2024Activity reportProject-TeamABS
RNSR: 200818987H- Research center Inria Centre at Université Côte d'Azur
- Team name: Algorithms - Biology - Structure
- Domain:Digital Health, Biology and Earth
- Theme:Computational Biology
Keywords
Computer Science and Digital Science
- A2.5. Software engineering
- A3.3.2. Data mining
- A3.4.1. Supervised learning
- A3.4.2. Unsupervised learning
- A6.1.4. Multiscale modeling
- A6.2.4. Statistical methods
- A6.2.8. Computational geometry and meshes
- A8.1. Discrete mathematics, combinatorics
- A8.3. Geometry, Topology
- A8.7. Graph theory
- A9.2. Machine learning
Other Research Topics and Application Domains
- B1.1.1. Structural biology
- B1.1.5. Immunology
- B1.1.7. Bioinformatics
1 Team members, visitors, external collaborators
Research Scientists
- Frédéric Cazals [Team leader, INRIA, Senior Researcher]
- Dorian Mazauric [INRIA, Researcher]
- Edoardo Sarti [INRIA, Researcher]
PhD Students
- Guillaume Carriere [INRIA]
- Simon Queric [INRIA, from Dec 2024]
- Ercan Seckin [INRAE]
Technical Staff
- Come Le Breton [INRIA, Engineer]
Interns and Apprentices
- Stéphanie Bottex [UNIV COTE D'AZUR, Intern, until Jan 2024]
- Vincent Chaye [CNRS, Apprentice]
- Hamadi Daghar [INRIA, Apprentice, from Sep 2024]
- Destiny Hanna [UNIV COTE D'AZUR, Apprentice]
- Akshat Jha [INRIA, Intern, from May 2024 until Jul 2024]
- Mael Riviere [UNIV COTE D'AZUR, Apprentice, from Sep 2024]
- Mael Riviere [INRIA, Apprentice, until Aug 2024]
- Abhinav Rajesh Shripad [INRIA, Intern, from May 2024 until Jul 2024]
- Amir Snouci [CNRS, Intern, from Apr 2024 until Sep 2024]
Administrative Assistant
- Florence Barbara [INRIA]
Visiting Scientist
- Markus Schreiber [INSTITUT MAX-PLANCK, from Sep 2024 until Oct 2024]
External Collaborators
- Caroline Chollet [CNRS, from Jul 2024]
- Alix Lhéritier [AMADEUS]
- David Wales [UNIV CAMBRIDGE]
2 Overall objectives
Biomolecules and their function(s).
Computational Structural Biology (CSB) is the scientific domain concerned with the development of algorithms and software to understand and predict the structure and function of biological macromolecules. This research field is inherently multi-disciplinary. On the experimental side, biology and medicine provide the objects studied, while biophysics and bioinformatics supply experimental data, which are of two main kinds. On the one hand, genome sequencing projects give supply protein sequences, and ~200 millions of sequences have been archived in UniProtKB/TrEMBL – which collects the protein sequences yielded by genome sequencing projects. On the other hand, structure determination experiments (notably X-ray crystallography, nuclear magnetic resonance, and cryo-electron microscopy) give access to geometric models of molecules – atomic coordinates. Alas, only ~150,000 structures have been solved and deposited in the Protein Data Bank (PDB), a number to be compared against the sequences found in UniProtKB/TrEMBL. With one structure for ~1000 sequences, we hardly know anything about biological functions at the atomic/structural level. Complementing experiments, physical chemistry/chemical physics supply the required models (energies, thermodynamics, etc). More specifically, let us recall that proteins with atoms has Cartesian coordinates, and fixing these (up to rigid motions) defines a conformation. As conveyed by the iconic lock-and-key metaphor for interacting molecules, Biology is based on the interactions stable conformations make with each other. Turning these intuitive notions into quantitative ones requires delving into statistical physics, as macroscopic properties are average properties computed over ensembles of conformations. Developing effective algorithms to perform accurate simulations is especially challenging for two main reasons. The first one is the high dimension of conformational spaces – see above, typically several tens of thousands, and the non linearity of the energy functionals used. The second one is the multiscale nature of the phenomena studied: with biologically relevant time scales beyond the millisecond, and atomic vibrations periods of the order of femto-seconds, simulating such phenomena typically requires conformations/frames, a (brute) tour de force rarely achieved 37.
Computational Structural Biology: three main challenges.
The first challenge, sequence-to-structure prediction, aims to infer the possible structure(s) of a protein from its amino acid sequence. While recent progress has been made recently using in particular deep learning techniques 36, the models obtained so far are static and coarse-grained.
The second one is protein function prediction. Given a protein with known structure, i.e., 3D coordinates, the goal is to predict the partners of this protein, in terms of stability and specificity. This understanding is fundamental to biology and medicine, as illustrated by the example of the SARS-CoV-2 virus responsible of the Covid19 pandemic. To infect a host, the virus first fuses its envelope with the membrane of a target cell, and then injects its genetic material into that cell. Fusion is achieved by a so-called class I fusion protein, also found in other viruses (influenza, SARS-CoV-1, HIV, etc). The fusion process is a highly dynamic process involving large amplitude conformational changes of the molecules. It is poorly understood, which hinders our ability to design therapeutics to block it.
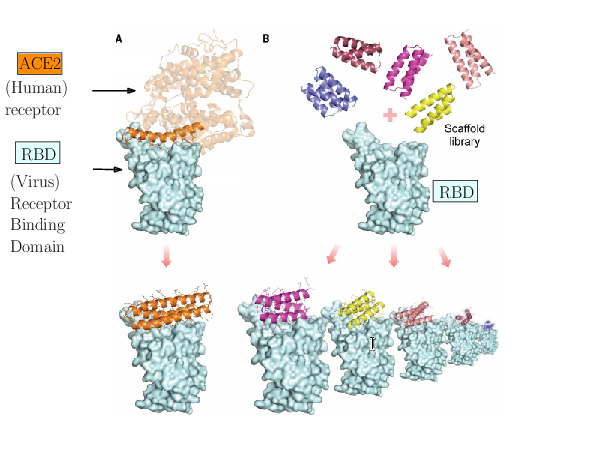
The spike of SARS-CoV-2 is responsible for the infection. Molecules can be engineered to compete with the receptor of the spike, and block infection.
