2024Activity reportProject-TeamMAASAI
RNSR: 202023544J- Research centerInria Centre at Université Côte d'Azur
- In partnership with:Université Côte d'Azur
- Team name: Models and Algorithms for Artificial Intelligence
- In collaboration with:Laboratoire informatique, signaux systèmes de Sophia Antipolis (I3S), Laboratoire Jean-Alexandre Dieudonné (JAD)
- Domain:Applied Mathematics, Computation and Simulation
- Theme:Optimization, machine learning and statistical methods
Keywords
Computer Science and Digital Science
- A3.1. Data
- A3.1.10. Heterogeneous data
- A3.1.11. Structured data
- A3.4. Machine learning and statistics
- A3.4.1. Supervised learning
- A3.4.2. Unsupervised learning
- A3.4.6. Neural networks
- A3.4.7. Kernel methods
- A3.4.8. Deep learning
- A9. Artificial intelligence
- A9.2. Machine learning
Other Research Topics and Application Domains
- B3.6. Ecology
- B6.3.4. Social Networks
- B7.2.1. Smart vehicles
- B8.2. Connected city
- B9.6. Humanities
1 Team members, visitors, external collaborators
Research Scientists
- Pierre-Alexandre Mattei [INRIA, Researcher]
- Remy Sun [INRIA, ISFP, from Oct 2024]
Faculty Members
- Charles Bouveyron [Team leader, UNIV COTE D'AZUR & INRIA, Professor]
- Marco Corneli [UNIV COTE D'AZUR, Associate Professor]
- Damien Garreau [UNIV COTE D'AZUR, Associate Professor, until Mar 2024]
- Diane Lingrand [UNIV COTE D'AZUR, Associate Professor, until Aug 2024]
- Frederic Precioso [UNIV COTE D'AZUR, Professor]
- Michel Riveill [UNIV COTE D'AZUR, Professor]
- Vincent Vandewalle [UNIV COTE D'AZUR, Professor]
Post-Doctoral Fellows
- Aude Sportisse [UNIV COTE D'AZUR, until Sep 2024]
- Remy Sun [CNRS, Post-Doctoral Fellow, until Sep 2024]
PhD Students
- Davide Adamo [CNRS]
- Kilian Burgi [UNIV COTE D'AZUR]
- Gatien Caillet [UNIV COTE D'AZUR, until May 2024]
- Antoine Collin [INRIA, from Aug 2024]
- Antoine Collin [UNIV COTE D'AZUR, from Mar 2024]
- Antoine Collin [UNIV COTE D'AZUR, until Jan 2024]
- Célia Dcruz [UNIV COTE D'AZUR]
- Mariam Grigoryan [UNIV COTE D'AZUR]
- Gianluigi Lopardo [UNIV COTE D'AZUR, until Sep 2024]
- Kevin Mottin [UNIV COTE D'AZUR, until Mar 2024]
- Seydina Ousmane Niang [UNIV COTE D'AZUR]
- Louis Ohl [UNIV COTE D'AZUR, until Jul 2024]
- Hugo Schmutz [UNIV COTE D'AZUR, until Jul 2024]
- Julie Tores [UNIV COTE D'AZUR]
Technical Staff
- Lucas Boiteau [INRIA, Engineer, until Jan 2024]
- Leonie Borne [INRIA, Engineer, until Aug 2024]
- Amosse Edouard [INSTANT SYSTEM , until Jan 2024]
- Stephane Petiot [UNIV COTE D'AZUR, Engineer, until Jan 2024]
- Li Yang [CNRS, Engineer]
- Mansour Zoubeirou A Mayaki [PRO BTP, Engineer, until Jun 2024]
Interns and Apprentices
- Brahim Akhyate [INRIA, Intern, from May 2024 until Sep 2024]
- Prabal Ghosh [INRIA, Intern, from May 2024 until Aug 2024]
- Gaël Guillot [CNRS, Intern, from May 2024 until Sep 2024]
- Maya Guy [UNIV COTE D'AZUR, from Sep 2024 until Oct 2024]
- Maya Guy [CNRS, Intern, from Mar 2024 until Aug 2024]
- Habeeb Olawale Hammed [INRIA, Intern, from May 2024 until Sep 2024]
- Ishfaaq Illahibuccus Sona [INRIA, Intern, from Apr 2024 until Jul 2024]
- Leelou Lebrun [CNRS, Intern, from Jul 2024 until Aug 2024]
- Ludwig Hagen Letzig [INRIA, Intern, from May 2024 until Jun 2024]
- Federico Raspanti [Univ Côte d'Azur, from Mar 2024 until Jul 2024]
- Raphael Razafindralambo [INRIA, from Nov 2024]
- Raphael Razafindralambo [INRIA, Intern, from May 2024 until Sep 2024]
- Vedang Bhupesch Shenvi Nadkarni [INRIA, Intern, from Nov 2024]
- Pradeep Singh [IIITB INDE, from Sep 2024 until Nov 2024]
- Juliana Varela Quintero [INRIA, Intern, from May 2024 until Aug 2024]
Administrative Assistant
- Claire Senica [INRIA]
Visiting Scientists
- Arnaud Droit [INRIA, from Dec 2024]
- Félix Mejia Cajica [Univ Santander, from Nov 2024]
External Collaborators
- Alexandre Destere [CHU NICE]
- Pierre Latouche [UNIV CLERMONT AUVERG]
- Diane Lingrand [UNIV COTE D'AZUR, from Sep 2024]
- Hans Ottosson [IBM, until Jul 2024]
2 Overall objectives
Artificial intelligence has become a key element in most scientific fields and is now part of everyone life thanks to the digital revolution. Statistical, machine and deep learning methods are involved in most scientific applications where a decision has to be made, such as medical diagnosis, autonomous vehicles or text analysis. The recent and highly publicized results of artificial intelligence should not hide the remaining and new problems posed by modern data. Indeed, despite the recent improvements due to deep learning, the nature of modern data has brought new specific issues. For instance, learning with high-dimensional, atypical (networks, functions, …), dynamic, or heterogeneous data remains difficult for theoretical and algorithmic reasons. The recent establishment of deep learning has also opened new questions such as: How to learn in an unsupervised or weakly-supervised context with deep architectures? How to design a deep architecture for a given situation? How to learn with evolving and corrupted data?
To address these questions, the Maasai team focuses on topics such as unsupervised learning, theory of deep learning, adaptive and robust learning, and learning with high-dimensional or heterogeneous data. The Maasai team conducts a research that links practical problems, that may come from industry or other scientific fields, with the theoretical aspects of Mathematics and Computer Science. In this spirit, the Maasai project-team is totally aligned with the “Core elements of AI” axis of the Institut 3IA Côte d’Azur. It is worth noticing that the team hosts three 3IA chairs of the Institut 3IA Côte d’Azur, as well as several PhD students funded by the Institut.
3 Research program
Within the research strategy explained above, the Maasai project-team aims at developing statistical, machine and deep learning methodologies and algorithms to address the following four axes.
Unsupervised learning
The first research axis is about the development of models and algorithms designed for unsupervised learning with modern data. Let us recall that unsupervised learning — the task of learning without annotations — is one of the most challenging learning challenges. Indeed, if supervised learning has seen emerging powerful methods in the last decade, their requirement for huge annotated data sets remains an obstacle for their extension to new domains. In addition, the nature of modern data significantly differs from usual quantitative or categorical data. We ambition in this axis to propose models and methods explicitly designed for unsupervised learning on data such as high-dimensional, functional, dynamic or network data. All these types of data are massively available nowadays in everyday life (omics data, smart cities, ...) and they remain unfortunately difficult to handle efficiently for theoretical and algorithmic reasons. The dynamic nature of the studied phenomena is also a key point in the design of reliable algorithms.
On the one hand, we direct our efforts towards the development of unsupervised learning methods (clustering, dimension reduction) designed for specific data types: high-dimensional, functional, dynamic, text or network data. Indeed, even though those kinds of data are more and more present in every scientific and industrial domains, there is a lack of sound models and algorithms to learn in an unsupervised context from such data. To this end, we have to face problems that are specific to each data type: How to overcome the curse of dimensionality for high-dimensional data? How to handle multivariate functional data / time series? How to handle the activity length of dynamic networks? On the basis of our recent results, we ambition to develop generative models for such situations, allowing the modeling and the unsupervised learning from such modern data.
On the other hand, we focus on deep generative models (statistical models based on neural networks) for clustering and semi-supervised classification. Neural network approaches have demonstrated their efficiency in many supervised learning situations and it is of great interest to be able to use them in unsupervised situations. Unfortunately, the transfer of neural network approaches to the unsupervised context is made difficult by the huge amount of model parameters to fit and the absence of objective quantity to optimize in this case. We therefore study and design model-based deep learning methods that can handle unsupervised or semi-supervised problems in a statistically grounded way.
Finally, we also aim at developing explainable unsupervised models that can ease the interaction with the practitioners and their understanding of the results. There is an important need for such models, in particular when working with high-dimensional or text data. Indeed, unsupervised methods, such as clustering or dimension reduction, are widely used in application fields such as medicine, biology or digital humanities. In all these contexts, practitioners are in demand of efficient learning methods which can help them to make good decisions while understanding the studied phenomenon. To this end, we aim at proposing generative and deep models that encode parsimonious priors, allowing in turn an improved understanding of the results.
Understanding (deep) learning models
The second research axis is more theoretical, and aims at improving our understanding of the behavior of modern machine learning models (including, but not limited to, deep neural networks). Although deep learning methods and other complex machine learning models are obviously at the heart of artificial intelligence, they clearly suffer from an overall weak knowledge of their behavior, leading to a general lack of understanding of their properties. These issues are barriers to the wide acceptance of the use of AI in sensitive applications, such as medicine, transportation, or defense. We aim at combining statistical (generative) models with deep learning algorithms to justify existing results, and allow a better understanding of their performances and their limitations.
We particularly focus on researching ways to understand, interpret, and possibly explain the predictions of modern, complex machine learning models. We both aim at studying the empirical and theoretical properties of existing techniques (like the popular LIME), and at developing new frameworks for interpretable machine learning (for example based on deconvolutions or generative models). Among the relevant application domains in this context, we focus notably on text and biological data.
Another question of interest is: what are the statistical properties of deep learning models and algorithms? Our goal is to provide a statistical perspective on the architectures, algorithms, loss functions and heuristics used in deep learning. Such a perspective can reveal potential issues in exisiting deep learning techniques, such as biases or miscalibration. Consequently, we are also interested in developing statistically principled deep learning architectures and algorithms, which can be particularly useful in situations where limited supervision is available, and when accurate modeling of uncertainties is desirable.
