M.A. Benadjaoud (IRSN/SERAMED, Fontenay-aux-Roses)).
-
>Lisa Balsollier (PhD in progress): "Spatiotemporal stochastic models for the comprehension of membrane trafficking at the single cell scale" (started in October 2021, supervised by F. Lavancier, C. Kervrann, and J. Salamero).
-
>Quentin Rapilly (PhD in progress): "Hybrid CNN-Snake algorithms for quantitative analysis of 3D+time live-cell images" (started in December 2022, supervised by A. Badoual and C. Kervrann).
-
>
-
–
Postdoc supervision
-
>Vincent Briane, since March 2023, supervised C. Kervrann.
-
>Ilyes Hamitouche, since June 2023, supervised by M. Shafaq-Zadah, E. Dransart, and L. Johannes.
-
>
-
–
Master 2 supervision
-
>Yasmine Hachani, March-September 2023, supervised by P. Bouthemy and E. Fromont (Lacodam Team, IRISA, Rennes).
-
>Sarra Khairi, March-September 2023, supervised by P. Bouthemy and E. Meunier.
-
>Maelys Risset, March-September 2023, supervised by P. Bouthemy and E. Meunier.
-
>Manuel Sanchez Laguardia, March-September 2023, supervised by C. Kervrann, E. Moebel, and S. Herbreteau.
-
>
11.2.3 Juries
Participants: Charles Kervrann, Ludger Johannes.
-
–
Charles Kervrann:
-
>President of the PhD thesis committee of S. Ichbiah (PSL University, Collège de France, Paris, supervised by H. Turlier), R. Fermanian (University of Rennes, Centre Inria, supervised by C. Guillemot).
-
>Reviewer of the HdR of A. Chessel (Ecole Polytechnique, Palaiseau), T. Lagache (University Paris Cité, Institut Pasteur, Paris), M. El-Beheiry (University Paris-Saclay).
-
>Reviewer for the PhD theses of J. Roos (University of Bordeaux, supervised by J.-B. Sibarita), I. Hamitouche (Sorbonne University, supervised by S. Jonic).
-
>Member of the PhD thesis committee of J. Bai ((Sorbonne University, Institut Pasteur, Paris, supervised by C. Zimmer).
-
>
-
–
Ludger Johannes:
-
>Member of the PhD thesis committee of A. Salomon (University of Rennes, supervised by C. Kervrann).
-
>
12 Scientific production
12.1 Publications of the year
International journals
International peer-reviewed conferences
Scientific book chapters
Doctoral dissertations and habilitation theses
Reports & preprints

