2024Activity reportProject-TeamEPIONE
RNSR: 201822641L- Research center Inria Centre at Université Côte d'Azur
- Team name: E-Patient: Images, Data & MOdels for e-MediciNE
- Domain:Digital Health, Biology and Earth
- Theme:Computational Neuroscience and Medicine
Keywords
Computer Science and Digital Science
- A3.3. Data and knowledge analysis
- A3.4. Machine learning and statistics
- A4.3. Cryptography
- A4.4. Security of equipment and software
- A4.8. Privacy-enhancing technologies
- A5.2. Data visualization
- A5.3. Image processing and analysis
- A5.4. Computer vision
- A5.6. Virtual reality, augmented reality
- A5.9. Signal processing
- A6.1. Methods in mathematical modeling
- A6.2. Scientific computing, Numerical Analysis & Optimization
- A6.3. Computation-data interaction
- A8.3. Geometry, Topology
- A9. Artificial intelligence
- A9.2. Machine learning
- A9.3. Signal analysis
- A9.6. Decision support
- A9.7. AI algorithmics
- A9.9. Distributed AI, Multi-agent
- A9.10. Hybrid approaches for AI
Other Research Topics and Application Domains
- B2.2. Physiology and diseases
- B2.3. Epidemiology
- B2.4. Therapies
- B2.6. Biological and medical imaging
- B2.6.1. Brain imaging
- B2.6.2. Cardiac imaging
- B2.6.3. Biological Imaging
1 Team members, visitors, external collaborators
Research Scientists
- Nicholas Ayache [Team leader, Inria, Senior Researcher, HDR]
- Irene Balelli [Inria, ISFP]
- Benjamin Billot [Inria, Researcher, from Dec 2024]
- Hervé Delingette [Inria, Senior Researcher, HDR]
- Marco Lorenzi [Inria, Researcher, HDR]
- Guillaume Olikier [Inria, Starting Research Position]
- Xavier Pennec [Inria, Senior Researcher, HDR]
- Maxime Sermesant [Inria, Senior Researcher, HDR]
Post-Doctoral Fellows
- Safaa Al Ali [Inria, Post-Doctoral Fellow]
- Francesco Cremonesi [Inria]
- Jia Guo [CHU Nice, from Sep 2024]
- Josquin Harrison [Inria, from Jul 2024]
- Ghiles Reguig [Inria, Post-Doctoral Fellow]
- Jesus Jairo Rodriguez Padilla [Inria]
- Alessandro Viani [Inria, Post-Doctoral Fellow, from Feb 2024]
- Yingyu Yang [Inria]
PhD Students
- Olivier Bisson [Inria]
- Alix De Langlais [Inria, from Jun 2024]
- Federica Facente [Inria]
- Camilla Ferrario [Inria, from Sep 2024]
- Sebastien Goffart [CHU Nice]
- Lisa Guzzi [Université Côte d’Azur]
- Josquin Harrison [Inria, until Jun 2024]
- Lucia Innocenti [Inria, until Jul 2024]
- Manasi Kattel [Inria]
- Wassila Khatir [Université Côte d’Azur, from Mar 2024]
- Huiyu Li [Inria, until Oct 2024]
- Maelis Morier [Inria]
- Huyen Trang Nguyen [Inria, from Nov 2024]
- Evariste Njomgue Fotso [Inria]
- Rafael Luis Soares Da Costa E Silva [Inria]
- Hari Sreedhar [Université Côte d’Azur, until Oct 2024]
- Tom Szwagier [Inria]
- Riccardo Taiello [Inria, until Sep 2024]
Technical Staff
- Lucie Chambon [Inria, from Feb 2024]
- Hye Lim Lee [Inria, Engineer, from Dec 2024]
- Marco Milanesio [Université Côte d’Azur, Engineer]
- Guillaume Robert-Siegwald [Inria, Engineer]
- Anaëlle Zanella [Inria, Engineer]
Interns and Apprentices
- Andrea Chierici [CHU Nice, until Oct 2024]
- Elena Da Pozzo [Inria, Intern, from Jun 2024 until Nov 2024]
- Nicolas Drettakis [Inria, Intern, from Jun 2024 until Nov 2024]
- Hye Lim Lee [Inria, Intern, from May 2024 until Sep 2024]
- Giuseppe Orlando [Inria, Intern, from Sep 2024]
Administrative Assistant
- Nathalie Nordmann [Inria]
Visiting Scientists
- Mihaela Pop [Sunnybrook Research Institute, until Apr 2024]
- Javier Villar Valero [Polytechnic University of Valencia, from Apr 2024 until Jul 2024]
External Collaborators
- Sébastien Frey [CHU Nice]
- Dimbihery Rabenoro [Université Paris 1]
- Cécile Rouzier [CHU Nice, from Sep 2024]
2 Overall objectives
2.1 Description
Our long-term goal is to contribute to the development of what we call the e-patient (digital patient) for e-medicine (digital medicine) (Fig. 1).
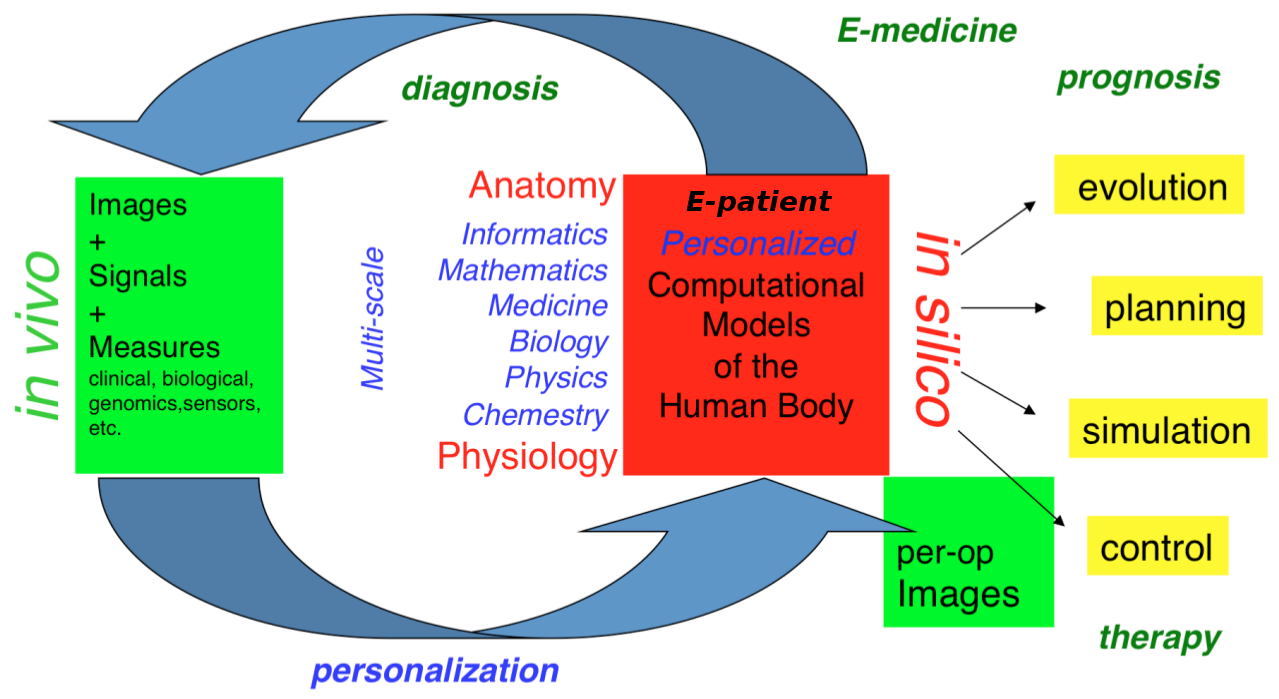
The e-patient for e-medicine
- the e-patient (or digital patient) is a set of computational models of the human body able to describe and simulate the anatomy and the physiology of the patient's organs and tissues, at various scales, for an individual or a population. The e-patient can be seen as a framework to integrate and analyze in a coherent manner the heterogeneous information measured on the patient from disparate sources: imaging, biological, clinical, sensors, ...