Adaptive and Robust Learning
The third research axis aims at designing new learning algorithms which can learn incrementally, adapt to new data and/or new context, while providing predictions robust to biases even if the training set is small.
For instance, we have designed an innovative method of so-called cumulative learning, which allows to learn a convolutional representation of data when the learning set is (very) small. The principle is to extend the principle of Transfer Learning, by not only training a model on one domain to transfer it once to another domain (possibly with a fine-tuning phase), but to repeat this process for as many domains as available. We have evaluated our method on mass spectrometry data for cancer detection. The difficulty of acquiring spectra does not allow to produce sufficient volumes of data to benefit from the power of deep learning. Thanks to cumulative learning, small numbers of spectra acquired for different types of cancer, on different organs of different species, all together contribute to the learning of a deep representation that allows to obtain unequalled results from the available data on the detection of the targeted cancers. This extension of the well-known Transfer Learning technique can be applied to any kind of data.
We also investigate active learning techniques. We have for example proposed an active learning method for deep networks based on adversarial attacks. An unlabelled sample which becomes an adversarial example under the smallest perturbations is selected as a good candidate by our active learning strategy. This does not only allow to train incrementally the network but also makes it robust to the attacks chosen for the active learning process.
Finally, we address the problem of biases for deep networks by combining domain adaptation approaches with Out-Of-Distribution detection techniques.
Learning with heterogeneous and corrupted data
The last research axis is devoted to making machine learning models more suitable for real-world, "dirty" data. Real-world data rarely consist in a single kind of Euclidean features, and are genereally heterogeneous. Moreover, it is common to find some form of corruption in real-world data sets: for example missing values, outliers, label noise, or even adversarial examples.
Heterogeneous and non-Euclidean data are indeed part of the most important and sensitive applications of artificial intelligence. As a concrete example, in medicine, the data recorded on a patient in an hospital range from images to functional data and networks. It is obviously of great interest to be able to account for all data available on the patients to propose a diagnostic and an appropriate treatment. Notice that this also applies to autonomous cars, digital humanities and biology. Proposing unified models for heterogeneous data is an ambitious task, but first attempts (e.g. the Linkage1 project) on combination of two data types have shown that more general models are feasible and significantly improve the performances. We also address the problem of conciliating structured and non-structured data, as well as data of different levels (individual and contextual data).
On the basis of our previous works (notably on the modeling of networks and texts), we first intend to continue to propose generative models for (at least two) different types of data. Among the target data types for which we would like to propose generative models, we can cite images and biological data, networks and images, images and texts, and texts and ordinal data. To this end, we explore modelings through common latent spaces or by hybridizing several generative models within a global framework. We are also interested in including potential corruption processes into these heterogeneous generative models. For example, we are developing new models that can handle missing values, under various sorts of missingness assumptions.
Besides the modeling point of view, we are also interested in making existing algorithms and implementations more fit for "dirty data". We study in particular ways to robustify algorithms, or to improve heuristics that handle missing/corrupted values or non-Euclidean features.
4 Application domains
The Maasai research team has the following major application domains:
Medicine
Most of team members apply their research work to Medicine or extract theoretical AI problems from medical situations. In particular, our main applications to Medicine are concerned with pharmacovigilance, medical imaging, and omics. It is worth noticing that medical applications cover all research axes of the team due to the high diversity of data types and AI questions. It is therefore a preferential field of application of the models and algorithms developed by the team.
Digital humanities
Another important application field for Maasai is the increasingly dynamic one of digital humanities. It is an extremely motivating field due to the very original questions that are addressed. Indeed, linguists, sociologists, geographers and historians have questions that are quite different than the usual ones in AI. This allows the team to formalize original AI problems that can be generalized to other fields, allowing to indirectly contribute to the general theory and methodology of AI.
Multimedia
The last main application domain for Maasai is multimedia. With the revolution brought to computer vision field by deep learning techniques, new questions have appeared such as combining subsymbolic and symbolic approaches for complex semantic and perception problems, or as edge AI to embed machine learning approaches for multimedia solutions preserving privacy. This domain brings new AI problems which require to bridge the gap between different views of AI.
Other application domains
Other topics of interest of the team include astrophysics, bioinformatics and ecology.
5 Highlights of the year
- Rémy Sun was hired as an Inria permanent researcher (ISFP).
- Pierre-Alexandre Mattei became Area Chair for the conferences NeurIPS and ICML.
- Cédric Vincent-Cuaz received the 2024 PhD award of the SFA PhD school.
- The 3IA Côte d'Azur Institute, leaded by Charles Bouveyron, was renewed for an additional 5 years under the "AI Cluster" label (France 2030 initiative).
6 New software, platforms, open data
For the Maasai research team, the main objective of the software implementations is to experimentally validate the results obtained and ease the transfer of the developed methodologies to industry. Most of the software will be released as R or Python packages that requires only a light maintaining, allowing a relative longevity of the codes. Some platforms are also proposed to ease the use of the developed methodologies by users without a strong background in Machine Learning, such as scientists from other fields. The list of maintained sofware and platforms is available on https://team.inria.fr/maasai/software/.
6.1 New platforms
The team is also proposing some SAAS (software as a service) platforms in order to allow scientists from other fields or companies to use our technologies. The team developed the following platforms:
DiagnoseNET: Automatic Framework to Scale Neural Networks on Heterogeneous Systems.
Website: https://diagnosenet.github.io/
- Software Family : transfer;
- Audience: community;
- Evolution and maintenance: basic;
- Free Description: DiagnoseNET is a platform oriented to design a green intelligence medical workflow for deploying medical diagnostic tools with minimal infrastructure requirements and low power consumption. The first application built was to automate the unsupervised patient phenotype representation workflow trained on a mini-cluster of Nvidia Jetson TX2. The Python code is available on Github and freely distributed.
Indago
- Software Family : transfer;
- Audience: partners;
- Evolution and maintenance: lts, long term support;
- Duration of the Development (Duration): 1.8 years;
-
Free Description: Indago implements a textual graph clustering method based on a joint analysis of the graph structure and the content exchanged between each nodes. This allows to reach a better segmentation than what could be obtained with traditional methods. Indago's main applications are built around communication network analysis, including social networks. However, Indago can be applied on any graph-structured textual network. Thus, Indago have been tested on various data, such as tweet corpus, mail networks, scientific paper co-publication network, etc.
The software is used as a fully autonomous SaaS platform with 2 parts :
- A Python kernel that is responsible for the actual data processing.
- A web application that handles collecting, pre-processing and saving the data, such as providing a set of visualization for the interpretation of the results.
Indago is deployed internally on the Inria network and used mainly by the development team for testing and research purposes. We also build tailored versions for industrial or academic partners that use the software externally (with contractual agreements).
Topix
Website: https://topix.mi.parisdescartes.fr/
- Software Family : research;
- Audience: universe;
- Evolution and maintenance: lts, long term support;
- Free Description: Topix is an innovative AI-based solution allowing to summarize massive and possibly extremely sparse data bases involving text. Topix is a versatile technology that can be applied in a large variety of situations where large matrices of texts / comments / reviews are written by users on products or addressed to other individuals (bi-partite networks). The typical use case consists in an e-commerce company interested in understanding the relationship between its users and the sold products thanks to the analysis of user comments. A simultaneous clustering (co-clustering) of users and products is produced by the Topix software, based on the key emerging topics from the reviews and by the underlying model. The Topix demonstration platform allows you to upload your own data on the website, in a totally secured framework, and let the AI-based software analyze them for you. The platform also provides some typical use cases to give a better idea of what Topix can do.
Cassiopy
Website: https://pypi.org/project/cassiopy/
- Software Family : vehicle;
- Audience: community;
- Evolution and maintenance: basic;
- Free Description: CassioPy is a python library for clustering using the Skew-t distribution. This model is designed to handle data with skewness, providing more accurate clustering results in many real-world scenarios where data may not follow a normal distribution.
7 New results
7.1 Unsupervised learning
7.1.1 Sparse GEMINI for Joint Discriminative Clustering and Feature Selection
Participants: Louis Ohl, Pierre-Alexandre Mattei, Charles Bouveyron, Arnaud Droit, Frédéric Precioso.
Keywords: Clustering, Deep learning, Sparsity, Feature Selection
Collaborations: Mickael Leclercq
Feature selection in clustering is a hard task which involves simultaneously the discovery of relevant clusters as well as relevant variables with respect to these clusters. While feature selection algorithms are often model-based through optimized model selection or strong assumptions on the data distribution, we introduce a discriminative clustering model trying to maximize a geometry-aware generalization of the mutual information called GEMINI with a simple penalty: the Sparse GEMINI 20. This algorithm avoids the burden of combinatorial feature subset exploration and is easily scalable to high-dimensional data and large amounts of samples while only designing a discriminative clustering model (see Figure 1). We demonstrate the performances of Sparse GEMINI on synthetic datasets and large-scale datasets. Our results show that Sparse GEMINI is a competitive algorithm and has the ability to select relevant subsets of variables with respect to the clustering without using relevance criteria or prior hypotheses.