Finally, the third one, large assembly reconstruction, aims at solving (coarse-grain) structures of molecular machines involving tens or even hundreds of subunits. This research vein was promoted about 15 years back by the work on the nuclear pore complex 25. It is often referred to as reconstruction by data integration, as it necessitates to combine coarse-grain models (notably from cryo-electron microscopy (cryo-EM) and native mass spectrometry) with atomic models of subunits obtained from X ray crystallography. Fitting the latter into the former requires exploring the conformation space of subunits, whence the importance of protein dynamics.
As an illustration of these three challenges, consider the problem of designing proteins blocking the entry of SARS-CoV-2 into our cells (Fig. 1). The first challenge is illustrated by the problem of predicting the structure of a blocker protein from its sequence of amino-acids – a tractable problem here since the mini proteins used only comprise of the order of 50 amino-acids (Fig. 1(A), 28). The second challenge is illustrated by the calculation of the binding modes and the binding affinity of the designed proteins for the RBD of SARS-CoV-2 (Fig. 1(B)). Finally, the last challenge is illustrated by the problem of solving structures of the virus with a cell, to understand how many spikes are involved in the fusion mechanism leading to infection. In 28, the promising designs suggested by modeling have been assessed by an array of wet lab experiments (affinity measurements, circular dichroism for thermal stability assessment, structure resolution by cryo-EM). The hyperstable minibinders identified provide starting points for SARS-CoV-2 therapeutics 28. We note in passing that this is truly remarkable work, yet, the designed proteins stem from a template (the bottom helix from ACE2), and are rather small.
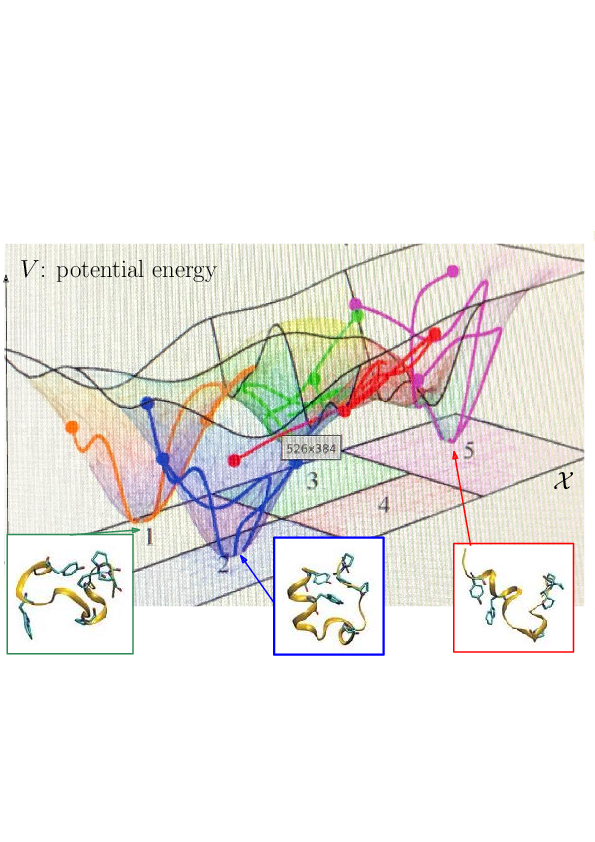
Prosaically, the energy landscape of a molecular system is the set of valleys and passes connecting them, corresponding to stable states and transitions between them.
Protein dynamics: core CS - maths challenges.
To present challenges in structural modeling, let us recall the following ingredients (Fig. 2). First, a molecular model with atoms is parameterized over a conformational space of dimension in Cartesian coordinates, or in internal coordinate–upon removing rigid motions, also called degree of freedom (d.o.f.). Second, recall that the potential energy landscape (PEL) is the mapping from to providing a potential energy for each conformation 38, 35. Example potential energies (PE) are CHARMM, AMBER, MARTINI, etc. Such PE belong to the realm of molecular mechanics, and implement atomic or coarse-grain models. They may embark a solvent model, either explicit or implicit. Their definition requires a significant number of parameters (up to ), fitted to reproduce physico-chemical properties of (bio-)molecules 39.
These PE are usually considered good enough to study non covalent interactions – our focus, even tough they do not cover the modification of chemical bonds. In any case, we take such a function for granted 1.
The PEL codes all structural, thermodynamic, and kinetic properties, which can be obtained by averaging properties of conformations over so-called thermodynamic ensembles. The structure of a macromolecular system requires the characterization of active conformations and important intermediates in functional pathways involving significant basins. In assigning occupation probabilities to these conformations by integrating Boltzmann's distribution, one treats thermodynamics. Finally, transitions between the states, modeled, say, by a master equation (a continuous-time Markov process), correspond to kinetics. Classical simulation methods based on molecular dynamics (MD) and Monte Carlo sampling (MC) are developed in the lineage of the seminal work by the 2013 recipients of the Nobel prize in chemistry (Karplus, Levitt, Warshel), which was awarded “for the development of multiscale models for complex chemical systems”. However, except for highly specialized cases where massive calculations have been used 37, neither MD nor MC give access to the aforementioned time scales. In fact, the main limitation of such methods is that they treat structural, thermodynamic and kinetic aspects at once 31. The absence of specific insights on these three complementary pieces of the puzzle makes it impossible to optimize simulation methods, and results in general in the inability to obtain converged simulations on biologically relevant time-scales.