- e-medicine (or digital medicine) is defined as the computational tools applied to the e-patient to assist the physician and the surgeon in their medical practice, to assess the diagnosis/prognosis, and to plan, control and evaluate the therapy.
The models that govern the algorithms designed for e-patients and e-medicine come from various disciplines: computer science, mathematics, medicine, statistics, physics, biology, chemistry, etc. The parameters of those models must be adjusted to an individual or a population based on the available images, signals and data. This adjustment is called personalization and usually requires solving difficult inverse problems. The overall picture of the construction of the personalized e-patient for e-medicine was presented at the College de France through an inaugural lecture and a series of courses and seminars (fr), concluded by an international workshop.
2.2 Organization
The research organization in our field is often built on a virtuous triangle (Fig. 2). On one vertex, academic research requires multidisciplinary collaborations associating informatics and mathematics to other disciplines: medicine, biology, physics, chemistry ... On a second vertex, a clinical partnership is required to help defining pertinent questions, to get access to clinical data, and to clinically evaluate any proposed solution. On the third vertex, an industrial partnership can be introduced for the research activity itself, and also to transform any proposed solution into a validated product that can ultimately be transferred to the clinical sites for an effective use on the patients.
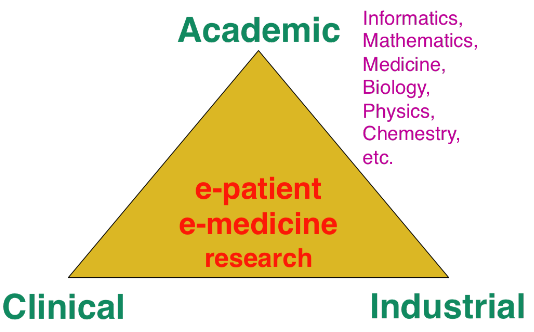
A pluridisciplinary research triangle
Keeping this triangle in mind, we choose our research directions within a virtuous circle: we look at difficult problems raised by our clinical or industrial partners, and then try to identify some classes of generic fundamental/theoretical problems associated to their resolution. We also study some fundamental/theoretical problems per se in order to produce fundamental scientific advances that can help in turn to promote new applications.
3 Research program
3.1 Introduction
Our research objectives are organized along 5 scientific axes (Fig. 3):
- Biomedical Image Analysis & Machine Learning
- Imaging & Phenomics, Biostatistics
- Computational Anatomy, Geometric Statistics
- Computational Physiology & Image-Guided Therapy
- Computational Cardiology & Image-Based Cardiac Interventions
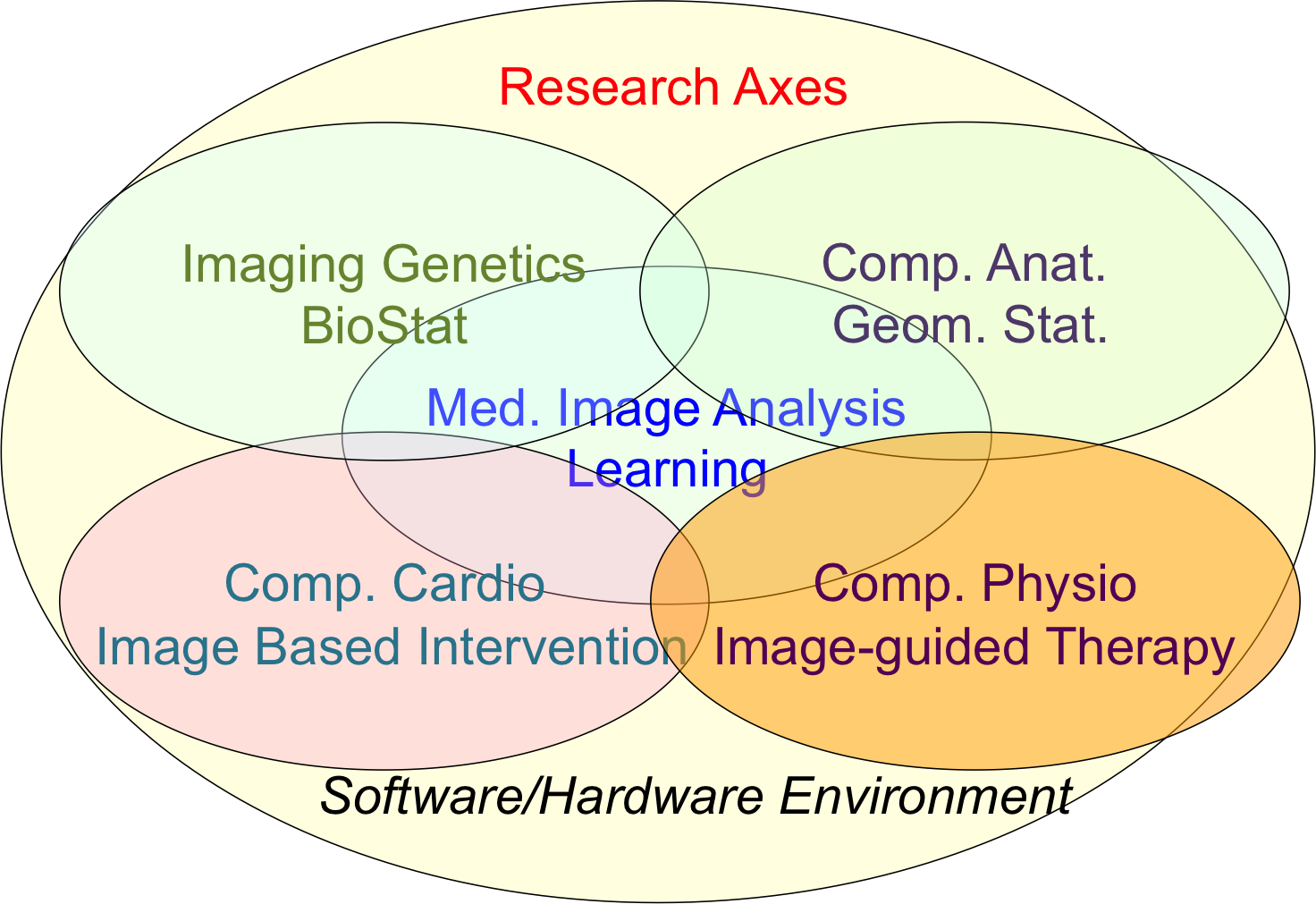
Epione's five main research axes
For each scientific axis, we introduce the context and the long term vision of our research.