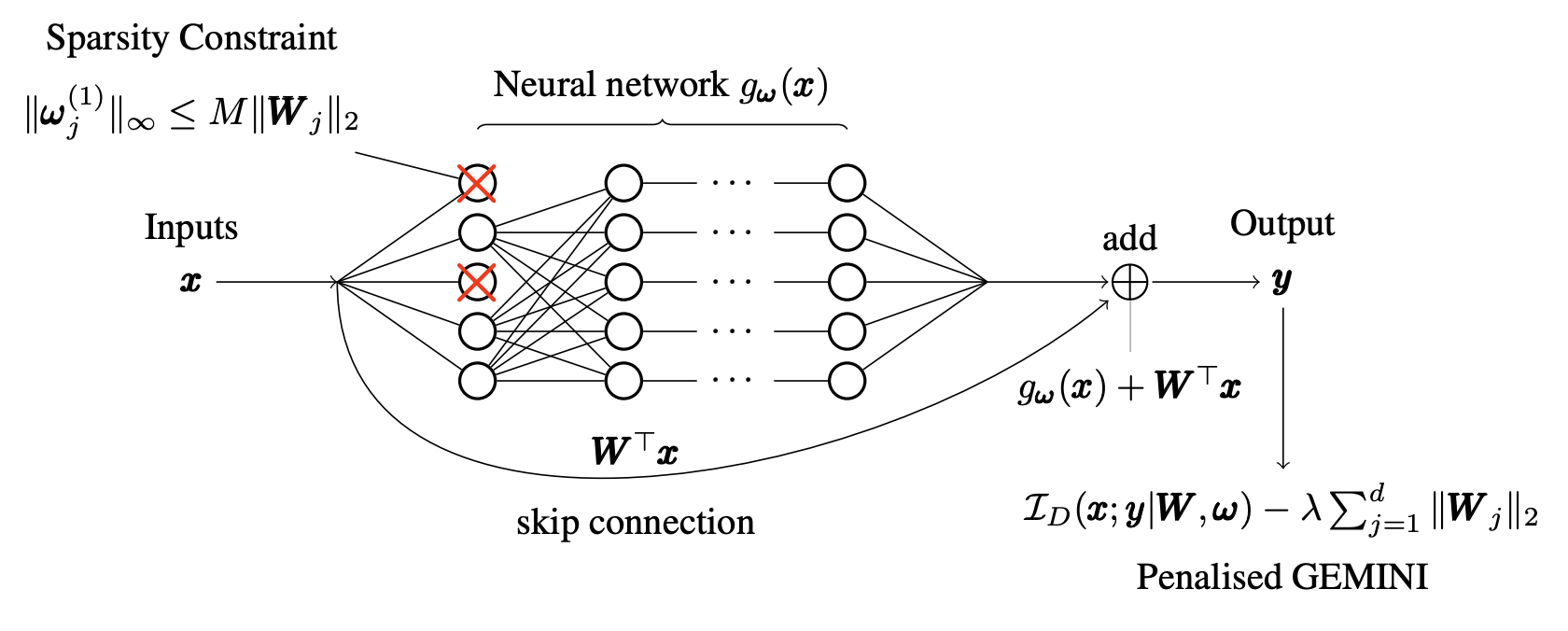
Description of the complete Sparse GEMINI model. For more details, see 7.1.1.
7.1.2 A Deep Dynamic Latent Block Model for the Co-clustering of Zero-Inflated Data Matrices
Participants: Giulia Marchello, Marco Corneli, Charles Bouveyron.
Keywords: Co-clustering, Latent Block Model, zero-inflated distributions, dynamic systems, VEM algorithm.
Collaborations: Regional Center of Pharmacovigilance (RCPV) of Nice.
The simultaneous clustering of observations and features of data sets (known as co-clustering) has recently emerged as a central machine learning application to summarize massive data sets. However, most existing models focus on continuous and dense data in stationary scenarios, where cluster assignments do not evolve over time. In 18, we introduce a novel latent block model for the dynamic co-clustering of data matrices with high sparsity. To properly model this type of data, we assume that the observations follow a time and block dependent mixture of zero-inflated distributions, thus combining stochastic processes with the time-varying sparsity modeling. To detect abrupt changes in the dynamics of both cluster memberships and data sparsity, the mixing and sparsity proportions are modeled through systems of ordinary differential equations. The inference relies on an original variational procedure whose maximization step trains fully connected neural networks in order to solve the dynamical systems. Numerical experiments on simulated data sets demonstrate the effectiveness of the proposed methodology in the context of count data. The proposed method, called -dLBM, was then applied to two real data sets. The first is the data set on the London Bike sharing system while the second is a pharmacovigilance data set, on adverse drug reaction (ADR) reported to the Regional Center of Pharmacovigilance (RCPV) in Nice, France. Figure 2 shows some of the main results obtained through the application of -dLBM on the pharmacovigilance data set.

Estimated Poisson intensities, each color represents a different drug (ADR) cluster. For more details see Section 7.1.2.
7.1.3 Deep Latent Position Topic Model for Clustering and Representation of Networks with Textual Edges
Participants: Charles Bouveyron, Rémi Boutin, Pierre Latouche.
Keywords: generative models, clustering, networks, text, topic modeling
Numerical interactions leading to users sharing textual content published by others are naturally represented by a network where the individuals are associated with the nodes and the exchanged texts with the edges. To understand those heterogeneous and complex data structures, clustering nodes into homogeneous groups as well as rendering a comprehensible visualization of the data is mandatory. To address both issues, we introduced Deep-LPTM, a model-based clustering strategy relying on a variational graph auto-encoder approach as well as a probabilistic model to characterize the topics of discussion. Deep-LPTM allows to build a joint representation of the nodes and of the edges in two embeddings spaces. The parameters are inferred using a variational inference algorithm. We also introduce IC2L, a model selection criterion specifically designed to choose models with relevant clustering and visualization properties. An extensive benchmark study on synthetic data is provided. In particular, we find that Deep-LPTM better recovers the partitions of the nodes than the state-of-the art ETSBM and STBM (see Figure 3). Eventually, the emails of the Enron company are analyzed and visualizations of the results are presented, with meaningful highlights of the graph structure.
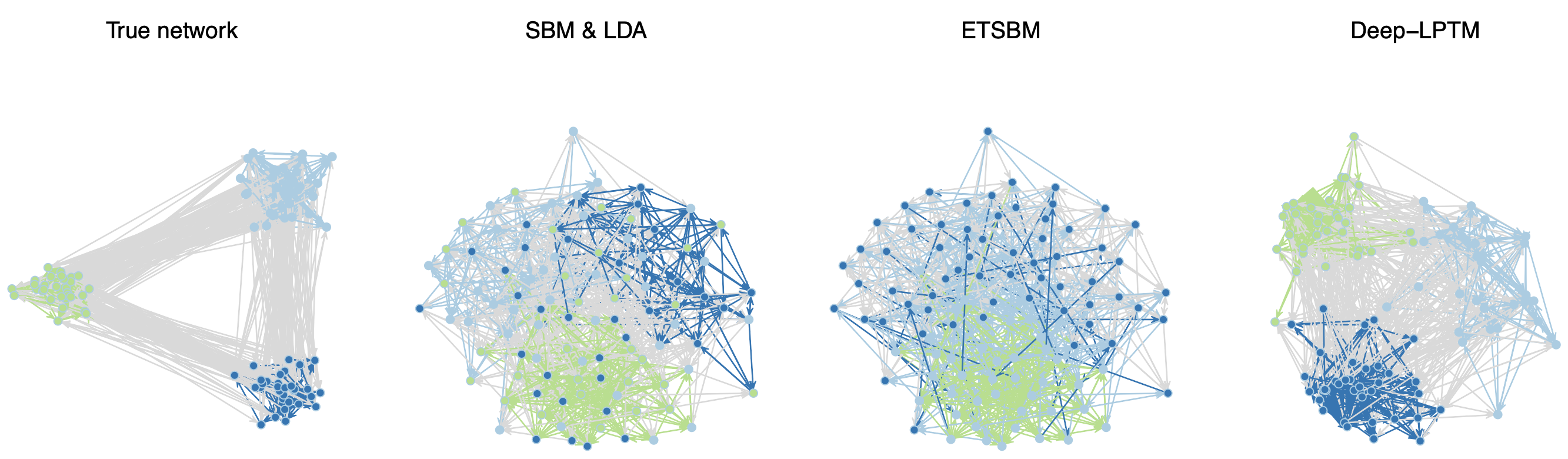
Illustration of Deep-LPTM main contributions on a synthetic network.
7.1.4 Kernel KMeans clustering splits for end-to-end unsupervised decision trees
Participants: Louis Ohl, Pierre-Alexandre Mattei, Arnaud Droit, Frédéric Precioso.
Keywords: Clustering, interpretability, k-means, kernel methods
Collaborations: Mickael Leclercq
Trees are convenient models for obtaining explainable predictions on relatively small datasets. Although there are many proposals for the end-to-end construction of such trees in supervised learning, learning a tree end-to-end for clustering without labels remains an open challenge. As most works focus on interpreting with trees the result of another clustering algorithm, we present in 41 a novel end-to-end trained unsupervised binary tree for clustering: Kauri. This method performs a greedy maximization of the kernel KMeans objective without requiring the definition of centroids. We compare this model on multiple datasets with recent unsupervised trees and show that Kauri performs identically when using a linear kernel. For other kernels, Kauri often outperforms the concatenation of kernel KMeans and a CART decision tree.
7.1.5 The Deep Latent Position Block Model for the Clustering of Nodes in Multi-Graphs
Participants: Seydina Ousmane Niang, Charles Bouveyron, Marco Corneli, Pierre Latouche.
Keywords: Stochastic block model, Latent position model, Clustering, Multi-Graphs, Graph convolutional network
Network data capture relationships among actors across multiple contexts, often forming clusters of individuals. These relationships frequently involve multiple types of interactions, necessitating the use of multidimensional networks, or multigraphs, to capture their full complexity. Latent position models (LPM) embed nodes based on connection probabilities, but cannot uncover heterogeneous clusters such as disassortative patterns. Stochastic block models (SBM), in contrast, excels at clustering but lack interpretative latent representations. To address these limitations, the deep latent position block model (Deep-LPBM) was introduced to provide clustering and continuous latent space representation simultaneously in unidimensional networks. In this paper, we extend this work to multidimensional networks by introducing the deep latent position block model for multidimensional networks (Deep-LPBMM). Deep-LPBMM integrates block modeling and latent embedding across multiple interaction types, allowing nodes to partially belong to several groups, which better captures overlapping clustering structures. Our model uses a deep variational autoencoder with graph convolutional networks (GCNs) for each layer and a multi-layer perceptron to merge latent representations into a unified latent embedding representing cluster partial membership probabilities and offering effective clustering and enhanced visualization.
7.1.6 Clustering by Deep Latent Position Model with Graph Convolutional Network
Participants: Charles Bouveyron, Marco Corneli, Pierre Latouche.
Keywords: Clustering, Random Graphs, Deep Learning, Latent Position Models
Collaborations: Dingge Liang
With the significant increase of interactions between individuals through numeric means, clustering of vertices in graphs has become a fundamental approach for analyzing large and complex networks. In 17, we propose the deep latent position model (DeepLPM), an end-to-end generative clustering approach which combines the widely used latent position model (LPM) for network analysis with a graph convolutional network (GCN) encoding strategy. Moreover, an original estimation algorithm is introduced to integrate the explicit optimization of the posterior clustering probabilities via variational inference and the implicit optimization using stochastic gradient descent for graph reconstruction. Numerical experiments on simulated scenarios highlight the ability of DeepLPM to self-penalize the evidence lower bound for selecting the intrinsic dimension of the latent space and the number of clusters, demonstrating its clustering capabilities compared to state-of-the-art methods. Finally, DeepLPM is further applied to an ecclesiastical network in Merovingian Gaul and to a citation network Cora to illustrate the practical interest in exploring large and complex real-world networks.
7.1.7 A Deep Dynamic Latent Block Model for the Co-clustering of Zero-Inflated Data Matrices
Participants: Charles Bouveyron, Marco Corneli.