The hardness of structural modeling owes to three intertwined reasons.
First, PELs of biomolecules usually exhibit a number of critical points exponential in the dimension 26; fortunately, they enjoy a multi-scale structure 29. Intuitively, the significant local minima/basins are those which are deep or isolated/wide, two notions which are mathematically qualified by the concepts of persistence and prominence. Mathematically, problems are plagued with the curse of dimensionality and measure concentration phenomena. Second, biomolecular processes are inherently multi-scale, with motions spanning 15 and 4 orders of magnitude in time and amplitude respectively 24. Developing methods able to exploit this multi-scale structure has remained elusive. Third, macroscopic properties of biomolecules, i.e., observables, are average properties computed over ensembles of conformations, which calls for a multi-scale statistical treatment both of thermodynamics and kinetics.
Validating models.
A natural and critical question naturally concerns the validation of models proposed in structural bioinformatics. For all three types of questions of interest (structures, thermodynamics, kinetics), there exist experiments to which the models must be confronted – when the experiments can be conducted.
For structures, the models proposed can readily be compared against experimental results stemming from X ray crystallography, NMR, or cryo electron microscopy. For thermodynamics, which we illustrate here with binding affinities, predictions can be compared against measurements provided by calorimetry or surface plasmon resonance. Lastly, kinetic predictions can also be assessed by various experiments such as binding affinity measurements (for the prediction of and ), or fluorescence based methods (for kinetics of folding).
3 Research program
Our research program ambitions to develop a comprehensive set of novel concepts and algorithms to study protein dynamics, based on the modular framework of PEL.
3.1 Modeling the dynamics of proteins
Keywords: Molecular conformations, conformational exploration, energy landscapes, thermodynamics, kinetics.
As noticed while discussing Protein dynamics: core CS - maths challenges, the integrated nature of simulation methods such as MD or MC is such that these methods do not in general give access to biologically relevant time scales. The framework of energy landscapes 38, 35 (Fig. 2) is much more modular, yet, large biomolecular systems remain out of reach.
To make a definitive step towards solving the prediction of protein dynamics, we will serialize the discovery and the exploitation of a PEL 4, 16, 3. Ideas and concepts from computational geometry/geometric motion planning, machine learning, probabilistic algorithms, and numerical probability will be used to develop two classes of probabilistic algorithms. The first deals with algorithms to discover/sketch PELs, i.e., enumerate all significant (persistent or prominent) local minima and their connections across saddles, a difficult task since the number of all local minima/critical points is generally exponential in the dimension. To this end, we will develop a hierarchical data structure coding PELs as well as multi-scale proposals to explore molecular conformations. (NB: in Monte Carlo methods, a proposal generates a new conformation from an existing one.) The second focuses on methods to exploit/sample PELs, i.e., compute so-called densities of states, from which all thermodynamic quantities are given by standard relations 2734. This is a hard problem akin to high-dimensional numerical integration. To solve this problem, we will develop a learning based strategy for the Wang-Landau algorithm 33–an adaptive Monte Carlo Markov Chain (MCMC) algorithm, as well as a generalization of multi-phase Monte Carlo methods for convex/polytope volume calculations 32, 30, for non convex strata of PELs.
3.2 Algorithmic foundations: geometry, optimization, machine learning
Keywords: Geometry, optimization, machine learning, randomized algorithms, sampling, optimization.
As discussed in the previous Section, the study of PEL and protein dynamics raises difficult algorithmic / mathematical questions. As an illustration, one may consider our recent work on the comparison of high dimensional distribution 7, statistical tests / two-sample tests 8, 13, the comparison of clustering 9, the complexity study of graph inference problems for low-resolution reconstruction of assemblies 12, the analysis of partition (or clustering) stability in large networks, the complexity of the representation of simplicial complexes 2. Making progress on such questions is fundamental to advance the state-of-the art on protein dynamics.
We will continue to work on such questions, motivated by CSB / theoretical biophysics, both in the continuous (geometric) and discrete settings. The developments will be based on a combination of ideas and concepts from computational geometry, machine learning (notably on non linear dimensionality reduction, the reconstruction of cell complexes, and sampling methods), graph algorithms, probabilistic algorithms, optimization, numerical probability, and also biophysics.
3.3 Software: the Structural Bioinformatics Library
Keywords: Scientific software, generic programming, molecular modeling.
While our main ambition is to advance the algorithmic foundations of molecular simulation, a major challenge will be to ensure that the theoretical and algorithmic developments will change the fate of applications, as illustrated by our case studies. To foster such a symbiotic relationship between theory, algorithms and simulation, we will pursue high quality software development and integration within the SBL, and will also take the appropriate measures for the software to be widely adopted.
Software in structural bioinformatics.
Software development for structural bioinformatics is especially challenging, combining advanced geometric, numerical and combinatorial algorithms, with complex biophysical models for PEL and related thermodynamic/kinetic properties. Specific features of the proteins studied must also be accommodated. About 50 years after the development of force fields and simulation methods (see the 2013 Nobel prize in chemistry), the software implementing such methods has a profound impact on molecular science at large. One can indeed cite packages such as CHARMM, AMBER, gromacs, gmin, MODELLER, Rosetta, VMD, PyMol, .... On the other hand, these packages are goal oriented, each tackling a (small set of) specific goal(s). In fact, no real modular software design and integration has taken place. As a result, despite the high quality software packages available, inter-operability between algorithmic building blocks has remained very limited.
The SBL.