3.2 Biomedical Image Analysis & Machine Learning
The long-term objective of biomedical image analysis is to extract, from biomedical images, pertinent information for the construction of the e-patient and for the development of e-medicine. This relates to the development of advanced segmentation and registration of images, the extraction of image biomarkers of pathologies, the detection and classification of image abnormalities, the construction of temporal models of motion or evolution from time-series of images, etc.
In addition, the growing availability of very large databases of biomedical images, the growing power of computers and the progress of machine learning (ML) approaches have opened up new opportunities for biomedical image analysis.
This is the reason why we decided to revisit a number of biomedical image analysis problems with ML approaches, including segmentation and registration problems, automatic detection of abnormalities, prediction of a missing imaging modality, etc. Not only those ML approaches often outperform the previous state-of-the-art solutions in terms of performances (accuracy of the results, computing times), but they also tend to offer a higher flexibility like the possibility to be transferred from one problem to another one with a similar framework. However, even when successful, ML approaches tend to suffer from a lack of explanatory power, which is particularly annoying for medical applications. We also plan to work on methods that can interpret the results of the ML algorithms that we develop.
3.3 Imaging & Phenomics, Biostatistics
The human phenotype is associated with a multitude of heterogeneous biomarkers quantified by imaging, clinical and biological measurements, reflecting the biological and patho-physiological processes governing the human body, and essentially linked to the underlying individual genotype. In order to deepen our understanding of these complex relationships and better identify pathological traits in individuals and clinical groups, a long-term objective of e-medicine is therefore to develop the tools for the joint analysis of this heterogeneous information, termed Phenomics, within the unified modeling setting of the e-patient.
To date the most common approach to the analysis of the joint variation between the structure and function of organs represented in medical images, and the classical -omics modalities from biology, such as genomics or lipidomics, is essentially based on the massive univariate statistical testing of single candidate features out of the many available. This is for example the case of genome-wide association studies (GWAS) aimed at identifying statistically significant effects in pools consisting of up to millions of genetics variants. Such approaches have known limitations such as multiple comparison problems, leading to underpowered discoveries of significant associations, and usually explain a rather limited amount of data variance. Although more sophisticated machine learning approaches have been proposed, the reliability and generalization of multivariate methods is currently hampered by the low sample size relatively to the usually large dimension of the parameters space.
To address these issues this research axis investigates novel methods for the integration of this heterogeneous information within a parsimonious and unified multivariate modeling framework. The cornerstone of the project consists in achieving an optimal trade-off between modeling flexibility and ability to generalize on unseen data by developing statistical learning methods informed by prior information, either inspired by "mechanistic" biological processes, or accounting for specific signal properties (such as the structured information from spatio-temporal image time series). Finally, particular attention will be paid to the effective exploitation of the methods in the growing Big Data scenario, either in the meta-analysis context, or for the application in large datasets and biobanks.
Federated learning in multi-centric studies. The current research scenario is characterized by medium/small scale (typically from 50 to 1000 patients) heterogeneous datasets distributed across centres and countries. The straightforward extension of learning algorithms successfully applied to big data problems is therefore difficult, and specific strategies need to be envisioned in order to optimally exploit the available information. To address this problem, we focus on learning approaches to jointly model clinical data localized in different centres. This is an important issue emerging from recent large-scale multi-centric imaging-genetics studies in which partners can only share model parameters (e.g. regression coefficients between specific genes and imaging features), as represented for example by the ENIGMA imaging-genetics study, led by the collaborators at University of Southern California. This problem requires the development of statistical methods for federated model estimation, in order to access data hosted in different clinical institutions by simply transmitting the model parameters, that will be in turn updated by using the local available data. This approach is extended to the definition of stochastic optimization strategies in which model parameters are optimized on local datasets, and then summarized in a meta-analysis context. Finally, this project studies strategies for aggregating the information from heterogeneous datasets, accounting for missing modalities due to different study design and protocols. The developed methodology finds important applications within the context of Big Data, for the development of effective learning strategies for massive datasets in the context of medical imaging (such as with the UK biobank), and beyond.
3.4 Computational Anatomy, Geometric Statistics
Computational anatomy is an emerging discipline at the interface of geometry, statistics and image analysis which aims at developing algorithms to model and analyze the biological shape of tissues and organs. The goal is not only to establish generative models of organ anatomies across diseases, populations, species or ages but also to model the organ development across time (growth or aging) and to estimate their variability and link to other functional, genetic or structural information. Computational anatomy is a key component to support computational physiology and is evidently crucial for building the e-patient and to support e-medicine.
Pivotal applications include the spatial normalization of subjects in neuroscience (mapping all the anatomies into a common reference system) and atlas to patient registration to map generic knowledge to patient-specific data. Our objectives will be to develop new efficient algorithmic methods to address the emerging challenges described below and to generate precise specific anatomical model in particular for the brain and the heart.
The objects of computational anatomy are often shapes extracted from images or images of labels (segmentation). The observed organ images can also be modeled using registration as the random diffeomorphic deformation of an unknown template (i.e. an orbit). In these cases as in many other applications, invariance properties lead us to consider that these objects belong to non-linear spaces that have a geometric structure. Thus, the mathematical foundations of computational anatomy rely on statistics on non-linear spaces.
Geometric Statistics aim at studying this abstracted problem at the theoretical level. Our goal is to advance the fundamental knowledge in this area, with potential applications to new areas outside of medical imaging. Beyond the now classical Riemannian spaces, we aim at developing the foundations of statistical estimation on affine connection spaces (e.g. Lie groups), quotient and stratified metric spaces (e.g. orbifolds and tree spaces). In addition to the curvature, one of the key problem is the introduction of singularities at the boundary of the regular strata (non-smooth and non-convex analysis).