Keywords: Dynamic Co-Clustering, Mixture Models, deep learning
Collaborations: Giulia Marchello
The simultaneous clustering of observations and features of data sets (a.k.a. co-clustering) has recently emerged as a central machine learning task to summarize massive data sets. However, most existing models focus on stationary scenarios, where cluster assignments do not evolve in time. In 18 we introduce a novel latent block model for the dynamic co-clustering of data matrices with high sparsity. The data are assumed to follow dynamic mixtures of block-dependent zeroinflated distributions. Moreover, the sparsity parameter as well as the cluster proportions are assumed to be driven by dynamic systems, whose parameters must be estimated. The inference of the model parameters relies on an original variational EM algorithm whose maximization step trains fully connected neural networks that approximate the dynamic systems. Due to the model ability to work with empty clusters, the selection of the number of clusters can be done in a (computationally) parsimonious way. Numerical experiments on simulated and real world data sets demonstrate the effectiveness of the proposed methodology in the context of count data.
7.1.8 The Multiplex Deep Latent Position Model for the Clustering of nodes in Multiview Networks
Participants: Charles Bouveyron, Marco Corneli, Pierre Latouche.
Keywords: multiview networks, latent position model, graph neural networks
Collaborations: Dingge Liang, Junping Yin
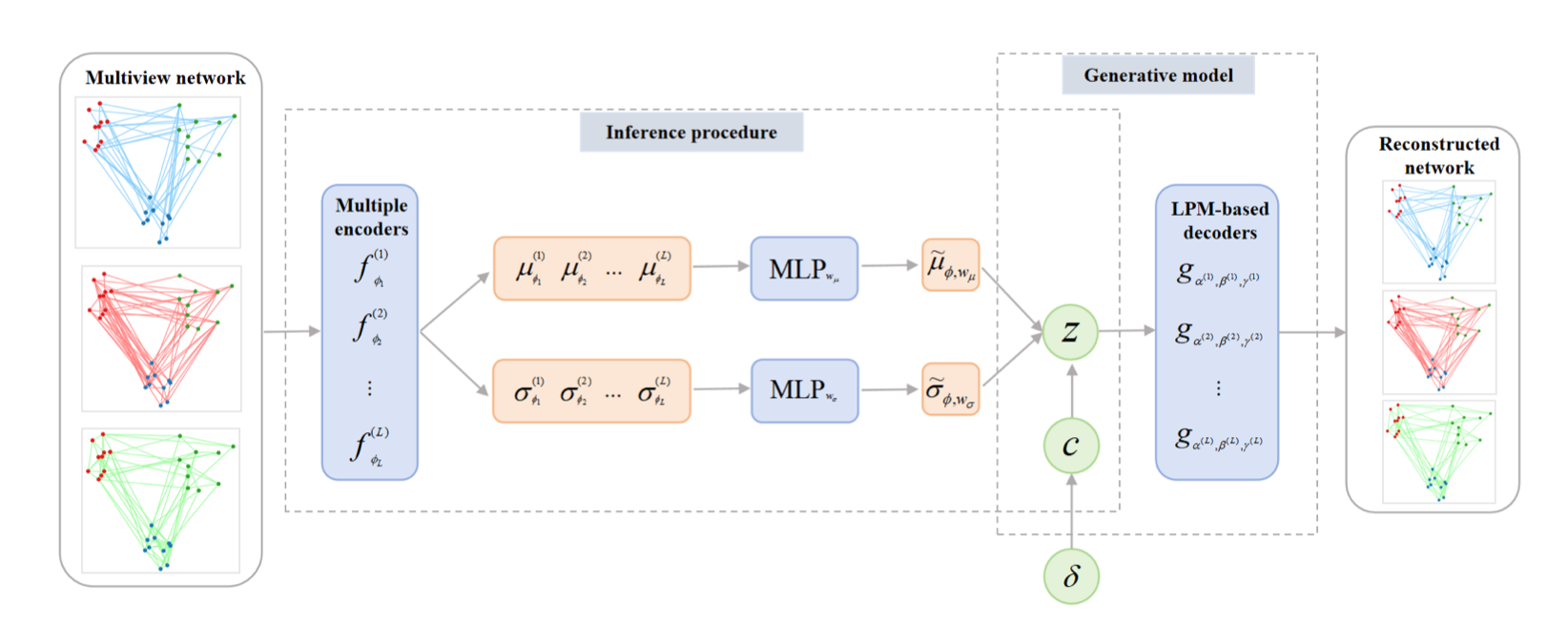
Multiplex networks capture multiple types of interactions among the same set of nodes, creating a complex, multi-relational framework. A typical example is a social network where nodes (actors) are connected by various types of ties, such as professional, familial, or social relationships. Clustering nodes in these networks is a key challenge in unsupervised learning, given the increasing prevalence of multiview data across domains. While previous research has focused on extending statistical models to handle such networks, these adaptations often struggle to fully capture complex network structures and rely on computationally intensive Markov chain Monte Carlo (MCMC) for inference, rendering them less feasible for effective network analysis. To overcome these limitations, in 37, we propose the multiplex deep latent position model (MDLPM), which generalizes and extends latent position models to multiplex networks. MDLPM combines deep learning with variational inference to effectively tackle both the modeling and computational challenges raised by multiplex networks. Unlike most existing deep learning models for graphs that require external clustering algorithms (e.g., k-means) to group nodes based on their latent embeddings, MDLPM integrates clustering directly into the learning process, enabling a fully unsupervised, end-to-end approach. This integration improves the ability to uncover and interpret clusters in multiplex networks without relying on external procedures. Numerical experiments across various synthetic data sets and two real-world networks demonstrate the performance of MDLPM compared to state-of-the-art methods, highlighting its applicability and effectiveness for multiplex network analysis.
7.2 Understanding (deep) learning models
7.2.1 A new perspective on optimizers: leveraging Moreau-Yosida approximation in gradient-based learning
Participants: Alessandro Betti, Frédéric Precioso, Kevin Mottin.
Keywords: optimization, deep learning
Collaborations: Gabriele Ciravegna, Marco Gori, Stefano Melacci
Machine Learning (ML) heavily relies on optimization techniques built upon gradient descent. Numerous gradient-based update methods have been proposed in the scientific literature, particularly in the context of neural networks, and have gained widespread adoption as optimizers in ML software libraries. This paper 11 introduces a novel perspective by framing gradient-based update strategies using the Moreau-Yosida (MY) approximation of the loss function. Leveraging a first-order Taylor expansion, we demonstrate the concrete exploitability of the MY approximation to generalize the model update process. This enables the evaluation and comparison of regularization properties underlying popular optimizers like gradient descent with momentum, ADAGRAD, RMSprop, and ADAM. The MY-based unifying view opens up possibilities for designing new update schemes with customizable regularization properties. To illustrate this potential, we propose a case study that redefines the concept of closeness in the parameter space using network outputs. We present a proof-of-concept experimental procedure, demonstrating the effectiveness of this approach in continual learning scenarios. Specifically, we employ the well-known permuted MNIST dataset, a progressively-permuted MNIST and CIFAR-10 benchmarks, and a non i.i.d. stream. Additionally, we validate the update scheme's efficacy in an offline-learning scenario. By embracing the MY-based unifying view, we pave the way for advancements in optimization techniques for machine learning.
7.2.2 Are Ensembles Getting Better all the Time?
Participants: Damien Garreau, Pierre-Alexandre Mattei.
Keywords: Ensembles, dropout, random forests
Ensemble methods combine the predictions of several base models. We study whether or not including more models always improves their average performance. This question depends on the kind of ensemble considered, as well as the predictive metric chosen. We focus on situations where all members of the ensemble are a priori expected to perform as well, which is the case of several popular methods such as random forests or deep ensembles. In this setting, we show in 39 that ensembles are getting better all the time if, and only if, the considered loss function is convex. More precisely, in that case, the average loss of the ensemble is a decreasing function of the number of models. When the loss function is nonconvex, we show a series of results that can be summarized as: ensembles of good models keep getting better, and ensembles of bad models keep getting worse. To this end, we prove a new result on the monotonicity of tail probabilities that may be of independent interest. We illustrate our results on a medical prediction problem (diagnosing melanomas using neural nets) and a "wisdom of crowds" experiment (guessing the ratings of upcoming movies).
7.2.3 The Risks of Recourse in Binary Classification
Participants: Damien Garreau.
Keywords: Interpretability, Recourse, Machine Learning Theory
Collaborations: Hidde Fokkema, Tim van Erven
Algorithmic recourse provides explanations that help users overturn an unfavorable decision by a machine learning system. But so far very little attention has been paid to whether providing recourse is beneficial or not. We introduce in 29 an abstract learning-theoretic framework that compares the risks (i.e. expected losses) for classification with and without algorithmic recourse. This allows us to answer the question of when providing recourse is beneficial or harmful at the population level. Surprisingly, we find that there are many plausible scenarios in which providing recourse turns out to be harmful, because it pushes users to regions of higher class uncertainty and therefore leads to more mistakes. We further study whether the party deploying the classifier has an incentive to strategize in anticipation of having to provide recourse, and we find that sometimes they do, to the detriment of their users. Providing algorithmic recourse may therefore also be harmful at the systemic level. We confirm our theoretical findings in experiments on simulated and real-world data. All in all, we conclude that the current concept of algorithmic recourse is not reliably beneficial, and therefore requires rethinking.
7.2.4 Visual Objectification in Films: Towards a New AI Task for Video Interpretation
Participants: Julie Tores, Frédéric Precioso.
Keywords: Objectification, Video analysis, Interpretability, Concept Bottleneck Models
Collaborations: Lucile Sassatelli, Hui-Yin Wu, Clement Bergman, Léa Andolfi, Victor Ecrement, Thierry Devars, Magali Guaresi, Virginie Julliard, Sarah Lecossais
In film gender studies the concept of "male gaze" refers to the way the characters are portrayed on-screen as objects of desire rather than subjects. In 31 we introduce a novel video-interpretation task to detect character objectification in films. The purpose is to reveal and quantify the usage of complex temporal patterns operated in cinema to produce the cognitive perception of objectification. We introduce the ObyGaze12 dataset made of 1914 movie clips densely annotated by experts for objectification concepts identified in film studies and psychology. We evaluate recent vision models show the feasibility of the task and where the challenges remain with concept bottleneck models. Our new dataset and code are made available to the community.
7.3 Adaptive and robust learning
7.3.1 Domain-Specific Long Text Classification from Sparse Relevant Information
Participants: Célia D'Cruz, Frédéric Precioso, Michel Riveill.