Predicting the dynamics of large molecular systems requires the integration of advanced algorithmic building blocks / complex software components. To achieve a sufficient level of integration, we undertook the development of the Structural Bioinformatics Library (SBL, SB) 5, a generic C++/python cross-platform library providing software to solve complex problems in structural bioinformatics. For end-users, the SBL provides ready to use, state-of-the-art applications to model macro-molecules and their complexes at various resolutions, and also to store results in perennial and easy to use data formats (SBL Applications). For developers, the SBL provides a broad C++/python toolbox with modular design (SBL Doc). This hybrid status targeting both end-users and developers stems from an advanced software design involving four software components, namely applications, core algorithms, biophysical models, and modules (SBL Modules). This modular design makes it possible to optimize robustness and the performance of individual components, which can then be assembled within a goal oriented application.
3.4 Applications: modeling interfaces, contacts, and interactions
Keywords: Protein interactions, protein complexes, structure/thermodynamics/kinetics prediction.
Our methods will be validated on various systems for which flexibility operates at various scales. Examples of such systems are antibody-antigen complexes, (viral) polymerases, (membrane) transporters.
Even very complex biomolecular systems are deterministic in prescribed conditions (temperature, pH, etc), demonstrating that despite their high dimensionality, all d.o.f. are not at play at the same time. This insight suggests three classes of systems of particular interest. The first class consists of systems defined from (essentially) rigid blocks whose relative positions change thanks to conformational changes of linkers; a Newton cradle provides an interesting way to envision such as system. We have recently worked on one such system, a membrane proteins involve in antibiotic resistance (AcrB, see 17). The second class consists of cases where relative positions of subdomains do not significantly change, yet, their intrinsic dynamics are significantly altered. A classical illustration is provided by antibodies, whose binding affinity owes to dynamics localized in six specific loops 14, 15. The third class, consisting of composite cases, will greatly benefit from insights on the first two classes. As an example, we may consider the spikes of the SARS-CoV-2 virus, whose function (performing infection) involves both large amplitude conformational changes and subtle dynamics of the so-called receptor binding domain. We have started to investigate this system, in collaboration with B. Delmas (INRAE).
In ABS, we will investigate systems in these three tiers, in collaboration with expert collaborators, to hopefully open new perspectives in biology and medicine. Along the way, we will also collaborate on selected questions at the interface between CSB and systems biology, as it is now clear that the structural level and the systems level (pathways of interacting molecules) can benefit from one another.
4 Application domains
The main application domain is Computational Structural Biology, as underlined in the Research Program.
5 Social and environmental responsibility
5.1 Footprint of research activities
A tenet of ABS is to carefully analyze the performances of the algorithms designed–either formally or experimentally, so as to avoid massive calculations. Therefore, the footprint of our research activities has remained limited.
5.2 Impact of research results
The scientific agenda of ABS is geared towards a fine understanding of complex phenomena at the atomic/molecular level. While the current focus is rather fundamental, as explained in Research program, an overarching goal for the current period (i.e. 12 years) is to make significant contributions to important problems in biology and medicine.
6 Highlights of the year
We wish to stress three elements.
On the scientific side, we have gained insights into important problems for structural biology analysis, in particular clustering algorithms used to model complex mixtures in flat torii, to encode joint distributions of dihedral angles in proteins. We also developed novel statistical analysis for AlphaFold predictions – cf the 2024 Nobel prize in chemistry, which shed light on these important predictions for biologists.
On the software side, we have continued with the development of the Structural Bioinformatics Library. In particular, we were awarded a project in the scope of the Programme Inria Quadrant (PIQ) funded in the scope of France 2030. This is an important step, given the difficulties we have been facing for software development and community animation.
Finally, on the teaching and dissemination side, we opened a class in the Master Vision Apprentissage program, to hopefully get a wider attraction basin for students.
7 New software, platforms, open data
7.1 New software
7.1.1 SBL
-
Name:
Structural Bioinformatics Library
-
Keywords:
Structural Biology, Biophysics, Software architecture
-
Functional Description:
The SBL is a generic C++/python cross-platform software library targeting complex problems in structural bioinformatics. Its tenet is based on a modular design offering a rich and versatile framework allowing the development of novel applications requiring well specified complex operations, without compromising robustness and performances.
More specifically, the SBL involves four software components (1-4 thereafter). For end-users, the SBL provides ready to use, state-of-the-art (1) applications to handle molecular models defined by unions of balls, to deal with molecular flexibility, to model macro-molecular assemblies. These applications can also be combined to tackle integrated analysis problems. For developers, the SBL provides a broad C++ toolbox with modular design, involving core (2) algorithms, (3) biophysical models, and (4) modules, the latter being especially suited to develop novel applications. The SBL comes with a thorough documentation consisting of user and reference manuals, and a bugzilla platform to handle community feedback.
-
Release Contributions:
In 2024, the following Application packages were integrated into the SBL: Kpax for structural alignments, and Spectraldom for the identification of quasi-rigid domains. A number of low-level algorithms were also developed / improved, in particular seeding methods for Kmeans and Expectation-Maximization, mixture design methods for torsion angles, as well as novel statistical analysis for AlphaFold (see contributions). These packages will be integrated to the release in 2025.
- URL:
- Publication:
-
Contact:
Frédéric Cazals
7.2 Open data
The ongoing collaboration with the Computational Structural Biology team of NINDS (NIH) in Bethesda, MD (USA) is continuing developing the EncoMPASS database for relating membrane protein structures and symmetries. EncoMPASS is the object of a novel recent publication 18 and is undergoing a significant expansion in view of the developing AI tools for structural molecular biology. We propose EncoMPASS as a very reliable source of information on membrane proteins, especially suitable for benchmarking and training prediction algorithms.
8 New results
Participants: Frédéric Cazals, Dorian Mazauric, Edoardo Sarti.
8.1 Modeling the dynamics of proteins
Keywords: Protein flexibility, protein conformations, collective coordinates, conformational sampling, loop closure, kinematics, dimensionality reduction.