A second objective is to develop parametric and non-parametric dimension reduction methods in non-linear space. An important issue is to estimate efficiently not only the model parameters (mean point, subspace, flag) but also their uncertainty. We also want to quantify the influence of curvature and singularities on non-asymptotic estimation theory since we always have a finite (and often too limited) number of samples. A key challenge in developing such a geometrization of statistics will not only be to unify the theory for the different geometric structures, but also to provide efficient practical algorithms to implement them.
A third objective is to learn the geometry from the data. In the high dimensional but low sample size (small data) setting which is the common situation in medical data, we believe that invariance properties are essential to reasonably interpolate and approximate. New apparently antagonistic notions like approximate invariance could be the key to this interaction between geometry and learning.
Beyond the traditional statistical survey of the anatomical shapes that is developed in computational anatomy above, we intend to explore other application fields exhibiting geometric but non-medical data. For instance, applications can be found in Brain-Computer Interfaces (BCI), tree-spaces in phylogenetics, Quantum Physics, etc.
3.5 Computational Physiology & Image-Guided Therapy
Computational Physiology aims at developing computational models of human organ functions, an important component of the e-patient , with applications in e-medicine and more specifically in computer-aided prevention, diagnosis, therapy planning and therapy guidance. The focus of our research is on descriptive (allowing to reproduce available observations), discriminative (allowing to separate two populations), and above all predictive models which can be personalized from patient data including medical images, biosignals, biological information and other available metadata. A key aspect of this scientific axis is therefore the coupling of biophysical models with patient data which implies that we are mostly considering models with relatively few and identifiable parameters. To this end, data assimilation methods aiming at estimating biophysical model parameters in order to reproduce available patient data are preferably developed as they potentially lead to predictive models suitable for therapy planning.
Previous research projects in computational physiology have led us to develop biomechanical models representing quasi-static small or large soft tissue deformations (e.g. liver or breast deformation after surgery), mechanical growth or atrophy models (e.g. simulating brain atrophy related to neurodegenerative diseases), heat transfer models (e.g. simulating radiofrequency ablation of tumors), and tumor growth models (e.g. brain or lung tumor growth).
To improve the data assimilation of biophysical models from patient data, a long term objective of our research will be to develop joint imaging and biophysical generative models in a probabilistic framework which simultaneously describe the appearance and function of an organ (or its pathologies) in medical images. Indeed, current approaches for the personalization of biophysical models often proceed in two separate steps. In a first stage, geometric, kinematic or/ functional features are first extracted from medical images. In a second stage, they are used by personalization methods to optimize model parameters in order to match the extracted features. In this process, subtle information present in the image which could be informative for biophysical models is often lost which may lead to limited personalization results. Instead, we propose to develop more integrative approaches where the extraction of image features would be performed jointly with the model parameter fitting. Those imaging and biophysical generative models should lead to a better understanding of the content of images, to a better personalization of model parameters and also better estimates of their uncertainty.
3.6 Computational Cardiology & Image-Based Cardiac Interventions
Computational Cardiology has been an active research topic within the Computational Anatomy and Computational Physiology axes of the previous Asclepios project, leading to the development of personalized computational models of the heart designed to help characterizing the cardiac function and predict the effect of some device therapies like cardiac resynchronization or tissue ablation . This axis of research has now gained a lot of maturity and a critical mass of involved scientists to justify an individualized research axis of the new project Epione, while maintaining many constructive interactions with the 4 other research axes of the project. This will develop all the cardiovascular aspects of the e-patient for cardiac e-medicine.
The new challenges we want to address in computational cardiology are related to the introduction of new levels of modeling and to new clinical and biological applications. They also integrate the presence of new sources of measurements and the potential access to very large multimodal databases of images and measurements at various spatial and temporal scales.
4 Application domains
The main applications of our research are in the field of healthcare and more precisely the domain of digital medicine and biomedical data analysis. The axes of research presented above are related to many branches of medicine including cardiology, oncology, urology, neurology, otology, pneumology, radiology, surgery, dermatology, nuclear medicine. Within those branches, the applications cover the following different stages of medicine : prevention, diagnosis, prognosis, treatment.
5 Social and environmental responsibility
5.1 Footprint of research activities
An important activity of Epione is to introduce priors from clinical knowledge within data analysis, through geometric information, biophysical models, causality, etc. This enables to develop AI method requiring less data and computations. Therefore with a positive impact on the environmental footprint of epione research activity.
6 Highlights of the year
6.1 Awards
- Xavier Pennec has won the “Grand prix Ampère de l'Électricité de France” from the French Académie des sciences. This prize rewards Xavier Pennec's research on statistical geometry and its applications to computational anatomy. More information available in Inria's article about this pretistigious award.
- Elodie Maignant was awarded the second ATSI prize of the EDSTIC Doctoral school of Université Côte d'Azur for her PhD work on "Barycentric embeddings for geometric manifold learning" under the supervision of Dr Xavier Pennec and Dr Alain Trouvé.
- Rafael Silva, Yingyu Yang, Maëlis Morier, Safaa Al Ali and Maxime Sermesant won the Best Poster Award at Computing in Cardiology Challenge 2024, and the Prix d'Excellence 2024 (Université Côte d'Azur), for their work on YOUR-Lead: YOLO and U-Net for Reconstruction of ECG Lead Signals 39.
- In December 2024, the Doctoral School ED STIC of Université Côte d'Azur awarded Paul Tourniaire with an exceptional scientific Prize for his PhD thesis work entitled "AI-based selection of imaging and biological markers predictive of therapy response in lung cancer".
- M. Lorenzi and O. Humbert received the UNICANCER Innovation Award for the project FEDERATED-PET for the deployment of the federated learning infrastructure Fed-BioMed in a real-world hospital setting.
- M. Lorenzi is editor with Prof. M.A. Zuluaga of the book Trustworthy AI in Medical Imaging, MICCAI Society book Series, Academic Press 46. The book features 22 chapters from world leading authors and institutes from the communities of medical imaging, machine learning and security. N. Ayache contributed to the preface 47; M. Lorenzi contributed to the introductory chapter 49; I. Balelli contributed with a chapter 48.
6.2 Recruitment
The Epione team has welcomed Benjamin Billot, who was appointed CRCN after successfully passing Inria's recruitment campaign. His project will focus on developing new data representation strategies for the analysis of clinical imaging data. Before joining Epione, Benjamin obtained his PhD at University College London and worked as a postdoc with Prof. Polina Golland at MIT. A complete record of his academic publications can be found on Google Scholar.
7 New software, platforms, open data
7.1 New software
7.1.1 CardiacSegmentationPropagation
-
Keywords:
3D, Segmentation, Cardiac, MRI, Deep learning
-
Functional Description:
Training of a deep learning model which is used for cardiac segmentation in short-axis MRI image stacks.