Keywords: Long Text Classification, Medical Report, Transformer, Information Extraction, Natural Language Processing
Collaborations: Jean-Marc Bereder
Large Language Models have undoubtedly revolutionized the Natural Language Processing field, the current trend being to promote one-model-for-all tasks (sentiment analysis, translation, etc.). However, the statistical mechanisms at work in the larger language models struggle to exploit the relevant information when it is very sparse, when it is a weak signal. This is the case, for example, for the classification of long domain-specific documents, when the relevance relies on a single relevant word or on very few relevant words from technical jargon. In the medical domain, it is essential to determine whether a given report contains critical information about a patient’s condition. This critical information is often based on one or few specific isolated terms. In 28, we propose a hierarchical model which exploits a short list of potential target terms to retrieve candidate sentences and represent them into the contextualized embedding of the target term(s) they contain. A pooling of the term(s) embedding(s) entails the document representation to be classified. We evaluate our model on one public medical document benchmark in English and on one private French medical dataset. We show that our narrower hierarchical model is better than larger language models for retrieving relevant long documents in a domain-specific context.
7.3.2 Detecting Brittle Decisions for Free: Leveraging Margin Consistency in Deep Robust Classifiers
Participants: Frédéric Precioso.
Keywords: adversarial robustness, empirical robustness estimation, classification, vulnerability detection
Collaborations: Jonas Ngnawé, Sabyasachi Sahoo, Yann Pequignot, Christian Gagné
Despite extensive research on adversarial training strategies to improve robustness, the decisions of even the most robust deep learning models can still be quite sensitive to imperceptible perturbations, creating serious risks when deploying them for high-stakes real-world applications. While detecting such cases may be critical, evaluating a model's vulnerability at a per-instance level using adversarial attacks is computationally too intensive and unsuitable for real-time deployment scenarios. The input space margin is the exact score to detect non-robust samples and is intractable for deep neural networks. In 30, we introduce the concept of margin consistency – a property that links the input space margins and the logit margins in robust models – for efficient detection of vulnerable samples. First, we establish that margin consistency is a necessary and sufficient condition to use a model's logit margin as a score for identifying non-robust samples. Next, through comprehensive empirical analysis of various robustly trained models on CIFAR10 and CIFAR100 datasets, we show that they indicate high margin consistency with a strong correlation between their input space margins and the logit margins. Then, we show that we can effectively and confidently use the logit margin to detect brittle decisions with such models. Finally, we address cases where the model is not sufficiently margin-consistent by learning a pseudo-margin from the feature representation. Our findings highlight the potential of leveraging deep representations to assess adversarial vulnerability in deployment scenarios efficiently. Figure 5 presents an overview of the MapEX method.
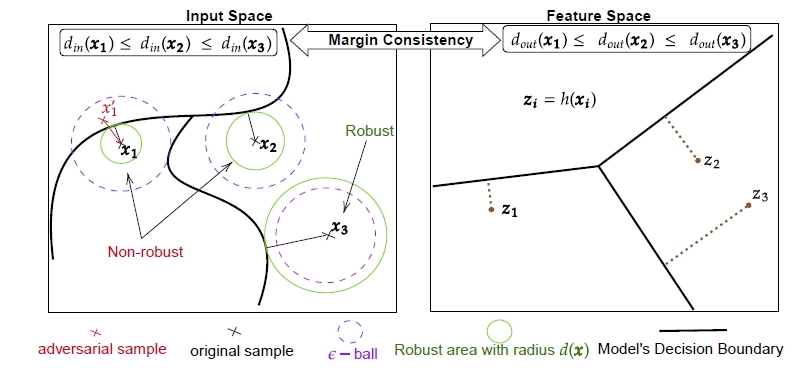
Overview of our MapEX method (see Sec. 7.3.2). Illustration of the input space margin, margin in the feature space and margin consistency. The model preserves the relative position of samples to the decision boundary in the input space to the feature space.
7.4 Learning with heterogeneous and corrupted data
7.4.1 Mind the map! Accounting for existing map information when estimating online HDMaps from sensor data
Participants: Rémy Sun, Li Yang, Diane Lingrand, Frédéric Precioso.
Keywords: Autonomous Driving, HDMaps, Online HDMap estimation
Collaborations: ANR Project MultiTrans
Online High Definition Map (HDMap) estimation from sensors offers a low-cost alternative to manually acquired HDMaps. As such, it promises to lighten costs for already HDMap-reliant Autonomous Driving systems, and potentially even spread their use to new systems. We proposed to improve online HDMap estimation by accounting for already existing maps. We identify 3 reasonable types of useful existing maps (minimalist, noisy, and outdated). We also introduce MapEX (see Fig. 6), a novel online HDMap estimation framework that accounts for existing maps. MapEX achieves this by encoding map elements into query tokens and by refining the matching algorithm used to train classic query based map estimation models. We demonstrate that MapEX brings significant improvements on the nuScenes dataset. For instance, given noisy maps, MapEX improves by 38% over the MapTRv2 detector it is based on and by 16% over the current SOTA (state-of-the-art).
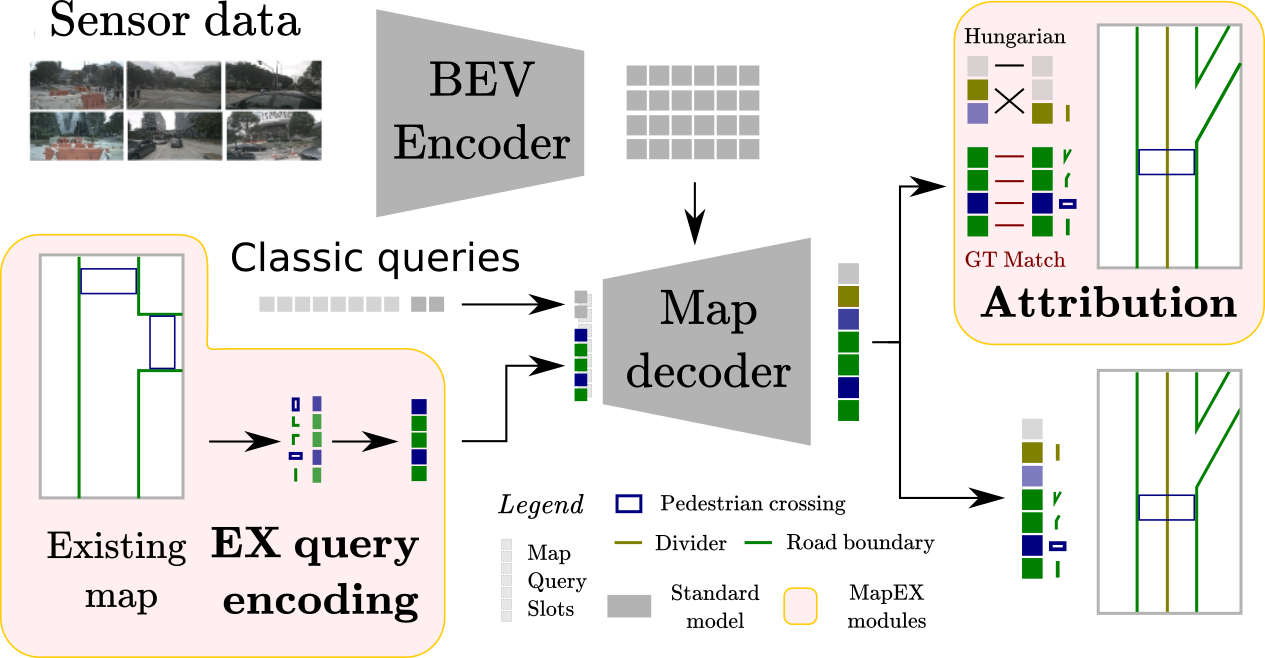
Overview of our MapEX method (see Sec. 7.4.1). We add two modules (EX query encoding, Attribution) to the standard query based map estimation pipeline (in gray on the figure). Map elements are encoded into EX queries, then decoded with a standard decoder.
7.4.2 Model-based clustering with Missing Not At Random Data
Participants: Aude Sportisse.
Keywords: model-based clustering, generative models, missing data
Collaborations: Christophe Biernacki (Inria Lille), Claire Boyer (Sorbonne Unviersité), Julie Josse (Inria Montpellier) Matthieu Marbac (Ensai Rennes)
Model-based unsupervised learning, as any learning task, stalls as soon as missing data occurs. This is even more true when the missing data are informative, or said missing not at random (MNAR). In 25, we propose model-based clustering algorithms designed to handle very general types of missing data, including MNAR data. To do so, we introduce a mixture model for different types of data (continuous, count, categorical and mixed) to jointly model the data distribution and the MNAR mechanism, remaining vigilant to the relative degrees of freedom of each. Several MNAR models are discussed, for which the cause of the missingness can depend on both the values of the missing variable themselves and on the class membership. However, we focus on a specific MNAR model, called MNARz, for which the missingness only depends on the class membership. We first underline its ease of estimation, by showing that the statistical inference can be carried out on the data matrix concatenated with the missing mask considering finally a standard MAR (missing at random) mechanism. Consequently, we propose to perform clustering using the Expectation Maximization algorithm, specially developed for this simplified reinterpretation. Finally, we assess the numerical performances of the proposed methods on synthetic data and on the real medical registry TraumaBase as well.
7.4.3 A Model-Based Clustering Approach for Chemical Toxicity Assessment Using Cell Painting Data
Participants: Grigoryan Mariam, Vincent Vandewalle.
Collaborations: David Rouquié (Bayer Crop Science)
Keywords: Model-based Clustering, Toxicity assessment, Anomaly detection
Risk assessment of chemicals relies heavily on toxicity studies conducted on laboratory animals. However, those studies are lengthy, costly, and are not always fully relevant to human physiology. This is where human-derived, cell-based assays, such as the Cell Painting technique, could play a crucial role. This high-content imaging approach allows for analyzing several cellular compartments, offering a comprehensive view of how these chemicals impact cell morphology and organelle structure.
In 44, we propose an answer to the complex dose-response analysis in high dimensions using an intermediate clustering step of the cells within each well of the assay plates from Cell Painting experiments. The clustering model is a Gaussian mixture model assuming that each well contains several classes of cells with class proportions depending on the well. That class-specific parameters are the same across wells. This enables to summarize the information of each well by its specific class proportions. In the second step, we use these estimated class proportions to compare wells treated with chemical compounds at different concentrations. Applying this approach to real data from Cell Painting experiments allows the identification of consistent patterns across experimental conditions and provides insights into how varying concentrations affect cell behavior. This model will be extended to consider information from several cell lines.
