2024Activity reportProject-TeamMICROCOSME
RNSR: 202124114Z- Research center Inria Centre at Université Grenoble Alpes
- In partnership with:Université de Grenoble Alpes
- Team name: Analysis, engineering, and control of microorganisms
- Domain:Digital Health, Biology and Earth
- Theme:Modeling and Control for Life Sciences
Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.4.5. Bayesian methods
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.4. Statistical methods
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.4.1. Deterministic control
Other Research Topics and Application Domains
- B1. Life sciences
- B1.1.2. Molecular and cellular biology
- B1.1.4. Genetics and genomics
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B1.1.10. Systems and synthetic biology
- B2.2.4. Infectious diseases, Virology
- B4.3.1. Biofuels
1 Team members, visitors, external collaborators
Research Scientists
- Delphine Ropers [Team leader, INRIA, Senior Researcher, Adjunct professor UGA]
- Eugenio Cinquemani [INRIA, Senior Researcher]
- Aline Marguet [INRIA, Researcher]
- Hidde de Jong [INRIA, Senior Researcher]
Faculty Members
- Johannes Geiselmann [UGA, Emeritus, from Dec 2024]
- Johannes Geiselmann [UGA, Professor, until Nov 2024]
Post-Doctoral Fellows
- Arnaud Belcour [INRIA, Post-Doctoral Fellow]
- Claudia Fonte Sanchez [UGA, Post-Doctoral Fellow]
PhD Students
- Rand Asswad [INRIA]
- Ignacia Cancino Aguirre [INRIA, until Nov 2024]
- Thibault Clavier [UGA, until Jun 2024]
- Eugene Ferragu [INRIA, from Oct 2024]
- Charles Medous [UGA]
- Emrys Reginato [LEAP Saint Gabriel Nantes Océan, from Apr 2024]
- Emrys Reginato [INRIA, until Mar 2024]
Technical Staff
- Soraya Arias [INRIA, Engineer]
- Ludovic Leau-Mercier [INRIA, Engineer]
Interns and Apprentices
- Eugene Ferragu [INRAE, Intern, from Apr 2024 until Aug 2024]
- Eugene Ferragu [INRIA, Intern, until Mar 2024]
- Gerardo Trimmer Lopez [INRIA, Intern, from May 2024 until Jul 2024]
Administrative Assistant
- Diane Courtiol [INRIA]
External Collaborators
- Muriel Cocaign-Bousquet [INRAe, Toulouse Biotechnology Institute, HDR]
- Antrea Pavlou [ETH ZURICH]
- Natale Scaramozzino [CNRS]
2 Overall objectives
MICROCOSME combines computational and experimental approaches for the analysis, engineering, and control of the growth of microorganisms. Understanding and controlling the dynamics of bacterial growth is vitally important in health, medicine, biotechnology, and food industries, for instance to halt the growth of pathogens or stimulate the growth of probiotics or industrial microorganisms.
We develop multiscale models of growth, where the macroscopic observable, growth of a microbial population or community, depends on various metabolic pathways and regulatory mechanisms operating at microscopic scales within the cells. We use our (deterministic or stochastic) models to interpret experimental data or to infer the underlying growth processes from the data. This requires developing a platform for the automation of experiments, as well as methods and software for model estimation and data analysis. The analysis of microbial growth calls for new methodologies at the interface of microbiology, control theory, applied mathematics, computer science, biophysics, and molecular biology, which also leads to contributions in all of these fields. Our workhorse for the realization of this research program is the bacterium Escherichia coli pictured in Figure 1. Part of the microbiota of the human gut, E. coli is the model organism par excellence in microbiology and a popular platform for bio-based chemical production. We intend to extend approaches developed in-house for this specific microbe to other microorganisms including pathogens.
MICROCOSME has been created on October 1st, 2021. A recomposition and follow-up of the former IBIS project-team, MICROCOSME joins researchers from Inria and the Laboratoire Interdisciplinaire de Physique (CNRS UMR 5588) at Université Grenoble Alpes.
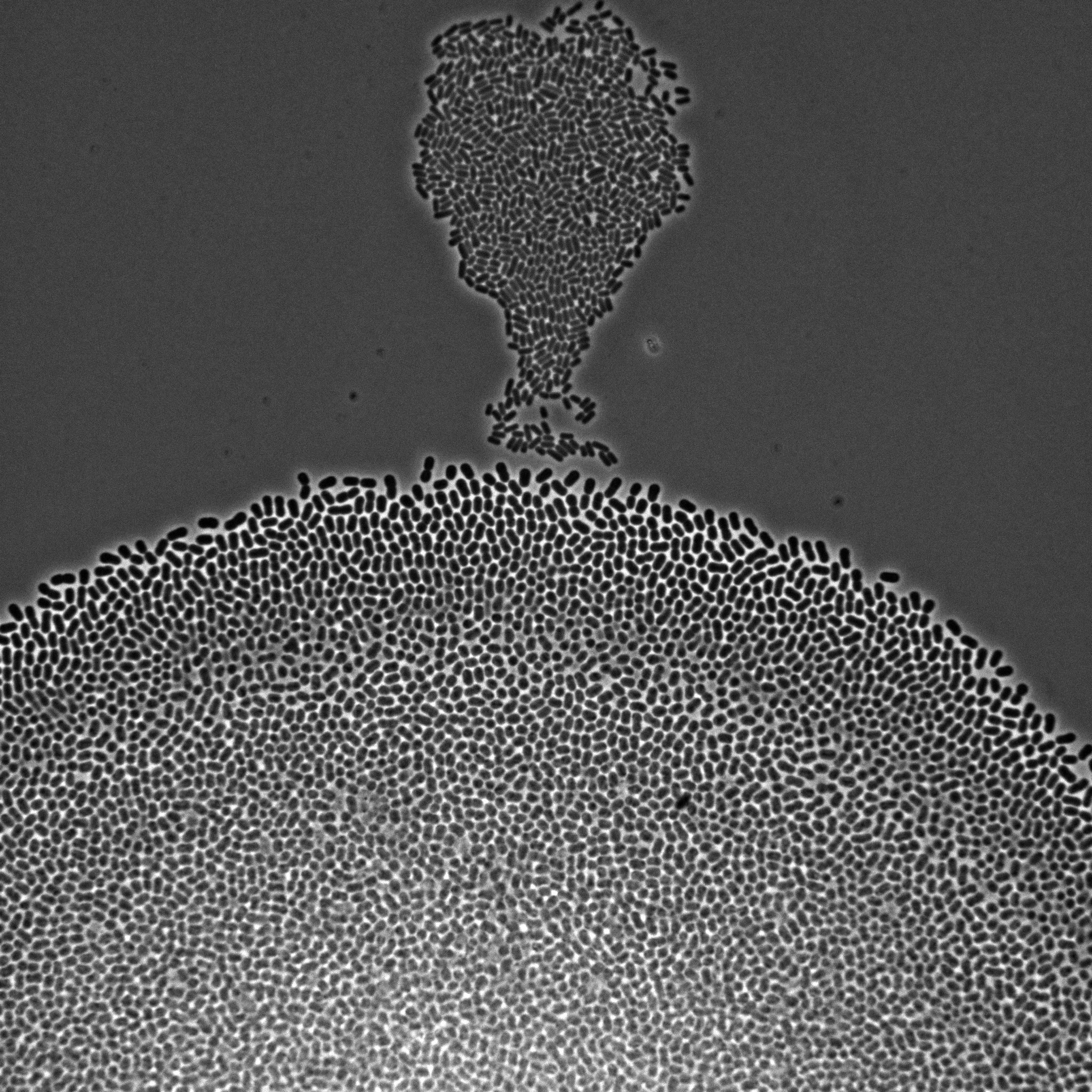
Figure 1: Microscopy image of Escherichia coli bacteria growing on a solid nutrient medium.
3 Research program
The research program of MICROCOSME is articulated around four research axes combining theory and experiments, which are illustrated in Figure 2 and detailed below.