- Publication:
-
Contact:
Qiao Zheng
7.1.2 CardiacMotionFlow
-
Keywords:
3D, Deep learning, Cardiac, Classification
-
Functional Description:
Creation of a deep learning model for the motion tracking of the heart, extraction of characteristic quantities of the movement and shape of the heart to classify a sequence of cine-MRI cardiac images in terms of the types of pathologies (infarcted heart, dilated , hypertrophied, abnormality of the right ventricle).
- Publication:
-
Contact:
Qiao Zheng
7.1.3 MedINRIA
-
Keywords:
Visualization, DWI, Health, Segmentation, Medical imaging
-
Scientific Description:
MedInria aims at creating an easily extensible platform for the distribution of research algorithms developed at Inria for medical image processing. This project has been funded by the D2T (ADT MedInria-NT) in 2010, renewed in 2012. A fast-track ADT was awarded in 2017 to transition the software core to more recent dependencies and study the possibility of a consortium creation.The Empenn team leads this Inria national project and participates in the development of the common core architecture and features of the software as well as in the development of specific plugins for the team's algorithm.
-
Functional Description:
medInria is a free software platform dedicated to medical data visualization and processing.
- URL:
-
Contact:
Florent Leray
-
Participants:
Maxime Sermesant, Olivier Commowick
-
Partners:
HARVARD Medical School, IHU - LIRYC, NIH
7.1.4 GP-ProgressionModel
-
Name:
GP progression model
-
Keywords:
Data modeling, Data visualization, Data integration, Machine learning, Biostatistics, Statistical modeling, Medical applications, Evolution, Brain, Uncertainly, Uncertainty quantification, Alzheimer's disease, Probability, Stochastic models, Stochastic process, Trajectory Modeling, Marker selection, Health, Statistic analysis, Statistics, Bayesian estimation
-
Functional Description:
Disease progression modeling (DPM) of Alzheimer's disease (AD) aims at revealing long term pathological trajectories from short term clinical data. Along with the ability of providing a data-driven description of the natural evolution of the pathology, DPM has the potential of representing a valuable clinical instrument for automatic diagnosis, by explicitly describing the biomarker transition from normal to pathological stages along the disease time axis.
In this software we reformulate DPM within a probabilistic setting to quantify the diagnostic uncertainty of individual disease severity in an hypothetical clinical scenario, with respect to missing measurements, biomarkers, and follow-up information. The proposed formulation of DPM provides a statistical reference for the accurate probabilistic assessment of the pathological stage of de-novo individuals, and represents a valuable instrument for quantifying the variability and the diagnostic value of biomarkers across disease stages.
This software is based on the publication:
Probabilistic disease progression modeling to characterize diagnostic uncertainty: Application to staging and prediction in Alzheimer's disease. Marco Lorenzi, Maurizio Filippone, Daniel C. Alexander, Sebastien Ourselin Neuroimage. 2019 Apr 15,190:56-68. doi: 10.1016/j.neuroimage.2017.08.059. Epub 2017 Oct 24. HAL Id : hal-01617750 https://hal.archives-ouvertes.fr/hal-01617750/
-
Release Contributions:
- New interface and output - Completely based on Pytorch
- URL:
- Publication:
-
Contact:
Marco Lorenzi
-
Participant:
Marco Lorenzi
7.1.5 Music
-
Name:
Multi-modality Platform for Specific Imaging in Cardiology
-
Keywords:
Medical imaging, Cardiac Electrophysiology, Computer-assisted surgery, Cardiac, Health
-
Functional Description:
MUSIC is a software developed by the Asclepios research project in close collaboration with the IHU LIRYC in order to propose functionalities dedicated to cardiac interventional planning and guidance. This includes specific tools (algorithms of segmentation, registration, etc.) as well as pipelines. The software is based on the MedInria platform.
- URL:
-
Contact:
Maxime Sermesant
-
Participants:
Florent Collot, Mathilde Merle, Maxime Sermesant
-
Partner:
IHU- Bordeau
7.1.6 SOFA
-
Name:
Simulation Open Framework Architecture
-
Keywords:
Real time, Multi-physics simulation, Medical applications
-
Functional Description:
SOFA is an Open Source framework primarily targeted at real-time simulation, with an emphasis on medical simulation. It is mostly intended for the research community to help develop new algorithms, but can also be used as an efficient prototyping tool. Based on an advanced software architecture, it allows the creation of complex and evolving simulations by combining new algorithms with algorithms already included in SOFA, the modification of most parameters of the simulation (deformable behavior, surface representation, solver, constraints, collision algorithm etc.) by simply editing an XML file, the building of complex models from simpler ones using a scene-graph description, the efficient simulation of the dynamics of interacting objects using abstract equation solvers, the reuse and easy comparison of a variety of available methods.
-
News of the Year:
The new version v20.06 has been released including new elements on SoftRobots + ModelOrderReduction integration, in addition to an improved architecture and lots of cleans and bugfixes.
- URL:
- Publication:
-
Contact:
Hugo Talbot
-
Participants:
Christian Duriez, François Faure, Hervé Delingette, Stephane Cotin, Hugo Talbot, Maud Marchal
-
Partners:
IGG, CRIStAL
7.1.7 geomstats
-
Name:
Computations and statistics on manifolds with geometric structures
-
Keywords:
Geometry, Statistic analysis
-
Scientific Description:
Geomstats is an open-source Python package for computations and statistics on manifolds. The package is organized into two main modules: “geometry“ and “learning“.
The module `geometry` implements concepts in differential geometry, and the module `learning` implements statistics and learning algorithms for data on manifolds.
The goal is to provide an easily accessible library for learning algorithms on Riemannian manifolds.
-
Functional Description:
GeomStats is a Python package that performs computations on manifolds such as hyperspheres, hyperbolic spaces, spaces of symmetric positive definite matrices and Lie groups of transformations. It provides efficient and extensively unit-tested implementations of these manifolds, together with useful Riemannian metrics and associated Exponential and Logarithm maps. The corresponding geodesic distances provide a range of intuitive choices of Machine Learning loss functions. The operations implemented in GeomStats are available with different computing backends such as numpy, autograd, pytorch, and tensorflow.
-
Release Contributions:
- addition of several metrics on the space of full-rank correlation matrices taking advantage of diffeomorphism class, existing Riemannian manifolds, and/or quotient space structure - refactoring of quotient structure in order to treat landmarks, curves, and shapes in an homogenized way, improvement of alignment algorithms in those spaces - addition of varifold metric (on surfaces) by leveraging pykeops - full refactoring of geodesic metric spaces: graph space, wald and BHV spaces, and spider (NB: only BHV explicitly takes advantage of quotient structure, so the renaming) - improvement of numerics: better objects to handle optimization, initial/boundary value problems, finite differences, and interpolation
-
News of the Year:
Presentation "Geomstats: a Python package for Riemannian geometry and geometric statistics" at the conference "Geometric Sciences in Action: from geometric statistics to shape analysis" (Mai 27-31, 2024) at CIRM, Luminy, FR.