7.4.4 Towards a fully automated underwater census for fish assemblages in the Mediterranean Sea
Participants: Kilian Bürgi, Charles Bouveyron, Diane Lingrand, Frédéric Precioso
Keywords: Diver operated video, Automated UVC, Deep learning, Object detection, Marine biology, Marine protected areas
Collaborations: Cecile Sabourault, Benoit Derijard
In marine biology and ecology the collection of data relies mostly on diver operations which is labour-intensive and financially costly to operate leading to reduced frequencies of data collection missions. Technological advances in the past decade have made it available for unmanned robots collecting data which resulted in numerous amounts of videos that need to be manually evaluated and analyzed which created a new bottleneck. In this study 35, we explored the possibilities and differences between a manual and automated analysis of collected videos by divers simulating a remotely operated vehicle (ROV). We discuss the difference between collecting data by diver and by videos and found that both methods added species to the overall species pool and that the automation was successful. This proof of concept will be used in future studies to fasten the process of data analysis and allow more frequent data collections creating more robust data for ecological decision making.
7.4.5 Automated Counting of Fish in Diver Operated Videos (DOV) for Biodiversity Assessments
Participants: Kilian Bürgi, Charles Bouveyron, Diane Lingrand, Frédéric Precioso.
Keywords: Underwater video, Fish count prediction, Temporal convolutional network, Object detection, Marine biology, Marine conservation
Collaborations: Cecile Sabourault, Benoit Derijard
Counting fish in moving underwater videos relies on labour-intensive manual counting or imprecise metrics from stationary cameras, while there is a great potential to use better methods to receive a better fish count. For this purpose, we explored traditional methods of counting fish as well as introduced three new methods to count fish from computer vision derived data (single frame detections). This resulted in a holistic and fully automated pipeline for fish abundance extraction 36. The following different methods are proposed on transect data of three Mediterranean species with different ecological niches: 1) traditional N, 2) 1d k-means clustering method, 3) an intuitive clustering approach N and 4) a Temporal Convolutional Neural Networks (TCN) counting method. Our results show evidence of underestimation by the traditional N while the other methods showed better overall results with the proposed N and TCN methods best representing the reality. With an absolute variation comparable to inter-observer variation, we demonstrated reliable methods for quantifying fish counts.
7.4.6 Topological data analysis and multiple kernel learning for species identification of modern and archaeological small ruminants
Participants: Marco Corneli, Davide Adamo.
Keywords: Shape Analysis, Point Clouds, Classification, Multiple Kernel Learning, Topological Data Analysis
Collaborations: Manon Vuillien, Emmanuelle Vila, Agraw Amane, Thierry Argant, et al
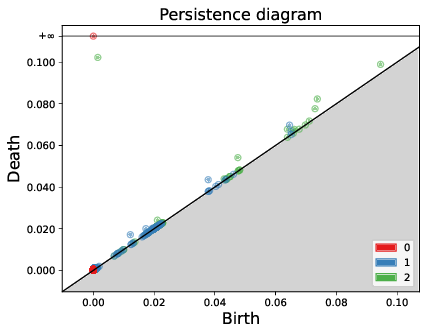
The faunal remains from numerous Holocene archaeological sites across southwest Asia frequently include the bones of several wild and domestic ungulates, such as sheep, goats, ibexes, roe deer and gazelles. These assemblages may provide insight into hunting and animal husbandry strategies and offer paleoecological information on ancient human societies. However, the skeletons of these taxa are highly similar in appearance, which presents a challenge for accurate identification based on their bones. This paper 43 presents a case study to test the potential of topological data analysis (TDA) and multiple kernel learning (MKL) for inter-specific identification of 150 3D astragali belonging to modern and archaeological specimens. The joint application of TDA and MKL demonstrated remarkable efficacy in accurately identifying wild species, with a correct identification rate of approximately 90%. In contrast, the identification of domestic species exhibited a lower success rate, at approximately 60%. The misidentification of sheep and goat species is attributed to the morphological variability of domestic breeds. Moreover, while these methods assist in clearly identifying wild taxa from one another, they also highlight their morphological diversity. In this context, TDA and MKL could be invaluable for investigating intra-specific variability in domestic and wild animals. These methods offer a means of expanding our understanding of past domestic animal selection practices and techniques. They also facilitate an investigation into the morphological evolution of wild animal populations over time (see Fig. 7).
| .2 | .2 | .3 |
| (a) | (b) | (c) |
7.4.7 A Bayesian approach for clustering and exact finite-sample model selection in longitudinal data mixtures
Participants: Marco Corneli.
Keywords: Clustering, Longitudinal Data, Mixture Models, Bayesian Model Selection
Collaborations: Elena Erosheva, Marco Lorenzi, Xunlei Qian
In 14, we consider mixtures of longitudinal trajectories, where one trajectory contains measurements over time of the variable of interest for one individual and each individual belongs to one cluster. The number of clusters as well as individual cluster memberships are unknown and must be inferred. We propose an original Bayesian clustering framework that allows us to obtain an exact finite-sample model selection criterion for selecting the number of clusters. Our finite-sample approach is more flexible and parsimonious than asymptotic alternatives such as Bayesian information criterion or integrated classification likelihood criterion in the choice of the number of clusters. Moreover, our approach has other desirable qualities: (i) it keeps the computational effort of the clustering algorithm under control and (ii) it generalizes to several families of regression mixture models, from linear to purely non-parametric. We test our method on simulated datasets as well as on a real world dataset from the Alzheimer’s disease neuroimaging initative database.
| .48 | .48 |
| (a) | (b) |
7.4.8 AI-Enhanced Prediction of Aortic Stenosis Progression: Insights From the PROGRESSA Study
Participants: Louis Ohl, Pierre-Alexandre Mattei, Frederic Precioso, Arnaud Droit.
Keywords: Cardiology, supervised learning
Collaborations: Melissa Sanabria, Lionel Tastet, Simon Pelletier, Mickael Leclercq, Lara Hermann, Nancy Coté, Philippe Pibarot
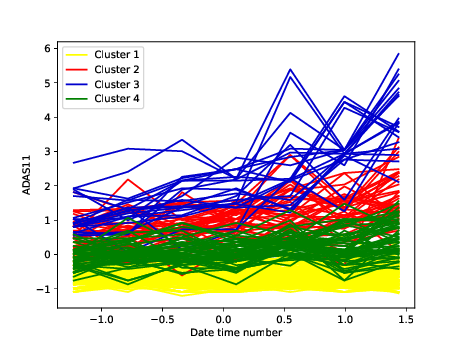
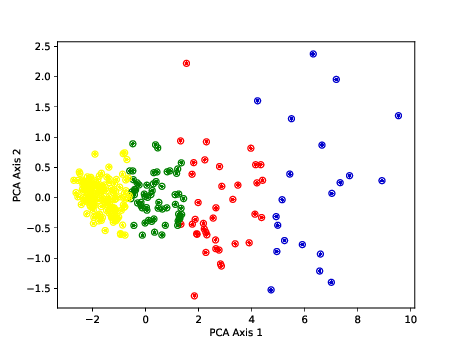
Aortic valve stenosis (AS) is a progressive chronic disease with progression rates that vary in patients and therefore difficult to predict. The aim of this study 24 was to predict the progression of AS using comprehensive and longitudinal patient data . Machine learning algorithms were trained on a data set of 303 patients enrolled in the PROGRESSA (Metabolic Determinants of the Progression of Aortic Stenosis) study who underwent clinical and echocardiographic follow-up on an annual basis. Performance of the models was measured to predict disease progression over long (next 5 years) and short (next 2 years) terms and was compared to a standard clinical model with usually used features in clinical settings based on logistic regression. For each annual follow-up visit including baseline, we trained various supervised learning algorithms in predicting disease progression at 2- and 5-year terms. At both terms, LightGBM consistently outperformed other models with the highest average area under curves across patient visits (0.85 at 2 years, 0.83 at 5 years). Recurrent neural network-based models (Gated Recurrent Unit and Long Short-Term Memory) and XGBoost also demonstrated strong predictive capabilities, while the clinical model showed the lowest performance. This study demonstrates how an artificial intelligence-guided approach in clinical routine could help enhance risk stratification of AS. It presents models based on multisource comprehensive data to predict disease progression and clinical outcomes in patients with mild-to-moderate AS at baseline.
7.4.9 BERNN: Enhancing classification of Liquid Chromatography Mass Spectrometry data with batch effect removal neural networks
Participants: Frederic Precioso, Arnaud Droit.
Keywords: Batch effect removal, Mass Spectrometry, omics
Collaborations: Melissa Sanabria, Lionel Tastet, Simon Pelletier, Mickael Leclercq, Lara Hermann, Nancy Coté, Philippe Pibarot
Liquid Chromatography Mass Spectrometry (LC-MS) is a powerful method for profiling complex biological samples. However, batch effects typically arise from differences in sample processing protocols, experimental conditions, and data acquisition techniques, significantly impacting the interpretability of results. Correcting batch effects is crucial for the reproducibility of omics research, but current methods are not optimal for the removal of batch effects without compressing the genuine biological variation under study. In 21, we propose a suite of Batch Effect Removal Neural Networks (BERNN) to remove batch effects in large LC-MS experiments, with the goal of maximizing sample classification performance between conditions. More importantly, these models must efficiently generalize in batches not seen during training. A comparison of batch effect correction methods across five diverse datasets demonstrated that BERNN models consistently showed the strongest sample classification performance. However, the model producing the greatest classification improvements did not always perform best in terms of batch effect removal. Finally, we show that the overcorrection of batch effects resulted in the loss of some essential biological variability. These findings highlight the importance of balancing batch effect removal while preserving valuable biological diversity in large-scale LC-MS experiments. Figure 9 shows the used architecture for the VAE.