3.1 Genome-scale analysis of microbial physiology
The molecular foundations of bacterial growth remain little understood today, because they involve large biochemical networks with physical and regulatory interactions across different levels of cellular organization. We investigate at the genome scale how the dynamics of gene expression and metabolism leads to microbial growth, using a combination of mathematical models and high-throughput data. The challenge is to integrate, in models of thousands of equations, multiple and heterogeneous datasets on the metabolic, transcriptomic, and proteomic level. We typically use constraint-based models to investigate the relations between microbial growth and metabolism, while the effect of growth on mRNA stability is analysed by means of non-linear mixed-effect models.
3.2 Natural and engineered resource allocation strategies in microorganisms
Microorganisms have evolved strategies to allocate their resources to different cellular functions and thus adjust their growth rate to fluctuating environments. We study these natural resource allocation strategies, by viewing cells as self-replicators that can be described using coarse-grained models and analysed by means of optimal and feedback control theory. The models take the form of systems of 5-10 nonlinear ordinary differential equations, with parameters estimated from published data or data obtained from dedicated experiments. Experimental work in the lab allows to validate model predictions on the single-cell and population level and to engineer new strategies for the reallocation of cell resources from growth to bioproduction.
3.3 Variability and robustness of microbial adaptation
The development of experimental techniques and the use of video-microscopy have led to a growing number of high-quality data showing the heterogeneity among cells in a population. We combine these single-cell data with models describing the stochastic dynamics of individual cells, such as birth-death processes, branching processes, and mixed-effect models. The models allow to investigate the origins of heterogeneity and its role in the adaptation of microorganisms to environmental changes, and to leverage population heterogeneity for biotechnological applications. In practice, this requires the extension of modelling approaches by taking into account the specificities of heterogeneity, as well as the development of appropriate methods and software for the inference of models and of biological quantities from quantitative time-course profiles of the microbial response to environmental changes.
3.4 Analysis and control of microbial communities
Heterogeneity also arises within communities consisting of different microbial species. Understanding microbial interactions is a challenging task that goes well beyond the characterization of single species, and offers great opportunities for applications, such as the control of the community for bioproduction. Indeed, suitably constructed microbial consortia carry the potential to outperform single species in the accomplishment of processes of societal interest, such as biofuel synthesis. On the theoretical side, we develop (deterministic or stochastic) models of microbial dynamics similar to those in the three other research axes, which can be used to investigate new control approaches for microbial communities. On the experimental side, the application of control strategies for biotechnological applications requires the engineering of microbial strains and the automation of experiments. To that aim we have been developing a platform for feedback control experiments allowing the real-time monitoring, data processing, evaluation, and application of control laws.
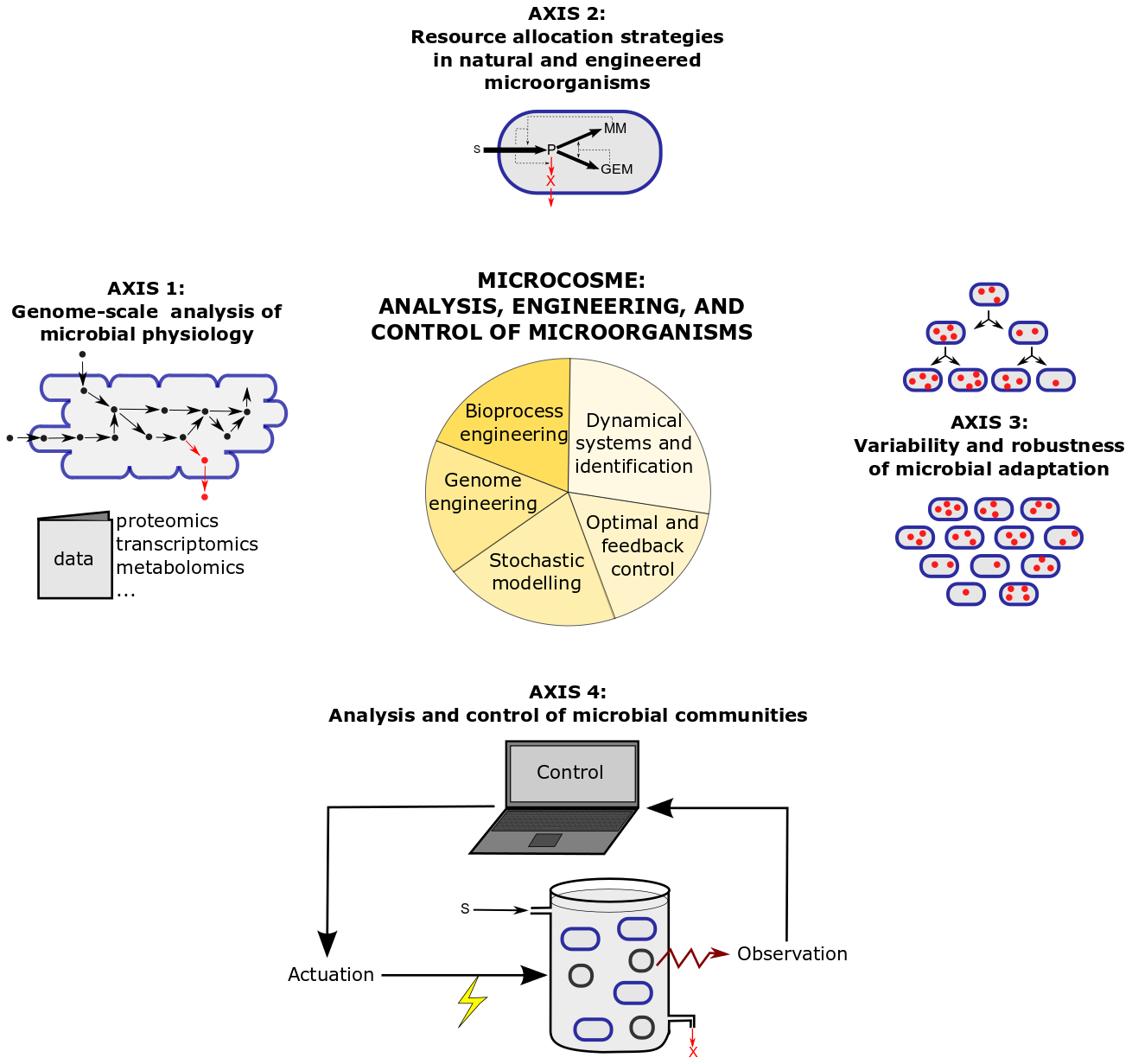
Figure 2: Research axes and methods in the MICROCOSME project-team.
4 Application domains
The research agenda of MICROCOSME is interdisciplinary in nature, driven by fundamental questions in biology, which we address by a combination of mathematical, computational, and experimental tools. This enables us to develop and share with partners a know-how useful to address challenging problems in health, bioeconomy, biotechnology, and environmental microbiology.
4.1 Biotechnology and bioeconomy
Bioproduction imposes a strong metabolic burden on microorganisms, detrimental to their growth and the production yield. Our studies of natural resource allocation strategies lead us to explore and engineer various reallocation strategies to improve bioproduction through growth control. For instance, in the past, we have successfully implemented a growth switch in E. coli bacteria, aiming at shuttling resources, away from protein synthesis (key for bacterial growth) to the high-yield production of a metabolite of interest (glycerol) 737. We also develop and test control strategies for synthetic microbial communities, composed of populations of different E. coli strains or in consortia with other species. In the wake of our studies on the relation between growth and metabolism, we develop bioeconomy strategies for the transformation of vegetal waste into value-added product.
4.2 Health
Numerous Mycobacteria species pose serious threats to human and animal health. Mycobacteria tuberculosis strains are also known to withstand several of the antibiotics used to treat the infection. We have started to extend our microbial physiology analyses by means of constraint-based models to understand the molecular control of mycobacterial growth and characterize the relations between metabolism, pathogenicity, and growth phenotype of mycobacterial species. This may lead, in the long term, to the development of new treatments for curing tuberculosis and other mycobacterial infections.