A new version (v2.8.0) was released in 2024, whose main improvements are detailed in the release description.
- URL:
- Publications:
-
Contact:
Xavier Pennec
-
Participants:
Olivier Bisson, Xavier Pennec, Yann Thanwerdas, Luis Pereira, Anna Calissano, Elodie Maignant, Nina Miolane, Alice Le Brigant
-
Partners:
University of California Santa Barbara, Université Panthéon-Sorbonne
7.1.8 MC-VAE
-
Name:
Multi Channel Variational Autoencoder
-
Keywords:
Machine learning, Artificial intelligence, Medical applications, Dimensionality reduction, High Dimensional Data, Unsupervised learning, Heterogeneity
-
Scientific Description:
Interpretable modeling of heterogeneous data channels is essential in medical applications, for example when jointly analyzing clinical scores and medical images. Variational Autoencoders (VAE) are powerful generative models that learn representations of complex data. The flexibility of VAE may come at the expense of lack of interpretability in describing the joint relationship between heterogeneous data. To tackle this problem, this software extends the variational framework of VAE to introduce sparsity of the latent representation, as well as interpretability when jointly accounting for latent relationships across multiple channels. In the latent space, this is achieved by constraining the variational distribution of each channel to a common target prior. Parsimonious latent representations are enforced by variational dropout. Experiments on synthetic data show that our model correctly identifies the prescribed latent dimensions and data relationships across multiple testing scenarios. When applied to imaging and clinical data, our method allows to identify the joint effect of age and pathology in describing clinical condition in a large scale clinical cohort.
-
Functional Description:
This software implements the work published in the paper "Sparse Multi-Channel Variational Autoencoder for the Joint Analysis of Heterogeneous Data" presented at the conference ICML 2019 (Long Beach, California, USA).
The software extends classical variational autoencoders by identifying a joint latent code associated to heterogeneous data represented in different channels. The software is implemented in Python and is based on Pytorch. It can be applied to any kind of data arrays, and provides functions for optimization, visualization and writing of the modeling results.
-
Release Contributions:
First release
- URL:
-
Contact:
Luigi Antelmi
-
Participants:
Luigi Antelmi, Marco Lorenzi, Nicholas Ayache
-
Partner:
CoBteK
7.1.9 SOFA-CardiacReduction
-
Keywords:
Simulation, 3D modeling, Model Order Reduction, Cardiac
-
Scientific Description:
Modification of a finite element deformation model : meshless approach and frame-based description, reduction in the number of affine degrees of freedom and integration points.
-
Functional Description:
This SOFA plugin is intented to build a reduced model for deformable solids (especially cardiac simulations).
- Publication:
-
Contact:
Gaetan Desrues
-
Participants:
Gaetan Desrues, Hervé Delingette, Maxime Sermesant
7.1.10 Fed-BioMed
-
Name:
A general software framework for federated learning in healthcare
-
Keywords:
Federated learning, Medical applications, Machine learning, Distributed Applications, Deep learning
-
Scientific Description:
While data in healthcare is produced in quantities never imagined before, the feasibility of clinical studies is often hindered by the problem of data access and transfer, especially regarding privacy concerns. Federated learning allows privacy-preserving data analyses using decentralized optimization approaches keeping data securely decentralized. There are currently initiatives providing federated learning frameworks, which are however tailored to specific hardware and modeling approaches, and do not provide natively a deployable production-ready environment. To tackle this issue, Fed-BioMed proposes an open-source federated learning frontend framework with application in healthcare. Fed-BioMed framework is based on a general architecture accommodating for different models and optimization methods
-
Functional Description:
The project is based on the development of a distributed software architecture, and the establishment of a server instance from which remote experiments are triggered on the clients sites. The software is distributed to the client's sites, allowing to run machine learning models on the local data. Model parameters are then transmitted to the server for federated aggregation.
Fed-BioMed finds application in all projects based on the development of learning models for multi-centric studies.
-
Release Contributions:
Fed-BioMed is based on Python and Pytorch. The software was recently revised (under an ADT) to adopt the libraries Pygrid and Pysyft.
- URL:
- Publication:
-
Contact:
Marco Lorenzi
7.1.11 EchoFanArea
-
Name:
delineation of the border of the fan in ultrasound
-
Keywords:
Ultrasound fan area, Deep learning, Statistics, Image processing
-
Functional Description:
This software allows the delimitation of the acquisition cone in ultrasound imaging and the inpainting of annotations (lines, characters) inside the cone. It allows both the perfect de-identification of the images but also to standardize the content of the images. It relies on a parametric probabilistic approach to generate a training dataset with region of interest (ROI) segmentation masks. This data will then be used to train a deep U-Net network to perform the same task in a supervised manner, thus considerably reducing the calculation time of the method, one hundred and sixty times faster. These images are then processed with existing filling methods to remove annotations present within the signal area.
- URL:
- Publication:
-
Contact:
Hind Dadoun
-
Partner:
Nhance
7.1.12 ProMFusion
-
Functional Description:
Code related to the paper "Robust Fusion of Probability Maps" by Benoît Audelan, Dimitri Hamzaoui, Sarah Montagne, Raphaële Renard-Penna and Hervé Delingette. The proposed approach allows to fuse probability maps in a robust manner with a spatial regularization of the consensus.
- URL:
- Publication:
-
Contact:
Benoit Audelan
-
Participants:
Benoit Audelan, Hervé Delingette, Dimitri Hamzaoui
7.1.13 SimulAD
-
Name:
Disease progression modeling for clinical intervention simulation
-
Keyword:
Automatic Learning
-
Scientific Description:
Recent failures of clinical trials in Alzheimer’s Disease underline the critical importance of identifying optimal intervention time to maximize cognitive benefit. While several models of disease progression have been proposed, we still lack quantitative approaches simulating the effect of treatment strategies on the clinical evolution. In this work, we present a data-driven method to model dynamical relationships between imaging and clinical biomarkers. Our approach allows simulating intervention at any stage of the pathology by modulating the progression speed of the biomarkers, and by subsequently assessing the impact on disease evolution.
-
Functional Description:
A machine-learning framework allowing to simulate the impact of intervention on the long-term progression of imaging and clinical biomarkers from collections of healthcare data
-
Release Contributions:
Most recent version available at https://gitlab.inria.fr/epione/simulad/-/tree/master/
- Publication:
-
Contact:
Marco Lorenzi
-
Participants:
Clément Abi Nader, Marco Lorenzi, Nicholas Ayache, Philippe Robert
8 New results
8.1 Medical Image Analysis & Machine Learning
8.1.1 Features extraction from 18-FDG CT/PET images for abnormal metabolic activity detection
This work was funded by the project FEDERATED-PET.