8.1.1 Simpler protein domain identification using spectral clustering
Participant: Frédéric Cazals, Edoardo Sarti.
The decomposition of a biomolecular complex into domains is an important step to investigate biological functions and ease structure determination. A successful approach to do so is the SPECTRUS algorithm, which provides a segmentation based on spectral clustering applied to a graph coding inter-atomic fluctuations derived from an elastic network model.
We present 20, which makes three straightforward and useful additions to SPECTRUS. For single structures, we show that high quality partitionings can be obtained from a graph Laplacian derived from pairwise interactions–without normal modes. For sets of homologous structures, we introduce a Multiple Sequence Alignment mode, exploiting both the sequence based information (MSA) and the geometric information embodied in experimental structures. Finally, we propose to analyze the clusters/domains delivered using the so-called D-Family matching algorithm, which establishes a correspondence between domains yielded by two decompositions, and can be used to handle fragmentation issues.
Our domains compare favorably to those of the original SPECTRUS, and those of the deep learning based method Chainsaw. Using two complex cases, we show in particular that is the only method handling complex conformational changes involving several sub-domains. Finally, a comparison of and Chainsaw on the manually curated domain classification ECOD as a reference shows that high quality domains are obtained without using any evolutionary related piece of information.
is provided in the Structural Bioinformatics Library, see SBL and Spectral domain explorer.
8.2 Algorithmic foundations
Keywords: Computational geometry, computational topology, optimization, graph theory, data analysis, statistical physics.
8.2.1 A mini-review of clustering algorithms and their theoretical properties, with applications to molecular science
Participant: Frédéric Cazals.
Clustering is a fundamental task, in particular to analyze potential and free energy landscapes in molecular science. In this survey 19, I review the key properties of three remarkable clustering algorithms (k-means ++, persistence-based clustering, and spectral clustering) with a double perspective. The first one is the specification of the main mathematical and algorithmic properties of the algorithms; the second one is the relevance of these methods for structural, thermodynamic, and kinetic analysis. Doing so provides a unique opportunity to mention important connexions between optimization, graph theory, geometry, and theoretical biophysics.
8.2.2 Improved seeding strategies for k-means and Gaussian mixture fitting with Expectation-Maximization
Participant: Guillaume Carrière, Frédéric Cazals.
k-means clustering and Gaussian Mixture model fitting are fundamental tasks in data analysis and statistical modeling. Practically, both algorithms follow a general iterative pattern, relying on (randomized) seeding techniques.
We revisit the previous seeding methods and formalize their key ingredients (metric used for seed sampling, number of seed candidates, metric used for seed selection). This analysis results in casting most of the previous methods into a coherent framework and, most importantly, yields novel families of initialization methods. Incidentally, these novel methods exploit a lookahead principle–conditioning the seed selection to an enhanced coherence with the final metric used to assess the algorithm, and a multipass strategy–using at least two selection passes to tame down the effect of randomization.
Experiments show a consistent constant factor improvement over classical contenders in terms of the final metric (sum of square error (SSE) for k-means, log-likelihood for Expectation-Maximization applied to Gaussian mixture model fitting), at the same cost. Roughly speaking, our improvement with respect to the greedy smart seeding of k-means++ matches that yielded by this greedy smart seeding with respect to the classical randomized smart seeding.
Remark. Due to the double blind review process of machine learning conferences, the tech report will be made public early 2025.
8.2.3 Subspace-Embedded Spherical Clusters: a novel cluster model for compact clusters of arbitrary dimension
Participant: Frédéric Cazals.
In collaboration with L. Goldenberg (former Inria intern), and with S. Suren (IIT Delhi).
Subspace clustering aims at selecting a small number of original coordinates (features) so that clusters are clearly identified in those subspaces. Subspace techniques rely on parametric cluster models including affine, spherical, Gaussian cluster models–to name a few. To go beyond fully dimensional spherical cluster models and affine clusters of arbitrary dimension, we introduce Subspace-embedded spherical clusters (SESC), a novel cluster model for compact clusters of arbitrary intrinsic dimension. The well posed nature of such clusters is established via the study of an optimization problem relying on an arrangement of hyper-spheres. This arrangement is used to exhibit a piecewise smooth strictly convex function, amenable to non smooth optimization.
We illustrate the merits of the SESC model via comparisons against projection medians and the distance to the measure, and for clustering.
Remark. Due to the double blind review process of machine learning conferences, the tech report will be made public early 2025.
8.3 Applications in structural bioinformatics and beyond
Keywords: Docking, scoring, interfaces, protein complexes, phylogeny, evolution.
8.3.1 AlphaFold predictions on whole genomes at a glance
Participant: Frédéric Cazals, Edoardo Sarti.
The 2024 Nobel prize in chemistry was awarded to David Baker (Univ. of Washington) for computational protein design, and to Demis Hassabis and John Jumper (Google DeepMind, London, UK), for protein structure prediction. The DeepMind software, called AlphaFold, plays a crucial role to help biologists understand protein functions. We designed novel statistical analysis to assess predictions 21.
For model organisms, AlphaFold predictions show that 30% to 40% of amino acids have a (very) low pLDDT (predicted local distance difference test) confidence score. This observation, combined with the method's high complexity, commands to investigate difficult cases, the link with IDPs (intrinsically disordered proteins) or IDRs (intrinsically disordered regions), and potential hallucinations. We do so via four contributions. First, we provide a multiscale characterization of stretches with coherent values along the sequence, an important analysis for model quality assessment. Second, we leverage the 3D atomic packing properties of predictions to represent a structure as a distribution. This distribution is then mapped into the so-called 2D arity map, which simultaneously performs dimensionality reduction and clustering, effectively summarizing all structural elements across all predictions. Third, using the database of domains ECOD , we study potential biases in AlphaFold predictions at the sequence and structural levels, identifying a specific region of the arity map populated with low quality 3D domains. Finally, with a focus on proteins with intrinsically disordered regions (IDRs), using DisProt and AIUPred, we identify specific regions of the arity map characterized by false positive and false negatives in terms of IDRs.