4.3 Environmental microbiology
Microbial subsurface ecology is poorly characterised. Current knowledge suggests that the subsurface is rich in microbial biodiversity, whose metabolic activity influences global biogeochemical cycles (e.g. carbon and nitrogen cycles). The metabolic potential of subsurface microbes can be inferred from high-throughput sequencing data, but this remains a difficult bioinformatics problem. We have started to extend our analyses of metabolism to the prediction of biogeochemical cycles from metabarcoding data. The approaches developed should help us to assess the microbial risk of underground hydrogen storage as part of our collaboration with BRGM and our partners in the European HyLife project.
5 Social and environmental responsibility
Several of our research activities have a direct societal impact. Our work on Mycobacteria addresses important questions of public health, while the project on the degradation and valorisation of vegetal waste meets European efforts in Circular Bioeconomy to replace fossil feedstock with renewable resources. Our recently accepted Clean Energy Transition Partnership project HyLife will allow us to address the issue of microbial risks associated with underground gas storage, as part of Europe's efforts to develop innovative energy system solutions towards net-zero by 2050.
6 Highlights of the year
Our paper “Single-cell data reveal heterogeneity of investment in ribosomes across a bacterial population” was accepted for Nature Communications and published early 2025 20. This work is the outcome of ten years of work in the framework of the ANR project Maximic, relating bacterial systems biology with (optimal) control theory, and has been the main subject of the PhD theses of Nils Giordano 34 and Antrea Pavlou 36.
7 New software, platforms, open data
7.1 New software
7.1.1 ODIN+
-
Name:
Platform for advanced monitoring, control and optimisation of bioprocesses
-
Keywords:
Systems Biology, Biotechnology, Automatic control, Monitoring
-
Functional Description:
This application proposes a framework for on-line supervision of bioreactors. It gathers the data sampled from different on-line and off-line sensors. ODIN+ is a distributed platform, enabling remote monitoring as well as remote data acquisition. More originally, it enables researchers and industrials to easily develop and deploy advanced control algorithms, optimisation strategies, together with estimates of state variables or process state. It also contains a process simulator which can be harnessed for experimentation and training purposes. It is modular in order to adapt to any plant and to run most of the algorithms, and it can handle the high level of uncertainties that characterises the biological processes. The architecture is based on Erlang, and communication between modules through a MQTT Broker with Python for running the algorithms. ODIN+ is developed in collaboration with the INRIA MICROCOSME research team.
-
News of the Year:
A diagnosis module was implemented to detect issues in the hardware or in the connection between the various modules. The calibration module was updated also allowing actuator calibration. Finally, the python module for managing the priorities accesses to the different elements of the platform was updated.
-
Contact:
Olivier Bernard
7.1.2 GNA
-
Name:
Genetic Network Analyzer
-
Keywords:
Model Checking, Bioinformatics, Gene regulatory networks, Qualitative simulation
-
Scientific Description:
Genetic Network Analyzer (GNA) is the implementation of methods for the qualitative modeling and simulation of gene regulatory networks developed in the IBIS (now MICROCOSME) project-team.
-
Functional Description:
The input of GNA consists of a model of the regulatory network in the form of a system of piecewise-linear differential equations (PLDEs), supplemented by inequality constraints on the parameters and initial conditions. From this information, GNA generates a state transition graph summarizing the qualitative dynamics of the system. In order to analyze large graphs, GNA allows the user to specify properties of the qualitative dynamics of a network in temporal logic, using high-level query templates, and to verify these properties on the state transition graph by means of standard model-checking tools, either locally installed or accessible through a remote web server.
-
Release Contributions:
(1) it supports the editing and visualization of regulatory networks, in an SBGN-compatible format, (2) it semi-automatically generates a prototype model from the network structure, thus accelerating the modeling process, and (3) it allows models to be exported in the SBML Qual standard.
- Publications:
-
Contact:
Hidde De Jong
-
Participants:
Hidde De Jong, Delphine Ropers
-
Partner:
UGA
7.1.3 WellInverter
-
Name:
WellInverter
-
Keywords:
Bioinformatics, Statistics, Data visualization, Data modeling
-
Scientific Description:
WellInverter is a web application that implements linear inversion methods for the reconstruction of gene expression profiles from fluorescent or luminescent reporter gene data. WellInverter makes the methods available to a broad audience of biologists and bioinformaticians. In particular, we have put in place a parallel computing architecture with a load balancer to distribute the analysis queries over several back-end servers, redesigned the graphical user interface, and developed a plug-in system for defining high-level routines for parsing data files produced by microplate readers from different manufacturers.
-
Functional Description:
As input, WellInverter reads the primary data file produced by a 96-well microplate reader, containing time-series measurements of the absorbance (optical density) as well as the fluorescence and luminescence intensities in each well (if available). Various modules exist to analyze the data, in particular for detecting outliers, subtracting background, estimating growth rates, promoter activities and protein concentrations, visualizing expression profiles, synchronizing replicate profiles, etc. The computational core of the web application consists of the Python library WellFARE.
- URL:
- Publications:
-
Contact:
Hidde De Jong
-
Participants:
Delphine Ropers, Hidde De Jong, Johannes Geiselmann
-
Partner:
UGA
7.1.4 WellFARE
-
Name:
WellFARE
-
Keywords:
Bioinformatics, Statistics, Data visualization, Data modeling
-
Scientific Description:
WellFARE is a Python library implementing linear inversion methods for the reconstruction of gene expression profiles from fluorescent or luminescent reporter gene data. WellFARE form the computational core of the WellInverter web application.
-
Functional Description:
As input, WellFARE reads the primary data file produced by a 96-well microplate reader, containing time-series measurements of the absorbance (optical density) as well as the fluorescence and luminescence intensities in each well (if available). Various functions exist to analyze the data, in particular for detecting outliers, subtracting background, estimating growth rates, promoter activities and protein concentrations, visualizing expression profiles, synchronizing replicate profiles, etc. WellFARE is the computational core of the web application WellInverter.
- URL:
- Publication:
-
Contact:
Hidde De Jong
-
Participants:
Delphine Ropers, Johannes Geiselmann, Hidde De Jong
-
Partner:
UGA
7.1.5 tabigecy
-
Keywords:
Taxonomies, Metabolism
-
Functional Description:
Analysis of microbial communities in their environment is crucial to understanding their environmental impact (e.g. on hydrogen or CO2 storage or the carbon cycle). Thanks to high-throughput sequencing, it is now possible to characterise microbial communities taxonomically. However, linking this taxonomic information to metabolic functions related to biogeochemical cycles (such as the carbon cycle) remains a challenging task. This has motivated the development of tabigecy, a Nextflow workflow that predicts metabolic functions linked to biogeochemical cycles using taxonomic affiliations. This workflow combines the tool EsMeCaTa to predict protein sequences from taxonomic affiliations with bigecyhmm to predict functions of biogeochemical cycles using protein sequences. tabigecy then produces multiple visualisations to facilitate the interpretation of the results. This makes it possible to identify the impact of the microbial community studied on carbon, nitrogen, and sulfur cycles.
-
Release Contributions:
First public release of the software.
- URL:
-
Contact:
Delphine Ropers
7.1.6 bigecyhmm
-
Keywords:
Proteins, Metabolism
-
Functional Description:
The Python package bigecyhmm is part of the tabigecy workflow. It aims at predicting metabolic functions linked to biogeochemical cycles using protein sequences as input. Hidden-Markov models associated with metabolic functions are used to search the input proteins. This makes it possible to identify the impact of the microbial community studied on carbon, nitrogen, and sulfur.
-
Release Contributions:
First public release of the software.
-
News of the Year:
First release of the software.
- URL:
-
Contact:
Delphine Ropers
7.2 New platforms
Participants: Soraya Arias, Eugenio Cinquemani, Johannes Geiselmann, Ludovic Léau-Mercier.