Keywords:
Participants: Lucie Chambon [Correspondant], Francesco Cremonesi, Olivier Humbert, Marco Lorenzi.
The goal of this project is to develop new methods, based on FDG CT/PET images (fludeoxyglucose-18 (FDG) positron emission tomography (PET) / computed tomography (CT)), for the extraction of biomarkers predictive of response to immunotherapy. We have developed organ-specific atlas of normal FDG uptake (Fig. 4):
- CT scans from a normal database have been segmented;
- PET scans and segmentations have been used to extract FDG uptake distributions;
- Normative atlas have been modeled by clustering methods;
- The atlas have been used to detect abnormal metabolic activity in pathological images.
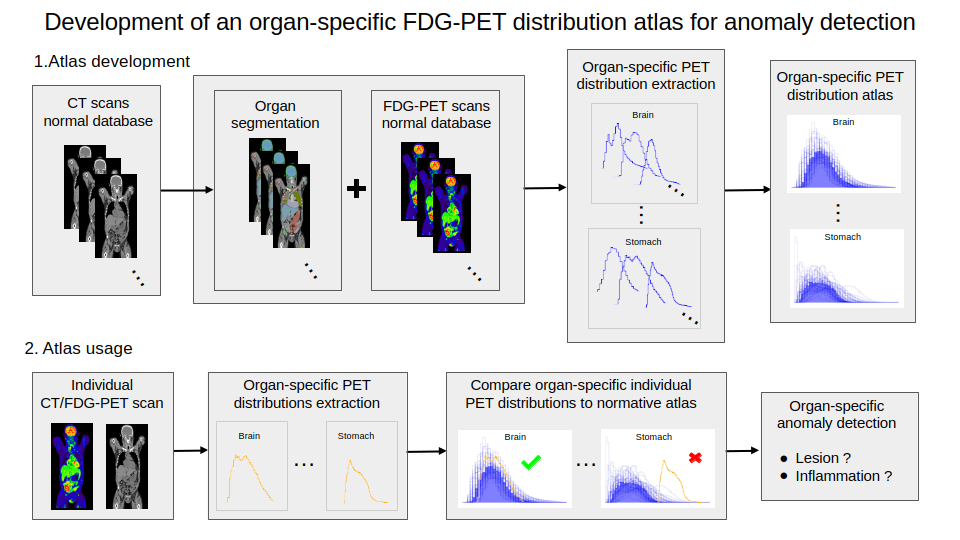
Illustration of the whole pipeline for the developement and usage of the organ-specific atlas of normal FDG uptake.
8.1.2 MRI-Ultrasound Image Registration for Prostate Cancer Care
This work was funded by 3IA Côte d'Azur.
Keywords:
Participants: Manasi Kattel [Correspondant], Hervé Delingette, Nicholas Ayache.
3D registration between magnetic resonance imaging (MRI) and transrectal ultrasound (TRUS) plays a crucial role in the management of prostate cancer, enabling precise targeting of the lesions during biopsy planning and radiotherapy. Fusing the two imaging modalities enhances registration accuracy as illustrated in Fig. 5, but faces many challenges due to the complex intensity differences and the potentially large rotations and deformations between both modalities.
In this work, we focus on rigid pre-alignment, which is essential for correcting large rotations. Specifically, we performed the following:
- Design of a reliable initialization approach that integrates anatomical information visible in both modalities by leveraging segmentation masks of the prostate and other anatomies around the prostate.
- Evaluation of intensity-based registration using commonly employed similarity metrics for MRI-TRUS registration, while highlighting their limitations for this application.
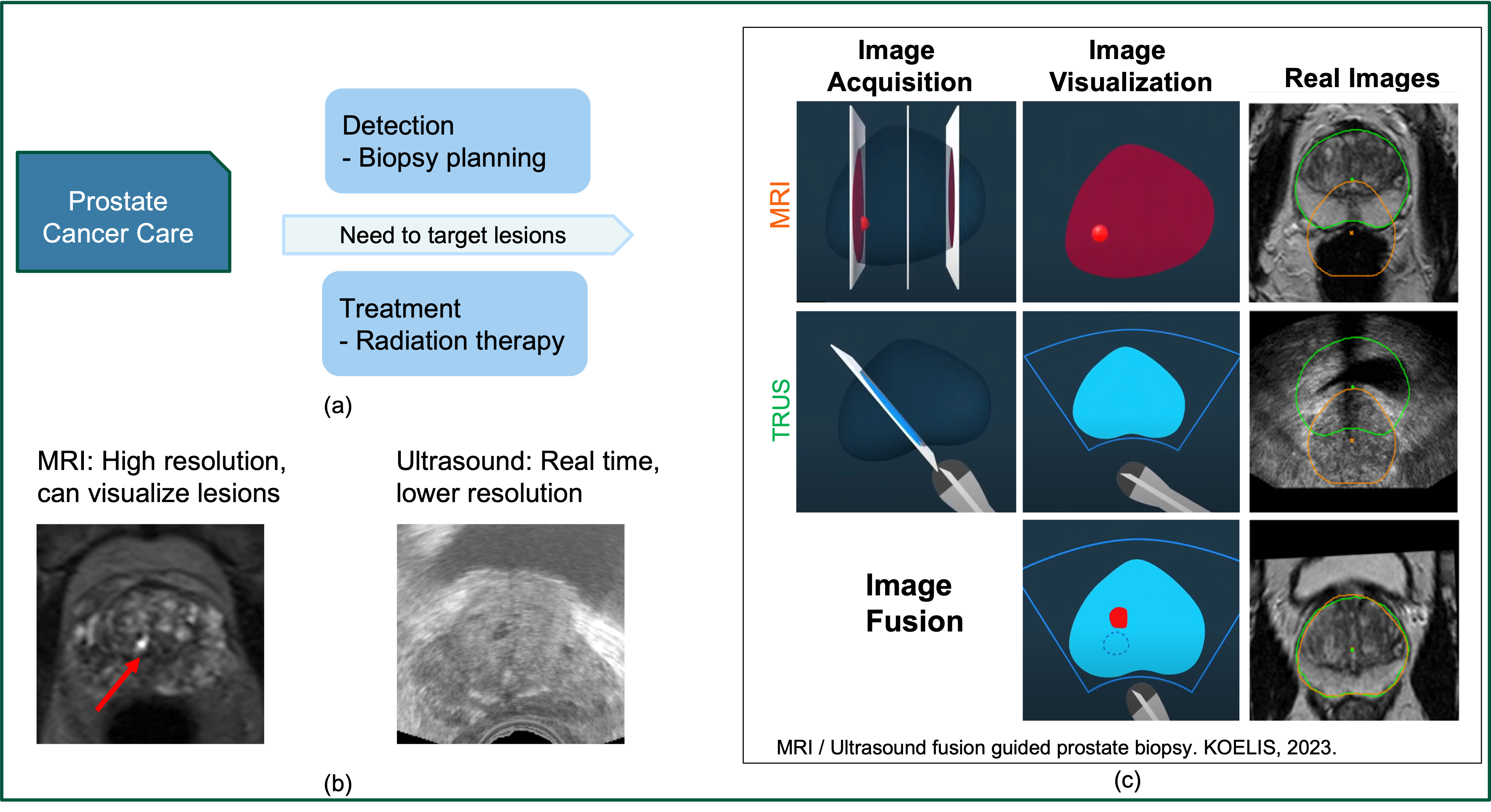
(a) Application of image registration for prostate cancer care. (b) Visualization of lesions on MRI. (c) Illustration of MRI-US registration.