Summarizing, the arity map sheds light on the accuracy of AlphaFold predictions, both in terms of 3D domains and IDRs.
8.3.2 EncoMPASS: a database for the analysis of membrane protein structures, and symmetries
Participant: Edoardo Sarti.
Membrane proteins (MPs) constitute about 30% of the proteome of each organisms, but they represent only 2% of the entries in the Protein Data Bank (PDB), as their three-dimensional structure is difficult to determine experimentally. Membrane protein structures differ from the rest of the proteome in two respects: 1) despite the great variety of functions performed, their structures are very similar, thus making structural classification more challenging and 2) although symmetric regions are common throughout the whole proteome, in MPs they are often essential for their functional mechanism.
Among the databases collecting and organizing experimental structures of MPs, EncoMPASS is the only one relating the structure and internal symmetry of experimentally determined membrane protein complexes. In this new publication 18, the pipeline and founding criteria for building the database are described along with a complete analysis of the available data. The quality and consistency checks regularly performed on EncoMPASS make it a high quality resource for membrane protein structure algorithms.
8.3.3 Detecting orphan proteins in a nematode's genome
Participant: Ercan Seçkin, Edoardo Sarti.
Protein classified in the same family are called homologs and are thought to share a common ancestor from which they have evolved. Proteins that cannot at present be classified in any known family are called orphan proteins, and their existence can be attributed to either the current limitations in protein classification (we talk then of distant homologs) or to genuinely novel proteins (de novo proteins). Determining whether a protein is orphan - or, even more, a distant homolog or a de novo - is particularly challenging due to the uncertainties and intricateness of homolog detection. In the poster 23 presented at JOBIM2024 by E. Seçkin, we show a new pipeline for determining orphan proteins, and its application to the genomes of the Meloidogyne genus of nematodes. This work is a fundamental step in preparation to the first ever algorithm for characterizing the structure of orphan proteins.
9 Partnerships and cooperations
Participants: David Wales, Markus Schreiber.
9.1 International research visitors
9.1.1 Visits of international scientists
Inria International Chair
- David Wales, Cambridge University, is endowed chair within 3IA 3IA Côte d'Azur / ABS.
Other international visits to the team
Markus Schreiber
-
Status
PhD student.
-
Institution of origin:
MPI Frankfurt.
-
Country:
Germany.
-
Dates:
01-30 2024.
-
Context of the visit:
PhD program mobility.
-
Mobility program/type of mobility:
internship.
9.2 National initiatives
ANR Innuendo.
This ANR project (01-2024 to 12-2027) is a joint project with INRAE Jouy-en-Josas (B. Delmas) and IBS Grenoble (W. Weissenhorn), and combines two goals: the first is methodological, and the second is applied.
Methods-wise, our project ambitions to advance the state-of-the-art of flexible computational protein design and binding affinity estimations, which raise difficult high dimensional geometric problems. The novel algorithms will make it possible to explore a larger design space, while at the same time reducing the experimental burden, via superior binding affinity estimates. All methods are made available to the community in the Structural Bioinformatics Library (SBL), a unique software environment providing both low level algorithms and applications for end-users.
Application-wise, we will develop high affinity neutralizing biosynthetic proteins, called α repeat proteins (Reps), with broad spectrum of recognition for circulating sarbecoviruses and limited sensitivity to immune escape mutations. This will be achieved by a virtuous cycle combining our novel computational protein design methods, as well as experiments for structure (cryoEM, X-ray crystallography) and thermodynamics (binding affinity measurements.)
Action Exploratoire Inria.
The AEx DEFINE, involving Inria ABS and Laboratory of Computational and Quantitative Biology (LCQB) from Sorbonne University started in September 2023, for a period of four years.
ABS develops novel methods to study protein structure and dynamics, using computational geom- etry/topology and machine learning. LCQB is a leading lab addressing core questions at the heart of modern biology, with a unique synergy between quantitative models and experiments. The goal of DEFINE is to provide a synergy between ABS and LCQB, with a focus on the prediction of protein functions, at the genome scale and for two specific applications (photosynthesis, DNA repair).
Co-supervised PhD thesis Inria-INRAE.
The PhD thesis of Ercan Seckin started in october 2023 is co-supervised by Etienne Danchin (supervisor) and Dominique Colinet at the INRAE GAME team and Edoardo Sarti at ABS.
The thesis title is: "Détection, histoire évolutive et relations structure - fonction des gènes orphelins chez les bioagresseurs des plantes". The two teams are closely collaborating for advancing current knowledge on the emergence of orphan genes/proteins in the Meloidogyne genus as well as their structural and functional characterization. Notably, the ABS team will focus on the structural and functional inference, and the interplay between structure and function in the process of gene formation.
10 Dissemination
Participants: Frédéric Cazals, Dorian Mazauric, Edoardo Sarti.
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
Frédéric Cazals was involved in the organization of:
- Winter School Algorithms in Structural Bioinformatics: Structural bioinformatics in the AlphaFold / Deep Learning era, Institute for Scientific Study of Cargese (IESC), November 17-22nd, 2024. Web: AlgoSB.
Edoardo Sarti was involved in the organization of:
- REBICA : Rencontre Annuelle de Bioinformatique à l'Université Côte d'Azur, Université Côte d'Azur (UniCA), July 1st, 2024. Web: REBICA.