7.2.1 Automated mini-bioreactor platform for (dynamical) monitoring and control of microbial cultures
Advanced dynamical experiments with microbial cultures require regular, complex measurement operations over several days or weeks. Manual execution by human operators is error-prone and exposed to weak reproducibility, besides being a poor utilisation of human resources. Reactive control experiments, in addition, necessitate online calculation of control actions in response to all acquired measurements. MICROCOSME is actively developing an automated platform for automated monitoring and reactive control experiments on microbial cultures. The platform consists of a system of mini-bioreactors connected to nutrient sources and measurement devices via a pump-based fluidic network, and it also supports optogenetic control. Computer-operated by software ODIN+ (Section 7.1) as well as via platform-specific software developments, it enables automated monitoring and online data processing, as already achieved in week-long experiments over several bioreactors 38, and it will be exploited for feedback control experiments as part of the ongoing (Section 9) and future research projects of the group.
7.3 Open data
Estimation of ribosome synthesis rate profiles from single-cell microfluidics data
-
Contributors:
Hidde de Jong , Eugenio Cinquemani , Antrea Pavlou
-
Description:
Estimation code, microfluidics dataset, and plotting script accompanying the Nat. Commun. paper of this year.
-
Dataset PID (DOI,...):
DOI 10.24433/CO.6310888.v1
- Project link:
- Publications:
-
Contact:
Hidde de Jong
-
Release contributions:
Hidde de Jong , Eugenio Cinquemani , Antrea Pavlou
EsMeCaTa precomputed database
-
Contributors:
Arnaud Belcour , Loris Mégy , Hidde de Jong , Delphine Ropers
-
Description:
EsMeCaTa 30 is a software application to infer consensus proteomes and metabolic functions from taxonomic affiliations. EsMeCaTa uses ETE3 and the NCBI Taxonomy database to parse the taxonomic affiliations and query the UniProt Proteomes database to find associated proteomes. These proteomes are clustered using MMseqs2 to create consensus proteomes, which are then annotated with eggNOG-mapper. EsMeCaTa can be time-consuming to run and requires a large number of resources to perform its various steps. A precomputed database has been created to facilitate its use.
-
Dataset PID (DOI,...):
DOI 10.5281/zenodo.13354072
- Project link:
-
Publications:
Paper under submission
-
Contact:
Arnaud Belcour , Delphine Ropers
-
Release contributions:
Arnaud Belcour , Loris Mégy , Hidde de Jong , Delphine Ropers
8 New results
8.1 Quantifying bacterial resource allocation on the single-cell level
Participants: E. Cinquemani, H. de Jong, J. Geiselmann, A. Pavlou.
Microbial growth involves the conversion of nutrients from the environment into biomass. The main component of biomass are proteins, which also play a major role in the synthesis of new biomass by functioning as enzymes in metabolism and by constituting the molecular machinery responsible for the synthesis of proteins and other macromolecules. Microbial growth therefore requires the coordinated investment of cellular resources in different categories of proteins. Ribosomes are probably the most important protein category for two reasons. First, they are responsible for the synthesis of all proteins in the cell. Second, they are themselves very costly to make: ribosomes constitute up to 40-50% of the total protein mass in Escherichia coli.
Very few studies have addressed the quantification of ribosomal resource allocation on the single-cell level. How do the resources allocated to the synthesis of ribosomal proteins, both during balanced and unbalanced growth, vary over the individual cells of an isogenic population? In order to answer this question, in the framework of the PhD thesis of Antrea Pavlou 36 and the ANR project Maximic (2017-2023), we constructed chromosomal reporter systems for monitoring ribosome expression in the model organism Escherichia coli. We measured the ribosome concentration in individual cells growing on a rich or a poor carbon source, as well as changes in ribosome concentration during upshifts and downshifts between these carbon sources, over extended periods of time (>80 generations). Moreover, we developed a method for the statistical inference of time-varying ribosome synthesis rates from the single-cell, time-course data thus acquired.
We found that, during balanced growth in a given medium, the bacteria display a wide variety of ribosome concentrations that are only weakly correlated with the single-cell growth rate. This would not be expected if bacteria had optimized costly ribosome expression to precisely match the protein synthesis rate required for a certain growth rate. During the upshift from a poor to a rich carbon source, we observed that cells with a higher pre-shift ribosome concentration more rapidly adapt their ribosome synthesis rate, but also the ribosome synthesis activity and the growth rate, to the new environment. We remark that these observations are consistent with the existence of a variable ribosome reserve which the bacterial cells may exploit to speed up adaptation to sudden changes in the environment.
In this study published in Nature Communications20, we thus quantified, using a combination of reporter genes and statistical inference algorithms, dynamic investment in ribosomes on the single-cell level. The results reveal a surprising variability in the allocation of resources to ribosomes, the most costly molecular machine in bacterial cells, during both balanced and unbalanced growth. This raises fundamental questions on the role of the variability of ribosome concentrations in shaping the growth of a bacterial population and its adaptation to changing environments. Given the importance of growth and adaptation in biomedical and biotechnological applications, we expect our findings to have practical implications as well.
8.2 Biotechnological applications of bacterial growth control
Participants: H. de Jong, J. Geiselmann, T. Clavier, D. Ropers.
The ability to experimentally control the growth rate is crucial for studying bacterial physiology. It is also of central importance for applications in biotechnology, where often the goal is to limit or even arrest growth. Growth-arrested cells with a functional metabolism open the possibility to channel resources into the production of a desired metabolite, instead of wasting nutrients on biomass production. In recent years we obtained a foundational result for growth control in bacteria 7, in that we engineered an E. coli strain where the transcription of a key component of the gene expression machinery, RNA polymerase, is under the control of an inducible promoter. By changing the inducer concentration in the medium, we can adjust the RNA polymerase concentration and thereby switch bacterial growth between zero and the maximal growth rate supported by the medium. The publication also presented a biotechnological application of the synthetic growth switch in which both the wild-type E. coli strain and our modified strain were endowed with the capacity to produce glycerol when growing on glucose. Cells in which growth has been switched off continue to be metabolically active and harness the energy gain to produce glycerol at a twofold higher yield than in cells with natural control of RNA polymerase expression, putting the yield very close to the theoretical maximum.
In the framework of the PhD thesis of Thibault Clavier, defended in March 2024 28, we addressed one of the limits of the growth switch that is inherent to the construction of synthetic networks in living microorganisms, namely that the latter evolve over time under the pressure of natural selection. In the case of the growth switch, the selection pressure is particularly high, since any spontaneously arising mutations disabling the inhibition of the expression of the RNA polymerase subunits will cause the population of growth-arrested cells to be taken over by cells that have resumed growth (at the expense of metabolite production). We improved the genetic stability of the growth switch by means of a redundant control mechanism of RNA polymerase expression, reducing the escape frequency to less than one in cells. We deposited a patent of this invention. An article describing the results obtained with the extended growth switch was published in Biotechnology and Bioengineering 16. Moreover, this work is the subject of the start-up project Switch2Prod, led by Thibault Clavier and to be submitted to the SATT Linksium.
8.3 Synthetic microbial communities for bioproduction processes: modelling, analysis and real-time monitoring
Participants: S. Arias, R. Asswad, E. Cinquemani, T. Clavier, H. de Jong, J. Geiselmann, L. Léau-Mercier.
Modelling, analysis and control of microbial community dynamics is a fast-developing subject with great potential implications in the understanding of natural processes and the enhancement of biotechnological processes. Within the now-ended project IPL COSY, we picked up the challenge to design and investigate the dynamics of synthetically engineered microbial communities with a consortium of Inria partners, and to test control strategies in vivo.
In MICROCOSME, we have addressed the design of a bacterial community of two E. coli strains, mimicking mutualistic relationships found in nature, and with the potential to outperform a producer strain working in isolation in the production of a heterologous protein. We previously developed an ODE model of the consortium, and analysed the model to characterize the conditions supporting coexistence and the tradeoffs involved in the production process 11, 35. The engineering of the consortium and its experimental characterization revealed a more complex picture as witnessed by partially unexpected results 38, which led us to formulate new hypotheses on the interactions among species and their intertwining with the E. coli overflow metabolism. The experimental scrutiny of these hypotheses and the achievement of stable coexistence with the studied consortium are object of an ongoing effort that involves several team members and that will be made value of by a journal paper submission. A subsequent aim for this research direction is the feedback control of the community for the robust stabilization of biomasses into the most desirable bioproduction regimes.