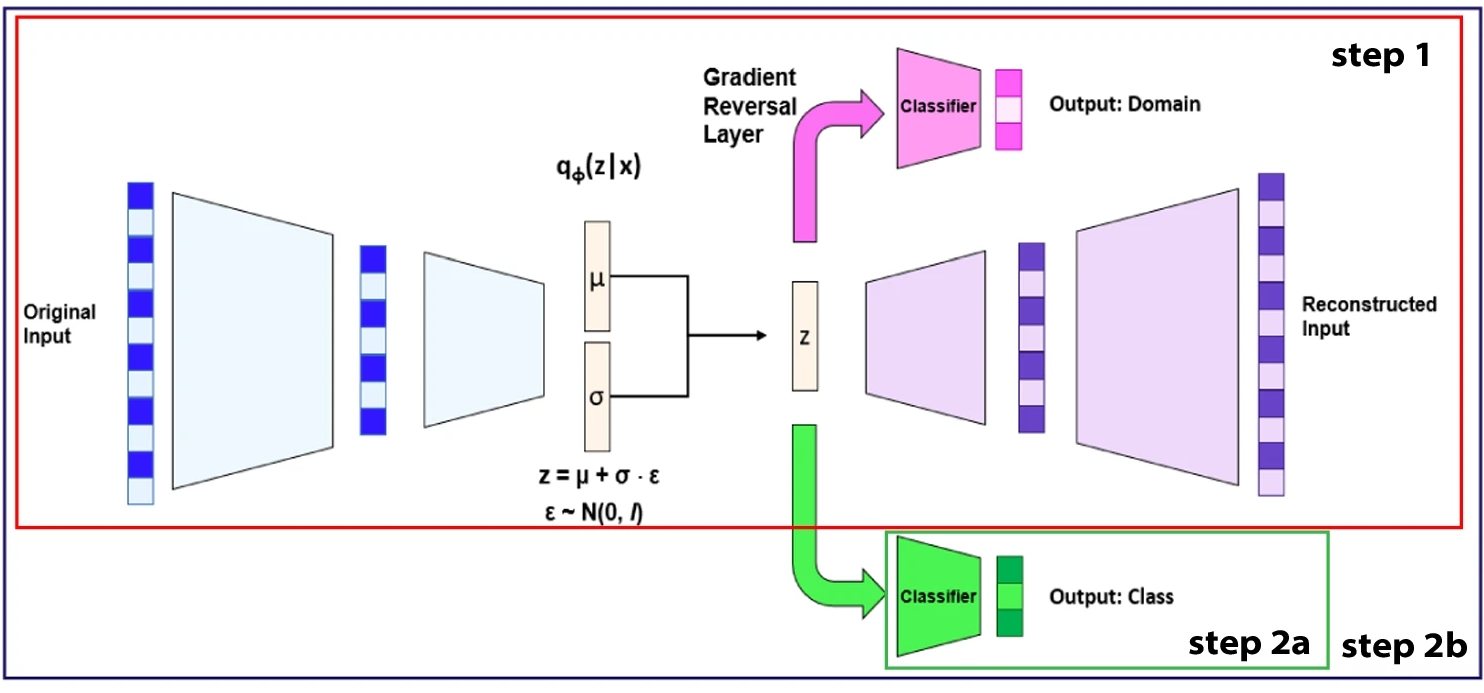
VAE-DANN is a variational autoencoder with a domain classifier trained with a Gradient Reversal Layer (GRL), where φ represents the parameters of the encoder (q), the parameters learned by the encoder are μ, σ, and ε, which correspond to the mean, variance and gaussian noise, respectively. (see Sec. 7.4.9).
7.4.10 An artificial intelligence algorithm for co-clustering to help in pharmacovigilance before and during the COVID-19 pandemic
Participants: Charles Bouveyron, Marco Corneli, Alexandre Destere.
Keywords: Pharmacovigilance, co-clustering, COVID-19
Collaborations: Giulia Marchello, Diane Merino, Nouha Ben Othman, Alexandre O. Gérard, Thibaud Lavrut, Delphine Viard, Fanny Rocher, Milou-Daniel Drici
Monitoring drug safety in real-world settings is the primary aim of pharmacovigilance. Frequent adverse drug reactions (ADRs) are usually identified during drug development. Rare ones are mostly characterized through post-marketing scrutiny, increasingly with the use of data mining and disproportionality approaches, which lead to new drug safety signals. Nonetheless, waves of excessive numbers of reports, often stirred up by social media, may overwhelm and distort this process, as observed recently with levothyroxine or COVID-19 vaccines. As human resources become rarer in the field of pharmacovigilance, we aimed to evaluate the performance of an unsupervised co-clustering method to help the monitoring of drug safety. In 15, a dynamic latent block model (dLBM), based on a time-dependent co-clustering generative method, was used to summarize all regional ADR reports () issued between January 1, 2012 and February 28, 2022. After analysis of their intra and extra interrelationships, all reports were grouped into different cluster types (time, drug, ADR). Our model clustered all reports in 10 time, 10 ADR and 9 drug collections. Based on such clustering, three prominent societal problems were detected, subsequent to public health concerns about drug safety, including a prominent media hype about the perceived safety of COVID-19 vaccines. The dLBM also highlighted some specific drug–ADR relationships, such as the association between antiplatelets, anticoagulants and bleeding. Co-clustering and dLBM appear as promising tools to explore large pharmacovigilance databases. They allow, without supervision, the detection, exploration and strengthening of safety signals, facilitating the analysis of massive upsurges of reports.
8 Bilateral contracts and grants with industry
The team is particularly active in the development of research contracts with private companies. The following contracts were active during 2024:
-
Participants: Pierre-Alexandre Mattei, Léonie Borne.
Pulse Audition This contract was the fruit of the "start it up" program of the 3IA Côte d'Azur. The goal is to work on semi-supervised learning for hearing glasses. A research engineer (Léonie Borne) was recruited via the "start it up" program. Amount: 15 000€.
-
Participants: Mansour Mayaki Zoubeirou, Michel Riveill.
NXP: This collaboration contract is a France Relance contract. Drift detection and predictive maintenance. Amount: 45 000€.
-
Participants: Vincent Vandewalle.
Orange: it is a CIFRE build upon the PhD of Gatien Caillet on decentralized and efficient federated AutoML learning for heterogeneous embedded devices. External participants: Tamara Tosic (Orange), Frédéric Guyard (Orange). Amount: 30 000€.
-
Participants: Pierre-Alexandre Mattei, Lucas Boiteau, Hugo Schmutz, Aude Sportisse.
Naval Group: The goal of this project is the development of an open-source Python library for semi-supervised learning, via the hiring of a research engineer, Lucas Boiteau. External participants: Alexandre Gensse, Quentin Oliveau (Naval Group). Amount: 125 000€.
-
Participants: Michel Riveill, Hugo Miralles.
Orange: it is a CIFRE contract built upon the PhD of Hugo Miralles on Distributed device-embedded classification and prediction in near-to-real time. External participants: Tamara Tosic (Orange), Thierry Nagellen (Orange). Amount: 45 000€.
-
Participants: Frédéric Precioso, Charles Bouveyron, Baptiste Pouthier.
NXP: This collaboration contract is a CIFRE contract built upon the PhD of Baptiste Pouthier on Deep Learning and Statistical Learning on audio-visual data for embedded systems. Participants: Frederic Precioso, Charles Bouveyron, Baptiste Pouthier. External participants: Laurent Pilati (NXP). Amount: 45 000€.
-
Participants: Frédéric Precioso, Michel Riveill, Amosse Edouard.
Instant System: This collaboration contract is a France Relance contract. The objective is to design new recommendation systems based on deep learning for multimodal public transport recommendations (e.g. combining on a same trip: bike, bus, e-scooter, metro, then bike again). Amount: 45 000€.
-
Participants: Charles Bouveyron.
EDF: In this project, we developed model-based clustering and co-clustering methods to summarize massive and multivariate functional data of electricity consumption. The data are coming from Linky meters, enriched by meteorological and spatial data. The developed algorithms were released as open source R packages. External participants: F. Simoes, J. Jacques. Amount: 50 000€.
9 Partnerships and cooperations
9.1 International initiatives
The Maasai team has informal relationships with the following international teams:
- Department of Statistics of the University of Washington, Seattle (USA) through collaborations with Elena Erosheva and Adrian Raftery,
- SAILAB team at Università di Siena, Siena (Italy) through collaborations with Marco Gori,
- School of Mathematics and Statistics, University College Dublin (Ireland) through the collaborations with Brendan Murphy, Riccardo Rastelli and Michael Fop,
- Department of Computer Science, University of Tübingen (Germany) through the collaboration with Ulrike von Luxburg,
- Université Laval, Québec (Canada) through the Research Program DEEL (DEpendable and Explainable Learning) with Christian Gagné, and through a FFCR funding with Arnaud Droit including the co-supervision of the PhD of Louis Ohl),
- DTU Compute, Technical University of Denmark, Copenhagen (Denmark), through collaborations with Jes Frellsen and his team (including the co-supervision of a PhD student in Denmark: Hugo Sénétaire).
9.2 International research visitors
9.2.1 Visits of international scientists
International Chairs
- Pr. Arnaud Droit, Université Laval, Canada (Inria Int. Chair)
- Pr. Marco Gori, Università di Siena, Italy (3IA Int. Chair)
Other international visits to the team
- Pr. Marc Scott, NYU, USA. He gave a seminar attended by members of several Inria teams.
- Pr. Brendan Murphy, University College Dublin, Ireland
- Pr. Masashi Sugiyama, RIKEN, The University of Tokyo. He gave a seminar in the Amphitheater Morgenstern attended by members of several Inria teams.
9.3 European initiatives
9.3.1 H2020 projects
Maasai is one of the 3IA-UCA research teams of AI4Media, one of the 4 ICT-48 Center of Excellence in Artificial Intelligence which has started in September 2020. There are 30 partners (Universities and companies), and 3IA-UCA received about 325k€.
9.4 National initiatives
Institut 3IA Côte d'Azur
Following the call of President Macron to found several national institutes in AI, we presented in front of an international jury our project for the Institut 3IA Côte d'Azur in April 2019. The project was selected for funding (50 M€ for the first 4 years, including 16 M€ from the PIA program) and started in September 2019. Charles Bouveyron and Marco Gori are two of the 29 3IA chairs which were selected ab initio by the international jury and Pierre-Alexandre Mattei was awarded a 3IA chair in 2021, and Vincent Vandewalle in 2022. Charles Bouveyron is also the Director of the institute since January 2021, after being the Deputy Scientific Director on 2019-2020. The research of the institute is organized around 4 thematic axes: Core elements of AI, Computational Medicine, AI for Biology and Smart territories. The Maasai reserch team is totally aligned with the first axis of the Institut 3IA Côte d'Azur and also contributes to the 3 other axes through applied collaborations. The team has several Ph.D. students and postdocs who are directly funded by the institute. The institute was renewed in 2024 for an additional period of 5 years, under the IA Cluster label.
Web site: 3ia.univ-cotedazur.eu
ANR Project MultiTrans
In MultiTrans project, we propose to tackle autonomous driving algorithms development and deployment jointly. The idea is to enable data, experience and knowledge to be transferable across the different systems (simulation, robotic models, and real-word cars), thus potentially accelerating the rate at which an embedded intelligent system can gradually learn to operate at each deployment stage. Existing autonomous vehicles are able to learn how to react and operate in known domains autonomously but research is needed to help these systems during the perception stage, allowing them to be operational and safer in a wider range of situations. MultiTrans proposes to address this issue by developing an intermediate environment that allows to deploy algorithms in a physical world model, by re-creating more realistic use cases that would contribute to a better and faster transfer of perception algorithms to and from a real autonomous vehicle test-bed and between multiple domains.