10.1.2 Scientific events: selection
Member of the conference program committees
Frédéric Cazals participated to the following program committees:
- Symposium on Solid and Physical Modeling
- Intelligent Systems for Molecular Biology (ISMB)
10.1.3 Invited talks
Frédéric Cazals:
- On the prediction of protein dynamics: should one be optimistic ?, Belgrade Bioinformatics Conference, June 2024.
- On the prediction of protein dynamics: should one be optimistic ?, MPI Frankfurt, June 2024.
- Generating backbone conformational changes with seven league boots, Joint Integrative Computational Biology workshop and CAPRI Meeting, Grenoble, February 2024.
- Generating backbone conformational changes with seven league boots, CNRS/Illinois Univ, LIA-IRP meeting. Hauteluce, January 2024.
Edoardo Sarti:
- Spectral partitioning into protein structural domains, Joint Integrative Computational Biology workshop and CAPRI Meeting, Grenoble, February 2024.
- Structural characterization of a nematode’s orphan proteome, SISSA Trieste (Italy), December 2024.
10.1.4 Leadership within the scientific community
Frédéric Cazals:
- 2010-...: Member of the steering committee of the GDR Bioinformatique Moléculaire, for the Structure and macro-molecular interactions theme.
- 2017-...: Co-chair, with Yann Ponty, of the working group / groupe de travail (GT MASIM - Méthodes Algorithmiques pour les Structures et Interactions Macromoléculaires), within the GDR de BIoinfor- matique Moléculaire (GDR BIM).
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
- 2014–...: Master Data Sciences Program (M2), Department of Applied Mathematics, Ecole Centrale-Supélec; Foundations of Geometric Methods in Data Analysis; F. Cazals and M. Carrière, Inria Sophia / (ABS, DataShape). Web: FGMDA.
- 2021–...: Master Data Sciences & Artificial Intelligence (M1), Université Côte d’Azur; Introduction to machine learning (course leader); E. Sarti; Web: IntroML
- 2021–...: Master Data Sciences & Artificial Intelligence (M2), Université Côte d’Azur; Geometric and topological methods in machine learning; F. Cazals, J-D. Boissonnat and M. Carrière, Inria Sophia / (ABS, DataShape, DataShape); Web: GTML.
- 2022–...: Master : Algorithmique avancée, 24h Cours et TD, niveau M1, Polytech Nice Sophia, Université Côte d'Azur, filière Sciences Informatiques, France; D. Mazauric (avec Éric Pascual)
- 2022–...: Bachelor Sciences de la Vie (L2), Université Côte d'Azur; Introduction à la programmation (course leader), E. Sarti; Web: IntroInfo
- 2021–...: Bachelor Informatique (L1), Université Côte d'Azur; Introduction aux Systemes Unix (practicals), E. Sarti
- Dizaine de formations (pour les enseignantes et enseignants, personnels de médiathèque, d'associations, etc.)
10.2.2 Supervision
Frédéric Cazals
- PhD thesis, ongoing, October 2023-...: Guillaume Carrière. Attention mechanisms for graphical models, with applications to protein structure analysis. Advisor: F. Cazals.
Edoardo Sarti
- PhD thesis, ongoing, October 2023-...: Ercan Seçkin. Detection, evolutionary history and structure-function relationships of orphan genes in plant parasitic nematodes. Advisor: E. Danchin (Inrae), D. Colinet, E. Sarti (co-supervision)
10.2.3 Juries
Frédéric Cazals participated to the following committees:
- David Loiseaux, Université Côte d'Azur, December 2024. Persistance Topologique Multi-Paramétrée pour l’Apprentissage Statistique. Advisor: Mathieu Carrière. (Official advisor / guarantor: Frédéric Cazals.)
- William Margerit, University of Toulouse, December 2024. Rapporteur for the PhD thesis Une approche interdisciplinaire pour la conception d'antimicrobiens efficaces à base de nanoparticules. Advisors: Juan Cortés and Nathalie Tarrat.
- Diego Alfredo Amaya Ramirez, University of Lorraine, September 2024. Rapporteur for the PhD thesis Data science approach for the exploration of HLA antigenicity based on 3D structures and molecular dynamics. Advisor: Marie-Dominique Devignes (CNRS), and Jean-Luc Taupin (Univ. Paris-Cité).
Edoardo Sarti participated to the following committees:
- Jury de Master Sciences du Vivant parcours Bioinformatique et Biologie Computationnelles, Université Côte d'Azur.
- Comité d'assignation des bourses doctorales interdisciplinaires EUR Life, Université Côte d'Azur.
Dorian Mazauric participated to the following committee:
- Taher Yacoub, Université Paris-Saclay, January 2024. Rapporteur for the PhD thesis Développement et implémentation d'une approche par fragments pour le design d'ARNs modifiés simple brin avec évaluation sur des protéines de liaison à l'ARN et un modèle d'étude la Bêta-Sécrétase 1. Advisors: Fabrice Leclerc, Yann Ponty.
10.3 Popularization
10.3.1 Specific official responsibilities in science outreach structures
Dorian Mazauric
- 2019-...: Coordinator of Terra Numerica – vers une Cité du Numérique, an ambitious scientific popularisation project. Its main goal is to create a "Dedicated Digital space" in the south of France, (in the spirit of the "Cité des Sciences" or "Palais de la découverte" in Paris). To do so, Terra Numerica is developing and structuring popularization activities, supports which are spread in different antennas throughout the territory (e.g., Espace Terra Numerica - Valbonne Sophia Antipolis, in schools, exhibition extensions...). This large-scale project involves (brings together) all the actors of research, education, industry, associations and collectivities... It is actually composed of more than one hundred people.
- Supervision of a bachelor student (apprenti) and two Master internships, in the scope of Terra Numerica.
- 2017-...: Member of projet de médiation Galéjade : Graphes et ALgorithmes : Ensemble de Jeux À Destination des Ecoliers... (mais pas que).