On the methodological side, in collaboration with BIOCORE, modelling, analysis and control problems are being addressed with the Ph.D. thesis of Rand Asswad. A simple ODE model of the above algal-bacterial consortium was developed based on literature knowledge, also integrating data from the partners for algal growth parameters. With reference to this model, toward model-based feedback control of the consortium, we first focused on the problem of real-time state estimation of microbial growth in a bioreactor. Several different Kalman-based approaches, tackling the challenge of the model nonlinearity by suitable “quasi-linear” problem reformulations, have been proposed in a paper that has been presented at and published as part of the proceedings of the 22nd European Control Conference (ECC 2024, Stockholm, Sweden) 24. Focused on a single species, the methods will be generalized and integrated into adaptive feedback control strategies currently being explored. We next focused on the problem of optimal productivity of algal biomass. We explored optimal control problems from a static and a dynamic viewpoint, and obtained intriguing results on the convexity of the static problem, and on the spontaneous onset of oscillatory control laws in the dynamic case leading to over-yielding. These results were presented at and published in the proceedings of the 63rd IEEE Conference on Decision and Control (CDC 2024, Milan, Italy) 25. A more complete set of results and a broader investigation are being arranged in the form of a journal paper to be submitted in the near future.
8.4 Modelling and inference of cellular metabolism
Participants: A. Belcour, I. Cancino-Aguirre, M. Cocaign-Bousquet, H. de Jong, D. Ropers.
Microorganisms are regularly exposed to environmental perturbations. This requires an adaptation of the microbial metabolism to thrive in new environments. In order to study the mechanisms of metabolic adaptation, we use genome-scale reconstructions of cellular metabolism, such as in 13. These networks are reconstructed using genomic annotations, in which genes encoding enzymes are linked to metabolic reactions. Such reconstructions are often available in public databases for well-studied microorganisms, but this is not the case for poorly studied species. As part of Ignacia Cancino Aguirre 's PhD thesis co-advised by Delphine Ropers and Hidde de Jong and the associate-team GERM (Section 9), we aim to develop such reconstructions from genome sequences for the Mycobacterium genus. This will allow us to analyse how differences in carbon metabolism account for the variability in growth rates of mycobacterial species, which include both dangerous pathogens and non-pathogenic bacteria. The results of this study are currently being prepared for publication.
Inferring metabolic function from sequenced genomes is a difficult problem, but even more so when dealing with microbial communities in natural environments. In collaboration with BRGM, the French geological survey, NORCE (Norway) and Isodetect Gmbh (Germany) in the framework of the European project HyLife (Section 9), Arnaud Belcour , Hidde de Jong , and Delphine Ropers have begun to address this issue. They developed a bioinformatics pipeline, Tabigecy (7.1.5), to use metabarcoding data in order to predict metabolic functions that make up biogeochemical cycles. A paper describing the approach has been submitted for publication. Tabigecy uses the tool EsMeCaTa to infer consensus proteomes and metabolic functions from taxonomic affiliations. EsMeCaTa is described in a recently submitted paper 30. This software can be time-consuming to run, which prompted us to develop a precomputed database of EsMeCaTa (Section 7.3) using the Gricad infrastructure supported by the Grenoble research community, in collaboration with Loris Mégy (Gricad, Inria, CNRS, Université Grenoble Alpes, Grenoble INP). As described in the proceedings of the Solution Mining Research Institute Spring conference (SMRI 2024, 23) and of the annual conference of the European Association of Geoscientists & Engineers (85th EAGE, 26), the next step will be to apply the pipeline Tabigecy to subsurface microbial communities to characterise their metabolic activity and possible detrimental effects on gas storage in underground reservoirs.
The functional predictions returned by the EsMeCaTa tool open up the possibility of classifying metabolic functions. Arnaud Belcour studied this question in collaboration with the Inria project-team DYLISS, the laboratory NuMeCan (INRAe, INSERM) and CHU Rennes. They developed an automated pipeline called SPARTA (Shifting Paradigms to Annotation Representation from Taxonomy to identify Archetypes) to discriminate between unhealthy and control samples in the study of the gut microbiome 21. An evolutionary perspective on metabolism was then adopted in a recent study 22. In this paper, Arnaud Belcour and his co-authors analysed possible mechanisms leading to metabolic plasticity in plant and algal lipid biosynthesis pathways.
8.5 Inference of parameters on lineage trees
Participants: E. Cinquemani, C. Fonte Sanchez, A. Marguet, E. Reginato.
Recent technological developments have made it possible to obtain single-cell measurements of gene expression and, in some cases, the associated lineage information. However, most of the existing methods for the identification of mathematical models of gene expression do not account for the fact that cells undergo divisions and are related to one another through parental relationships. Most methods developed for single-cell data make the simplifying assumptions that cells in a population are independent, thus ignoring cell lineages. The development of statistical tools taking into account the correlations between individual cells is needed in particular to enable the investigation of inheritance of traits in bacterial populations.
With the ending Ph.D. project of E. Reginato, we have advanced in the study of tree-structured single-cell gene expression models with mother-daughter inheritance that we had started in a previous publication 10. Contrary to this previous work, where model inference methods from single-cell gene expression data were developed assuming knowledge of the lineage of the observed cells, the thesis project has been focused on the case where lineage information is not available. We notably explored how well inheritance model parameters can be inferred depending on absence or presence of dynamics in the mean and variance data. In relation with certain literature datasets obtained by videomicroscopy, we developed statistically exact maximum-likelihood estimation methods leveraging correlation of empirical means along generations, and approximate methods also exploiting variance data that we showed to perform well even in absence of transient mean dynamics. Results have been presented at and published in the proceedings of the 22nd European Control Conference (ECC 2024, Stockholm, Sweden) 27. Demonstration on the reference datasets proved promising, however adaptations will be necessary for best exploiting real datasets.
While the above modelling approach to mother-daughter inheritance is of statistical nature, it can be related with mechanisms into play at cell division. For this, within the same Ph.D., we started looking at the regulation of the repartition of multicopy resistance plasmids at cell division and its impact on population growth in selective media. We have developed and compared several individual-based models, and obtained a first understanding of the role of plasmid repartition statistics and stochastic cell division in the emerging population dynamics. The study is being taken further with thorough simulation-based and mathematical characterization of the individual-based population model dynamics across selective and non-selective media, and comparison with real data.
Related to the above studies, Claudia Fonte Sanchez 's postdoc across projects AnaComBa (Persyval) and IMOCEP (PEPR MathVives; Section 9) is looking at individual-based models of population growth and gene expression, and the investigation of the relation between growth-rate variability across cells and gene expression distribution in population-snapshot data. A first set of results on the nonparametric statistical estimation of single-cell protein synthesis kernels from stationary distributions is being arranged in the form of a paper, for journal submission in the near future. Model-based analysis of population dynamics and identifiability of protein synthesis, cell division and repartition kernels from transient snapshot data will further be addressed, along with comparison with experimental data, for a possible second publication from this activity.
8.6 Mathematical analysis of structured branching populations
Participants: E. Ferragu, C. Fonte-Sanchez, Ch. Medous, A. Marguet.
The investigation of cellular populations at a single-cell level has already led to the discovery of important phenomena, such as the occurrence of different phenotypes in an isogenic population. Nowadays, several experimental techniques, such as microscopy combined with the use of microfluidic devices, enable one to take investigation further by providing time-profiles of the dynamics of individual cells over entire lineage trees. The development of models that take into account the genealogy is an important step in the study of inheritance in bacterial population. In particular, their mathematical analysis is essential for the efficient analysis of single cell data.