Web site: anr-multitrans.github.io
ANR Project FATE-MLOps
The MLOps movement adopts the DevOps objective of reducing the gaps between development and operations teams by integrating data scientist teams and Machine Learning (ML) models. In this project, we wish to apply and adapt good software engineering practices to strengthen both the overall quality of the ML model construction processes and the quality of the software systems produced, particularly in terms of extra-functional properties that will become crucial issues: Fairness, Accountability, Transparency, Ethics, and Security (FATES). The key concerns will tackle the study, formalization, measurement, and management of these properties throughout the continuous MLOps process. Indeed, more than traditional Key Performance Indicators (KPIs), such as precision and recall, are required to evaluate models' robustness in practical applications. Our project aims to study the FATES properties and, by refining proven software engineering concepts and tools, propose a systematic and tailored approach for considering those properties, particularly from the lens of ML Scientists or ML Engineers, throughout the lifecycle of the software developed following an MLOps approach.
Web site: fates-mlops.org
ANR Project PROFILE
In this project, we adopt a Software Engineering (SE) approach to this problem by proposing to link model engineering (MDE, here “model” in the SE sense) and statistical and static analyses to characterize these ML workflows by models (also in the SE sense), which we now call PROFILES to avoid confusion.
More specifically, our project aims to explore three complementary questions: (Q1) What information can and should be automatically extracted from a Notebook to build a profile for its analysis? (Q2) Is it possible to systematically identify typical errors from the profile (for example, the use of functions unsuited to the problem at hand) and to identify bad practices (for example, the use of test data for training)? (Q3) Can we exploit the profusion of Notebooks to accelerate ML research by encouraging, on the basis of extracted PROFILES, a pooling of knowledge and the elicitation of new good/bad practices? It is important to note that in this project, we are talking about verifying quality rules in the way a code linter would, and not in the sense of formally verifying properties.
9.5 Regional initiatives
Centre de pharmacovigilance, CHU Nice
Participants: Charles Bouveyron, Marco Corneli, Giulia Marchello, Michel Riveill, Xuchun Zhang
Keywords: Pharmacovigilance, co-clustering, count data, text data
Collaborators: Milou-Daniel Drici, Alexandre Destere
The team works very closely with the Regional Pharmacovigilance Center of the University Hospital Center of Nice (CHU) through several projects. The first project concerns the construction of a dashboard to classify spontaneous patient and professional reports, but above all to report temporal breaks. To this end, we are studying the use of dynamic co-classification techniques to both detect significant ADR patterns and identify temporal breaks in the dynamics of the phenomenon. The second project focuses on the analysis of medical reports in order to extract, when present, the adverse events for characterization. After studying a supervised approach, we are studying techniques requiring fewer annotations.
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
- GeMSS 2024: Pierre-Alexandre Mattei co-founded, co-organized, and co-taught the Generative Modelling Summer School (GeMSS) since 2023. The 2024 edition was held in Eindhoven (June 24th to 28th). More details on the GeMSS website.
- GenU 2024: Pierre-Alexandre Mattei co-founded in 2019 the workshop on Generative Models and Uncertainty Quantification (GenU). This small-scale workshop has been held physically in Copenhagen in the Fall. The 2024 edition was on September 18-19, 2023. More details on the GenU website.
- SophI.A Summit 2024: It is an AI conference that brings together researchers and companies doing AI, held every Fall in Sophia Antipolis. In 2024, Diane Lingrand and Pierre-Alexandre Mattei were members of the scientific committee, and Charles Bouveyron gave the opening speech. More details on the website.
- Deep learning school 2024: the team organized, under the guidance of F. Precioso, the 5th edition of this summer school on Deep Learning. The School took place from July 1st to July 5th 2024 and welcomed about 100 participants (School website).
10.1.2 Scientific publishing
Member of editorial boards
- Charles Bouveyron is Associate Editor for the Annals of Applied Statistics since 2016.
- Pierre-Alexandre Mattei is Area Chair since 2024 for the conferences NeurIPS and ICML.
Reviewing activities All permanent members of the team are serving as reviewers for the most important journals and conferences in statistical and machine learning, such as (non exhaustive list):
- International journals:
- Annals of Applied Statistics
- Statistics and Computing
- Journal of the Royal Statistical Society, Series C
- Journal of Machine Learning Research
- Transactions on Machine Learning Research
- International Conferences
- Neural Information Processing Systems (NeurIPS)
- International Conference on Machine Learning (ICML)
- International Conference on Learning Representations (ICLR)
- International Joint Conference on Artificial Intelligence (IJCAI)
- International Conference on Artificial Intelligence and Statistics (AISTATS)
- IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)
10.1.3 Leadership within the scientific community
- Charles Bouveyron is the Director of the Institut 3IA Côte d'Azur since January 2021 and of the EFELIA Côte d'Azur education program since September 2022.
- Vincent Vandewalle is the Deputy Scientific director of the EFELIA Côte d'Azur education program since September 2022.
10.1.4 Scientific expertise
- Charles Bouveyron is member of the Scientific Orientation Council of Centre Antoine Lacassagne, Unicancer center of Nice.
10.1.5 Research administration
Charles Bouveyron is the director the Institut 3IA Côte d'Azur and the EFELIA Côte d'Azur project. He also leaded the demand for renewing the Institute funding , that was granted by the French governement for an additional 5 years (budget 20M€).
10.2 Teaching - Supervision - Juries
10.2.1 Supervision
The team has 3 senior researchers with HDR that are able to supervise Ph.D. students. Usually, the supervision of the Ph.D. students of the team is jointly made by a senior and a junior researchers of the team. Two Ph.D. students of the team have defended their thesis in 2024.
10.2.2 Juries
C. Bouveyron, M. Riveill, F. Precioso and V. Vandewalle are full professors, D. Garreau and D. Lingrand are associate professors at Université Côte d'Azur and therefore handle usual teaching duties at the university. M. Corneli, P.-A. Mattei, and R. Sun are also teaching around 60h per year at Université Côte d'Azur. P.-A. Mattei is also teaching a graphical models course at the MVA masters from ENS Paris Saclay. M. Corneli has been hired in September 2022 on a “Chaire de Professeur Junior" on AI for Archeology and Historical Sciences.
M. Riveill is the current director of the Master of Science “Data Sciences and Artificial Intelligence" at Université Côte d'Azur, since September 2020. C. Bouveyron was the founder and first responsible (Sept. 2018 - Aug. 2020) of that MSc.
Since September 2022, C. Bouveyron and V. Vandewalle are respectively the Director and Deputy Scientific Director of the EFELIA Côte d'Azur program, funded by the French national plan “France 2030", through the “Compétences et Métiers d'Avenir" initiative (8M€ for 5 years). This program aims at enlarging the teaching capacities in AI of the Institut 3IA Côte d'Azur and developing new education programs for specialists and non-specialists.
All members of the team are also actively involved in the supervision of postdocs, Ph.D. students, interns and participate frequently to Ph.D. and HDR defenses. They are also frequently part of juries for the recruitment of research scientists, assistant-professors or professors.
10.3 Popularization
- F. Precioso, C. Bouveyron, F. Simoes and J. Torres Sanchez have developed a demonstrator for general public on the recognition and monitoring of wild species in the French National Park of Mercantour. This demonstrator has been exhibited during the “Fête des Sciences" in Antibes in October 2023. More details on the demo website.
- F. Precioso has developed an experimental platform both for research projects and scientific mediation on the topic of autonomous cars. This platform is currently installed in the “Maison de l'Intelligence Artificielle" where high school students have already experimented coding autonomous remote control cars.
- C. Bouveyron, F. Simoes and S. Bottini have developed an interactive software allowing to visualize the relationships between pollution and a health disease (dispnea) in the Région Sud. This platform is currently installed at the “Maison de l'Intelligence Artificielle".
- L. Ohl was interviewed in a podcast on AI and cardiology, and for a video published on the Youtube channel of the Health Data Hub.
- C. Bouveyron and F. Precioso participated in a TV documentary on Artificial Intelligence for TV Monaco and TV5 Monde Video replay.
11 Scientific production
11.1 Major publications
- 1 articleTowards a fully automated underwater census for fish assemblages in the Mediterranean Sea.Ecological Informatics85March 2025, 102959HALDOI
- 2 articleAn artificial intelligence algorithm for co‐clustering to help in pharmacovigilance before and during the COVID‐19 pandemic.British Journal of Clinical Pharmacology905February 2024, 1258-1267HALDOI
- 3 inproceedingsDomain-Specific Long Text Classification from Sparse Relevant Information.Frontiers in Artificial Intelligence and ApplicationsECAI 2024 - 27th european conference on artificial intelligence392: ECAI 2024cover 27th European Conference on Artificial Intelligence, 19–24 October 2024, Santiago de Compostela, Spain – Including 13th Conference on Prestigious Applications of Intelligent Systems (PAIS 2024)Santiago de Compostela, SpainIOS PressOctober 2024, 4003 - 4010HALDOI
- 4 inproceedingsThe Risks of Recourse in Binary Classification.Proceedings of Machine Learning Research,27th International Conference on Artificial Intelligence and Statistics (AISTATS)238Valencia (Espagne), Spain2024, 550-558HAL
- 5 articleA Deep Dynamic Latent Block Model for the Co-clustering of Zero-Inflated Data Matrices.Journal of Computational and Graphical Statistics334April 2024, 1224-1239HALDOI
- 6 inproceedingsDetecting Brittle Decisions for Free: Leveraging Margin Consistency in Deep Robust Classifiers.NeurIPS ProceedingsNeurIPS 2024 - Thirty-eighth Annual Conference on Neural Information Processing SystemsVancouver (CA), CanadaDecember 2024HAL
- 7 articleSparse and geometry-aware generalisation of the mutual information for joint discriminative clustering and feature selection.Statistics and Computing345July 2024, 155HALDOI
- 8 articleBERNN: Enhancing classification of Liquid Chromatography Mass Spectrometry data with batch effect removal neural networks.Nature Communications151May 2024, 3777HALDOI
- 9 articleModel-based Clustering with Missing Not At Random Data.Statistics and ComputingJune 2024HALDOI
- 10 inproceedingsVisual Objectification in Films: Towards a New AI Task for Video Interpretation.IEEE XploreCVPR 2024 - IEEE / CVF Computer Vision and Pattern Recognition Conference2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)Seattle (Washington), United StatesJune 2024, 10864-10874HALDOI
11.2 Publications of the year
International journals
Invited conferences
International peer-reviewed conferences
Doctoral dissertations and habilitation theses
Reports & preprints
Other scientific publications