10.3.2 Productions (articles, videos, podcasts, serious games, ...)
Dorian Mazauric contributed to the creation of more than 10 new resources in 2024. See dedicated website: Terra Numerica.
10.3.3 Participation in Live events
Frédéric Cazals
- Algorithmes et apprentissage pour la structure et la fonction des protéines, Fête de la Science, Octobre 2024, Antibes.
Dorian Mazauric participated and/or organized more than 300 popularization events in 2024. See Terra Numerica website: Terra Numerica.
11 Scientific production
11.1 Major publications
- 1 articleDistributed Link Scheduling in Wireless Networks.Discrete Mathematics, Algorithms and Applications1252020, 1-38HALDOI
- 2 articleOn the complexity of the representation of simplicial complexes by trees.Theoretical Computer Science617February 2016, 17HALDOIback to text
- 3 articleEnergy landscapes and persistent minima.The Journal of Chemical Physics14452016, 4URL: https://www.repository.cam.ac.uk/handle/1810/253412DOIback to text
- 4 articleConformational Ensembles and Sampled Energy Landscapes: Analysis and Comparison.J. of Computational Chemistry36162015, 1213--1231URL: https://hal.science/hal-01076317DOIback to text
- 5 articleThe Structural Bioinformatics Library: modeling in biomolecular science and beyond.Bioinformatics338April 2017HALDOIback to text
- 6 reportThe Structural Bioinformatics Library: modeling in biomolecular science and beyond.RR-8957InriaOctober 2016HAL
- 7 inproceedingsBeyond Two-sample-tests: Localizing Data Discrepancies in High-dimensional Spaces.IEEE/ACM International Conference on Data Science and Advanced AnalyticsIEEE/ACM International Conference on Data Science and Advanced AnalyticsIEEE/ACM International Conference on Data Science and Advanced AnalyticsParis, FranceMarch 2015, 29HALback to text
- 8 inproceedingsLow-Complexity Nonparametric Bayesian Online Prediction with Universal Guarantees.NeurIPS 2019 - Thirty-third Conference on Neural Information Processing SystemsVancouver, CanadaDecember 2019HALback to text
- 9 articleComparing Two Clusterings Using Matchings between Clusters of Clusters.ACM Journal of Experimental Algorithmics241December 2019, 1-41HALDOIback to text
- 10 inproceedingsEfficient computation of the volume of a polytope in high-dimensions using Piecewise Deterministic Markov Processes.AISTATS 2022 - 25th International Conference on Artificial Intelligence and StatisticsVirtual, FranceMarch 2022HAL
- 11 articleWang-Landau Algorithm: an adapted random walk to boost convergence.Journal of Computational Physics4102020, 109366HALDOI
- 12 articleComplexity dichotomies for the Minimum F -Overlay problem.Journal of Discrete Algorithms52-53September 2018, 133-142HALDOIback to text
- 13 articleA Sequential Non-Parametric Multivariate Two-Sample Test.IEEE Transactions on Information Theory645May 2018, 3361-3370HALback to text
- 14 reportHigh Resolution Crystal Structures Leverage Protein Binding Affinity Predictions.RR-8733InriaMarch 2015HALback to text
- 15 articleNovel Structural Parameters of Ig–Ag Complexes Yield a Quantitative Description of Interaction Specificity and Binding Affinity.Frontiers in Immunology8February 2017, 34HALDOIback to text
- 16 articleHybridizing rapidly growing random trees and basin hopping yields an improved exploration of energy landscapes.J. Comp. Chem.3782016, 739--752URL: https://hal.inria.fr/hal-01191028DOIback to text
- 17 articleStudying dynamics without explicit dynamics: A structure‐based study of the export mechanism by AcrB.Proteins - Structure, Function and BioinformaticsSeptember 2020HALDOIback to text
11.2 Publications of the year
International journals
Reports & preprints
Other scientific publications
11.3 Cited publications
- 24 articleMolecular dynamics: survey of methods for simulating the activity of proteins.Chemical reviews10652006, 1589--1615back to text
- 25 articleThe molecular architecture of the nuclear pore complex.Nature45071702007, 695--701back to text
- 26 articleDynamics on statistical samples of potential energy surfaces.The Journal of chemical physics11151999, 2060--2070back to text
- 27 bookThermodynamics and an Introduction to Thermostatistics.Wiley1985back to text
- 28 articleDe novo design of picomolar SARS-CoV-2 miniprotein inhibitors.Science37065152020, 426--431back to textback to textback to textback to text
- 29 articleEnergy landscapes and persistent minima.The Journal of Chemical Physics14452016, 4URL: https://www.repository.cam.ac.uk/handle/1810/253412DOIback to text
- 30 articleA practical volume algorithm.Mathematical Programming Computation822016, 133--160back to text
- 31 bookUnderstanding molecular simulation.Academic Press2002back to text
-
32
articleRandom walks and an
) volume algorithm for convex bodies.Random Structures & Algorithms1111997, 1--50back to text - 33 bookA guide to Monte Carlo simulations in statistical physics.Cambridge university press2014back to text
- 34 bookFree energy computations: A mathematical perspective.World Scientific2010back to text
- 35 articlePrediction, determination and validation of phase diagrams via the global study of energy landscapes.Int. J. of Materials Research10022009, 135back to textback to text
- 36 articleImproved protein structure prediction using potentials from deep learning.Nature2020, 1--5back to text
- 37 articleAtomic-level characterization of the structural dynamics of proteins..Science33060022010, 341--346URL: http://dx.doi.org/10.1126/science.1187409back to textback to text
- 38 bookEnergy Landscapes.Cambridge University Press2003back to textback to text
- 39 articleBuilding force fields: an automatic, systematic, and reproducible approach.The journal of physical chemistry letters5112014, 1885--1891back to text