Structured branching processes allow for the study of populations, where the lifecycle of each cell is governed by a given characteristic or trait, such as the internal concentration of proteins. The dependence on this characteristic of cellular mechanisms, like division or ageing, has been explored by Aline Marguet via the mathematical analysis of these processes. In collaboration with Charline Smadi (INRAE Grenoble), Aline Marguet investigated the long-time behavior of a parasite infection in a cell population. In this work, published in Stochastic Processes and their Applications 18, the dynamics of the cell population is modeled using a structured branching process, where the cell cycle depends on the dynamics of the parasites contained in the cell. The results obtained focus on the asymptotic behavior of the intracellular quantity of parasites. Based on criteria that depend on the comparison between the growth rate of the cell population and the parameters associated with the parasite dynamics, containment or explosion of the infection is established. The strategy of proof relies on the study of auxiliary processes that describe the level of infection of a typical cell in the population, using coupling techniques.
The fate of the infection appears to be very sensitive to the law of parasite sharing between the two daughter cells as they divide. In particular, the effect of different sharing strategies on the long-term behavior of the infection in the case of a constant division rate was investigated in a follow-up paper published in the Journal of Mathematical Biology 17. Perfectly symmetric sharing of the parasites between the two daughter cells proved to be the worst strategy for the survival of the cell population. Stochastic and deterministic strategies were also compared, highlighting that variability favors survival. Finally, we proved that there is a partitioning kernel that allows the cell population to survive the infection, regardless of the strength of the infection. The proofs are based on a spinal decomposition, which allows us to study the behavior of the whole population through the fate of a well-chosen lineage.
Spinal processes and many-to-one formulas have proved very useful for the study of complex structured branching processes, as they allow to reduce the problem to the study of a simpler lineage process. In the context of the project AnaComBa (Section 9), for the study of microbial communities, such tools appear to be needed for structured branching processes with interactions and were developed by Charles Medous during his PhD, defended in December 2024 29. Charles Medous established a spinal construction and a Girsanov-type result for branching processes describing structured, interacting populations in continuous time, where the dynamics of each individual can be influenced by the entire population. He also derived a modified continuous-time version of the Kesten-Stigum theorem that incorporates interactions, and proposed an alternative simulation approach for stochastic size-dependent populations using appropriate spine constructions. An article based on this work has been published in the Electronic Journal of Probability 19. In collaboration with Charline Smadi and Sylvain Billiard (Université de Lille), Charles Medous also extended the spinal construction to diffusive population to study the role of environmental noise in growing colonies. They have proved that the repartition of the population depends crucially on the comparison between the individual and the environmental noise. The results of this study are currently being prepared for publication. Charles Medous also used spine techniques to propose an exact simulation algorithm for individual-based population models. He provided many applications of this algorithm as well as its Python implementation. A paper on this new simulation algorithm is also in preparation.
The study of the asymptotic behavior of general semigroups is important for several aspects of branching processes, especially to prove the efficiency of statistical procedures. In this context, Claudia Fonte Sanchez , in collaboration with Pierre Gabriel (Université de Tours) et Stéphane Mischler (Université Paris-Dauphine) revisited the Krein-Rutman theory for semigroups of positive operators and provided some very general, efficient and practical results with constructive estimates on the existence of a solution to the first eigentriplet problem, the geometry of the principal eigenvalue problem, and the asymptotic stability of the first eigenvector with possible constructive rate of convergence 31.
9 Partnerships and cooperations
9.1 International initiatives
9.1.1 Inria associate team not involved in an IIL or an international program
GERM
Participants: A. Belcour, I. Cancino-Aguirre, H. de Jong, D. Ropers.
-
Title:
Growth-rate control in mycobacteria: Computational exploration of metabolic strategies
-
Duration:
2022 - 2024
-
Coordinator:
Delphine Ropers
-
Partners:
- Francis Crick Institute (Royaume-Uni)
-
Inria contact:
Delphine Ropers
-
Summary:
Numerous Mycobacterium species pose serious threats to human and animal health. Genome-scale mathematical models of Mycobacterium metabolism are promising avenues to uncover bottlenecks explaining the growth rate variability and pathogenicity observed across the genus. We will employ these models to integrate and analyze diverse types of experimental data, including measurements of doubling times and metabolite concentrations. The results will allow us to formulate hypotheses on molecular mechanisms responsible for growth rate variability and pathogenicity observed across mycobacteria. The hypotheses will be tested by targeted experiments.
9.1.2 Informal international partners
H. de Jong and D. Ropers collaborate with T. Gedeon (Montana State University), former invited researcher in our former team IBIS, on research allocation strategies in microorganisms. The collaboration has already resulted in a paper published in eLife in 2023 32.
9.1.3 Visits to international teams
Research stays abroad
Ignacia Cancino Aguirre
- Visited institution: Francis Crick Institute
- Country: United Kingdom
-
Dates:
03/01/2024 - 09/02/2024
-
Context of the visit:
visit to the Carvalho's laboratory in the context of the associate-team GERM
-
Mobility program/type of mobility:
research stay
Rand Asswad
- Visited institution: Institute of Automatic Control (IRT) - Leibniz University Hannover
- Country: Germany
-
Dates:
30/08/2024 - 30/11/2024
-
Context of the visit:
visit to Internship in the group of Prof. Dr.-Ing. Matthias Müller for the "Development of data-driven control strategies for synthetic microbial consortia"
-
Mobility program/type of mobility:
(German Academic Exchange Service), with additional funding from the outgoing international mobility program of IDEX Formation, Université Grenoble Alpes
9.2 European initiatives
9.2.1 Horizon Europe
| Project name | HyLife: Optimal control of microbial cells by natural and synthetic strategies |
| Coordinator | N. Dopffel (NORCE, Norway) |
| MICROCOSME participants | A. Belcour , H. de Jong , D. Ropers |
| Type | Clean Energy Transition Co-funded Partnership (CETP; 2023-2026) |
| Web page | Link to project description |
9.3 National initiatives
| Project name | Ctrl-AB : Optimization and control of the productivity of an algal-bacterial consortium |
| Coordinator | J.-L. Gouzé |
| MICROCOSME participants | R. Asswad , S. Arias , E. Cinquemani , Th. Clavier, H. de Jong , J. Geiselmann, L. Léau-Mercier, A. Marguet |
| Type | ANR project (2020-2025) |
| Web page | Link to project description |
| Project name | PlugNBio: A plug-and-play platform for reproducible microbial culture control experiments |
| Coordinator | E. Cinquemani |
| MICROCOSME participants | S. Arias, E. Cinquemani , J. Geiselmann, L. Léau-Mercier |
| Type | Inria ADT (2022-2024) |
| Project name | ARBOREAL: Branching resource allocation processes for the analysis and inference of phenotypic growth variability |
| Coordinator | A. Marguet |
| MICROCOSME participants | E. Cinquemani , J. Geiselmann, H. de Jong , A. Marguet |
| Type | ANR (2024-2029) |
| Web page | Link to project description |
| Project name | RECOM: Competition of RNAs for RNase E, a mechanism regulating their degradation and the energy and carbon metabolism in the cell |
| Coordinator | M. Cocaign-Bousquet |
| MICROCOSME participants | E. Cinquemani , M. Cocaign-Bousquet , D. Ropers |
| Type | ANR (2023-2027) |
| Web page | Link to project description |
| Project name | IMOCEP: Innovations for modeling of growth : from a cellular level to pediatric development. |
| Coordinator | A. Leclercq Samson, J. Stirnemann |
| MICROCOSME participants | E. Cinquemani , C. Fonte-Sanchez, A. Marguet |
| Type | ANR, PEPR Mathématiques en interaction, pour le vivant, l'environnement et la société (MathVives; 2024-2029) |
| Web page | Link to project description |
The following project has just been accepted and will start in spring 2025:
| Project name | MuSiHC: Multi-size Hybrid Cell Models |
| Coordinator | A. Tonda (INRAE, Palaiseau) |
| MICROCOSME participants | E. Cinquemani , H. de Jong , N. Scaramozzino |
| Type | ANR, PEPR Biomasses, biotechnologies et technologies durables pour la chimie et les carburants (B-BEST; 2025-2029) |
| Web page |
In addition to the above projects, A. Marguet contributes to the ANR JCJC NOLO of Bertrand Cloez (INRAE Montpellier) on non-local branching processes.
9.4 Regional initiatives
| Project name | AnaComBa: Analyse de Communautés Bactériennes : modélisation stochastique |
| Coordinator | A.Marguet & L. Coquille |
| MICROCOSME participants | E. Cinquemani , C. Fonte Sanchez , A. Marguet , C. Medous |
| Type | Equipe-Action du LABEX Persyval (2021 – 2024) |
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
Member of organizing committees
| MICROCOSME members | Conference, workshop, school | Date |
| Ignacia Cancino Aguirre | Inria PhD seminar, Montbonnot | 2024 |
| Hidde de Jong | Summer school on Economic Principles in Cell Physiology, Paris | Jul 2024 |
| Aline Marguet | GDT Math-Bio Sud-Est, Lyon-Grenoble | 2024 |
10.1.2 Scientific events: selection
Chair of conference program committees
| MICROCOSME member | Conference, workshop, school | Role |
| Eugenio Cinquemani | European Control Conference (ECC2024, ECC2025) | Associate editor |
| Eugenio Cinquemani | Special Issue of the 19th International Conference on Computational Methods in Systems Biology. BMC Bioinformatics Supplements, 24(1) 33 | Supplement editor (with Loïc Paulevé) |
Member of conference program committees
| MICROCOSME member | Conference, workshop, program |
| Eugenio Cinquemani | CMSB 2024 |
| Hidde de Jong | ECCB 2024 Proceedings program committee |
| Hidde de Jong | FOSBE 2024 |
| Delphine Ropers | ECCB 2024 Proceedings program committee |
| Delphine Ropers | BiGre Days program committee |
10.1.3 Journal
Member of editorial boards
| MICROCOSME member | Journal |
| Hidde de Jong | Journal of Mathematical Biology |
10.1.4 Invited talks and other presentations
Rand Asswad
| Title | Event and location | Date |
| Kalman-based approaches for online estimation of bioreactor dynamics from fluorescent reporter measurements | ECC 2024, Stockholm, Sweden | Jun 2024 |
| Optimization of microalgae biosynthesis via controlled algal-bacterial symbiosis | CDC 2024, Milan, Italy | Dec 2024 |
Eugenio Cinquemani
| Title | Event and location | Date |
| Single-cell data reveal heterogeneity of resource allocation across a bacterial population | 6th BactoGre Scientific Day, St Martin d'Hères | Jun 2024 |
| Inference of tree-structured auto-regressive models of gene expression parameters from generation-snapshot data | ECC 2024, Stockholm, Sweden | Jun 2024 |
Hidde de Jong
| Title | Event and location | Date |
| Modeling gene regulatory networks: from transcription factors to global cell physiology | Invited talk Frontiers in Mathematical Modeling: Celebrating Roderick Edwards' Contributions, Victoria, Canada (remote) | Feb 2024 |
| Understanding microbial growth by means of simple models and experiments | Lab seminar Laboratoire Interdisciplinaire de Physique (LIPhy) | Apr 2024 |
Claudia Fonte Sanchez
| Title | Event and location | Date |
| On the voltage conductance kinetic model | Kinetic Equation, Mathematical Physics and Probability Conference, Bilbao (Spain) | Jul 2024 |
| On the consequences of the Krein-Rutman theorem for growth-fragmentation equations | Non-local Branching Processes Conference, Marseille | Sept 2024 |
| Modeling the relationship between cell growth and gene expression | Séminaire Merge, Palaiseau | Nov 2024 |
| On the asymptotic behavior of weakly connected neuronal networks | Séminaire Modélisation, Analyse et Calcul, Toulouse | Nov 2024 |
Aline Marguet
| Title | Event and location | Date |
| Stochastic gene expression - Modelling and inference from lineage trees | Séminaire virtuel du GT BIOSS | Jan 2024 |
Delphine Ropers
| Title | Event and location | Date |
| Doing a PhD, good practice and pitfalls to avoid | Matinée des doctorants, Centre Inria de l'Université Grenoble Alpes | Oct 2024 |
| Model-based interpretation of high-throughput biological data to understand RNA degradation | Lab seminar Laboratoire Interdisciplinaire de Physique (LIPhy) | Sept 2024 |
10.1.5 Scientific evaluation and expertise
| MICROCOSME member | Organism | Role |
| Eugenio Cinquemani | Inria - Univ Grenoble Alpes | Membre du comité Concours idées innovantes 2024 |
| Johannes Geiselmann | UMR5240 CNRS-UCBL-INSA-BayerCropScience | Member scientific council |
| Hidde de Jong | Microbiology and Food Chain Department, INRAE | Member scientific council |
| Hidde de Jong | Univ Grenoble Alpes | Member of Vice-Présidence Recherche et Innovation élargie |
| Hidde de Jong | Univ Grenoble Alpes | Member of Collège des Ecoles Doctorales |
| Hidde de Jong | Univ Grenoble Alpes | Member advisory board of LabEX PERSYVAL 3 |
| Hidde de Jong | Univ Grenoble Alpes | Member of comité des directeurs du pôle MSTIC |
| Delphine Ropers | Microbiology and Food Chain Department, INRAE | Member scientific council (since Dec 2024) |
10.1.6 Research administration
| MICROCOSME member | Committee | Role |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité des Emplois Scientifiques (CES) |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité des Utilisateurs des Moyens Informatiques (CUMI) |
| Eugenio Cinquemani | Inria - Univ. Grenoble Alpes | Member Comité Développement Technologique (CDT) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | Member of Direction du centre |
| Hidde de Jong | Inria - Univ Grenoble Alpes | President Comité des Equipes Projets (CEP) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | Member Comité des Emplois Scientifiques (CES) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | Member Comité Développement Technologique (CDT) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | Member Comité des Etudes Doctorales (CED) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | President scientific council (COS) |
| Hidde de Jong | Inria | Member Commission d'évaluation (CE) |
| Hidde de Jong | Inria | Member Comité scientifique interne (COSI) |
| Aline Marguet | Inria - Univ Grenoble Alpes | Member Comité du centre (until Feb. 2024) |
| Aline Marguet | Inria - Univ. Grenoble Alpes | Member Comité des études doctorales |
| Aline Marguet | GDR Branchement | Member of Scientific comitee |
| Delphine Ropers | Inria - Univ Grenoble Alpes | Référente chercheurs |
10.1.7 Recruitment committees
| MICROCOSME member | Organism | Recruitment |
| Eugenio Cinquemani | Univ Grenoble Alpes | Assistant-ingénieur LIPhy |
| Eugenio Cinquemani | Inria | DR2 (jury d'admission) |
| Hidde de Jong | Inria | DR2 (jury d'admissibilité) |
| Hidde de Jong | Inria - Lyon | CRCN/ISFP (jury d'admissibilité) |
| Hidde de Jong | Inria - Univ Grenoble Alpes | ISFP (jury d'admission) |
| Delphine Ropers | Inria - Univ Côte d'Azur | CRCN/IFSP (présidence jury d'admissibilité) |
| Delphine Ropers | Inria - Univ Côte d'Azur | IFSP (jury d'admission) |
| Delphine Ropers | Univ Grenoble Alpes | Assistant-ingénieur LIPhy |
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
Delphine Ropers received the title of Full Professor ("Professeur attaché") at Univ Grenoble Alpes for 3 years (2023 - 2026) in recognition of her teaching activity.
Before retiring in December 2024, Johannes Geiselmann was a full professor at Univ Grenoble Alpes. He therefore had a full teaching service (at least 192 hours per year) and administrative duties related to the organization and evaluation of the university course programs on all levels.
Delphine Ropers organizes a module on the mathematical modelling of biological systems at Grenoble INP - Phelma, UGA and a module on the modelling of cell systems at the Faculty of Pharmacy (Univ Grenoble Alpes). Hidde de Jong organizes a module on the modelling of genetic and metabolic networks at INSA de Lyon.
The following people have also contributed to courses last year:
Eugenio Cinquemani
- Course: Modelling and identification of metabolic networks, M1, Phelma, INP Grenoble (4 h)
Hidde de Jong
- Course and practicals: Modeling and simulation of gene regulatory networks, M2, BIM, INSA de Lyon (32 h)
Eugene Ferragu
- Practicals: Applied linear algebra, L1, Licence Phsyique, Chimie, Mécanique et Mathématiques, Univ Grenoble Alpes (31.5 h)
- Practicals: Cell systems biology and modelling cell functions, M1, Master ingéniérie de la santé, Univ Grenoble Alpes (10 h)
Claudia Fonte Sanchez
- Course and practicals: introduction to mathematical modelling in biology, M1, Univ La Havane, Cuba (18 h)
Aline Marguet
- Practicals: Biostatistics, M2, Univ Grenoble Alpes (27 h)
Delphine Ropers
- Course and practicals: Modelling in systems biology, M1, Phelma, INP Grenoble (16 h)
- Course and practicals: Cell systems biology and modelling cell functions, M1, Master ingéniérie de la santé, Univ Grenoble Alpes (24 h)
- Course: Modelling and simulation of genetic regulatory networks, M2, INSA de Toulouse (4 h)
- Course: Metabolic modelling with omics data, M2, IA4 Health International master course, Univ Grenoble Alpes (6 h)
10.2.2 Supervision
- PhD in progress: Rand Asswad, Development of control strategies for synthetic microbial consortia. Supervisors: Eugenio Cinquemani and Jean-Luc Gouzé (Inria - Univ Côte d'Azur)
- PhD in progress: Ignacia Cancino Aguirre, Computational analysis of metabolic strategies in pathogenic bacteria. Supervisors: Delphine Ropers and Hidde de Jong
- PhD completed: Thibault Clavier Genetic growth control to maximize the bioproduction in E. coli. Supervisors: Johannes Geiselmann and Hidde de Jong
- PhD completed: Charles Medous, Interacting populations, spinal constructions and stochastic simulations. Supervisors: Loren Coquille (Institut Fourier, Grenoble), Aline Marguet and Charline Smadi (INRAE Grenoble)
- PhD in progress: Eugene Ferragu, Stochastic models of host-pathogen dynamics, Supervisors: Aline Marguet and Charline Smadi (INRAE Grenoble)
- PhD in progress: Emrys Reginato, Development, analysis, and inference of stochastic models of gene expression in growing cell populations. Supervisors: Eugenio Cinquemani and Aline Marguet
10.2.3 Juries
PhD thesis committees
| MICROCOSME member | Role | PhD student | University | Date |
| Eugenio Cinquemani | Rapporteur | Joel Ignacio Fierro Ulloa | Université Côte d'Azur | Oct 2024 |
| Aline Marguet | Co-encadrante | Charles Medous | Univ Grenoble Alpes | Dec 2024 |
| Delphine Ropers | Présidente | Amandine Paulay | Univ Paris-Saclay | Jul 2024 |
| Delphine Ropers | Rapporteur | Mathieu Bolteau | Univ Nantes | Oct 2024 |
| Delphine Ropers | Examinatrice | Delphine Nègre | Univ Nantes | Dec 2024 |
Habilitation (HDR) committees
| MICROCOSME member | Role | HDR candidate | University | Date |
| Delphine Ropers | Rapporteur | Simon Labarthe | Univ Bordeaux | Jul 2024 |
PhD advisory committees
| MICROCOSME member | PhD student | University |
| Delphine Ropers | Paul Ahavi | Univ Paris Saclay |
| Delphine Ropers | Mathilde Burck | Univ Paul Sabatier, Toulouse |
| Delphine Ropers | Marvin Ramos | Univ Paul Sabatier, Toulouse |
| Delphine Ropers | Sthyve Tatho | Univ Bordeaux |
10.3 Popularization
Hidde de Jong supervised a one-week lab internship of two highschool students participating in the U-Talent program of Utrecht University (the Netherlands). Delphine Ropers took part in the "Journée Filles, Math et Info" at ENSIMAG. The aim of this event is to make high-school girls aware of careers in computer science and mathematics.
11 Scientific production
11.1 Major publications
- 1 articleResource allocation accounts for the large variability of rate-yield phenotypes across bacterial strains.eLife12May 2023, 1-29HALDOI
- 2 articleEstimation of time-varying growth, uptake and excretion rates from dynamic metabolomics data.Bioinformatics33142017, i301-i310HALDOI
- 3 articleStochastic reaction networks with input processes: Analysis and application to gene expression inference.Automatica1012019, 150-156HALDOI
- 4 articleCompetitive effects in bacterial mRNA decay.Journal of Theoretical Biology504November 2020HALDOI
- 5 articleDynamical allocation of cellular resources as an optimal control problem: Novel insights into microbial growth strategies.PLoS Computational Biology123March 2016, e1004802HALDOI
- 6 articleStatistical estimation in a randomly structured branching population.Stochastic Processes and their Applications129122019, 5236-5277HALDOI
- 7 articleA synthetic growth switch based on controlled expression of RNA polymerase.Molecular Systems Biology1111November 2015, 840HALback to textback to text
- 8 articleWhat population reveals about individual cell identity: Single-cell parameter estimation of models of gene expression in yeast.PLoS Computational Biology122February 2016, e1004706HALDOI
- 9 articleOptimal proteome allocation and the temperature dependence of microbial growth laws.npj Systems Biology and Applications7142021HALDOI
- 10 articleInheritance and variability of kinetic gene expression parameters in microbial cells: modeling and inference from lineage tree data.Bioinformatics35142019, i586-i595HALDOIback to text
- 11 articleEnhanced production of heterologous proteins by a synthetic microbial community: Conditions and trade-offs.PLoS Computational Biology1642020, e1007795HALDOIback to text
- 12 articleThe Csr System Regulates Escherichia coli Fitness by Controlling Glycogen Accumulation and Energy Levels.mBio85October 2017, 1-14HALDOI
- 13 articleThe post-transcriptional regulatory system CSR controls the balance of metabolic pools in upper glycolysis of Escherichia coli.Molecular Microbiology1004May 2016, 686-700HALDOIback to text
- 14 articleSingle-cell data reveal heterogeneity of investment in ribosomes across a bacterial population.Nature Communications161January 2025, 285HALDOI
- 15 articleAcetate metabolism and the inhibition of bacterial growth by acetate.Journal of Bacteriology20113July 2019, 147 - 166HALDOI
11.2 Publications of the year
International journals
Invited conferences
International peer-reviewed conferences
Doctoral dissertations and habilitation theses
Reports & preprints
11.3 Cited publications
- 32 articleResource allocation accounts for the large variability of rate-yield phenotypes across bacterial strains.eLife12May 2023, 1-29HALDOIback to text
- 33 miscSpecial Issue of the 19th International Conference on Computational Methods in Systems Biology.May 2023HALback to text
- 34 phdthesisMicrobial growth control in changing environments : Theoretical and experimental study of resource allocation in Escherichia coli.Université Grenoble AlpesMarch 2017HALback to text
- 35 articleOptimal protein production by a synthetic microbial consortium: Coexistence, distribution of labor, and syntrophy.Journal of Mathematical Biology8712023, 1-37HALDOIback to text
- 36 phdthesisQuantification of bacterial resource allocation in changing environments on the single-cell level.Université Grenoble Alpes [2020-....]July 2022HALback to textback to text
- 37 articleMultiomics Study of Bacterial Growth Arrest in a Synthetic Biology Application.ACS Synthetic Biology1011November 2021, 2910-2926HALDOIback to text
- 38 phdthesisDevelopment, characterization and control of E. coli communities on an automated experimental platform.Université Grenoble AlpesMay 2023HALback to textback to text