8.1.3 Automatic generation of three-dimensional models of proximal humerus extremity fractures for preoperative planning and intraoperative assistance in mixed reality
This work was supported by an Inserm-Inria funding.
Keywords:
Participants: Alix de Langlais [Correspondant], Hervé Delingette, Marc-Olivier Gauci.
In absence of ground-truth segmentation, quality control of input images and generated masks is needed for evaluating segmentation algorithms. We developed a tool that generates an HTML report with a synthetic view of CT datasets of fractured and healthy humeri, along with segmentation masks from TotalSegmentator model. The report includes:
- Statistical data on image resolution and field of view.
- 3D visualization of CT images and masks (Fig. 6).
- Segmentation inconsistencies detected with UnSegQC algorithm.
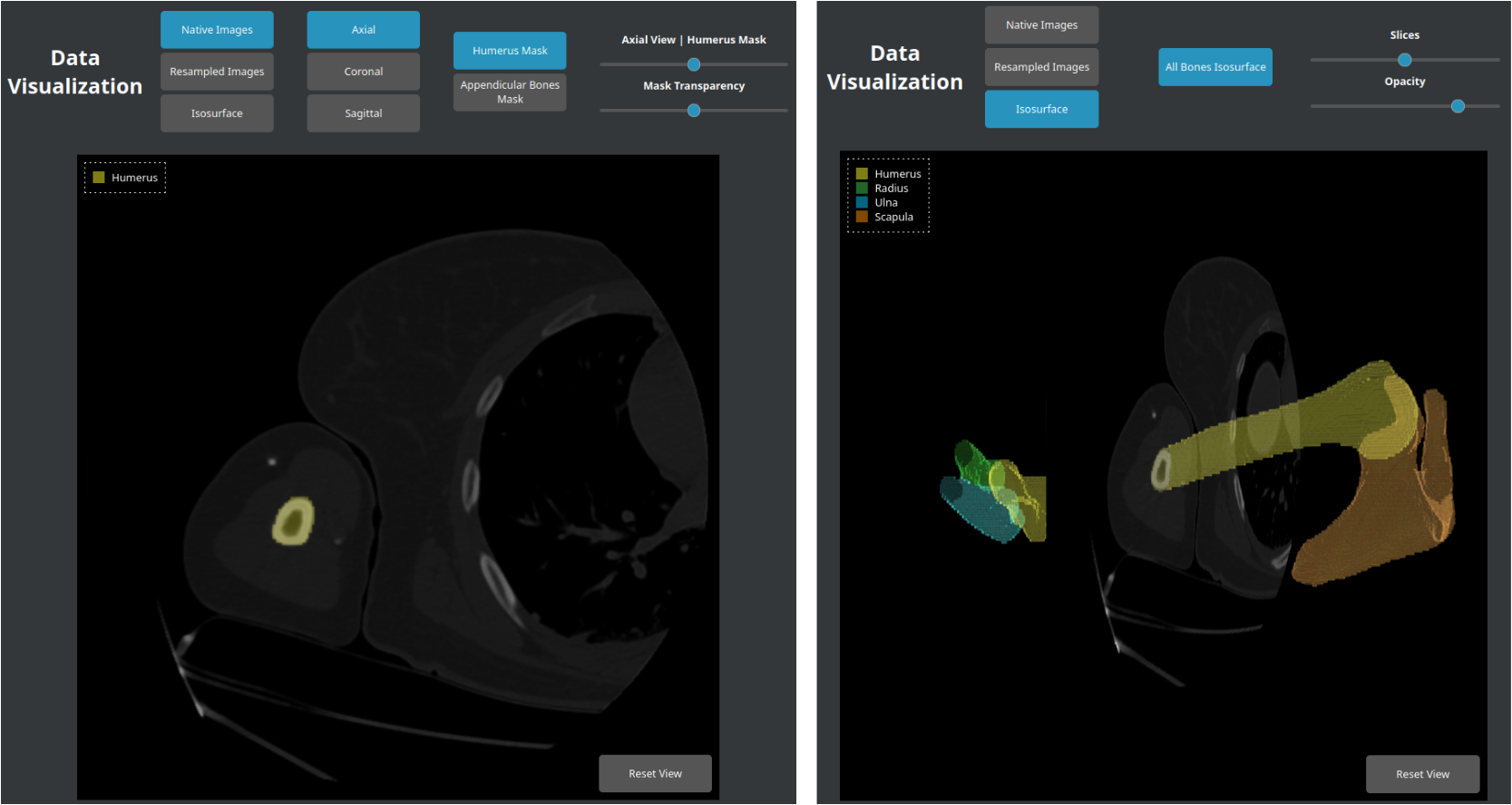
Interactive viewer of CT scans and their proposed segmentation masks developed in HTML with the X_ITE library. This tool allows users to select axial (Left), coronal or sagittal planes, switch between native images or those resampled along the humerus diaphysis axis, add humerus (Left) or appendicular bone masks (radius, ulna, scapula), adjust mask opacity, display isosurfaces of bone segmentations (Right).
8.1.4 Optimizing Intraoperative AI: Evaluation of YOLOv8 for Real-Time Recognition of Robotic and Laparoscopic Instruments
This work was funded by 3IA Côte d'Azur.
Keywords:
Participants: Sébastien Frey [Correspondant], Federica Facente, Wen Wei, Eric Séjor, Patrick Baqué, Matthieu Durand, Hervé Delingette, Pierre Berthet-Rayne, François Bremond, Nicholas Ayache.
YOLOv8 was tested for recognizing robotic and laparoscopic instruments in robot-assisted surgeries 60. Trained on a dataset of 7,400 images and 17,175 annotations, it produced useful results that are quantified in Fig. 7.
- The model achieved strong performances in both detection and segmentation tasks.
- YOLOv8 also demonstrated impressive inference speed, highlighting its potential for real-time clinical applications.
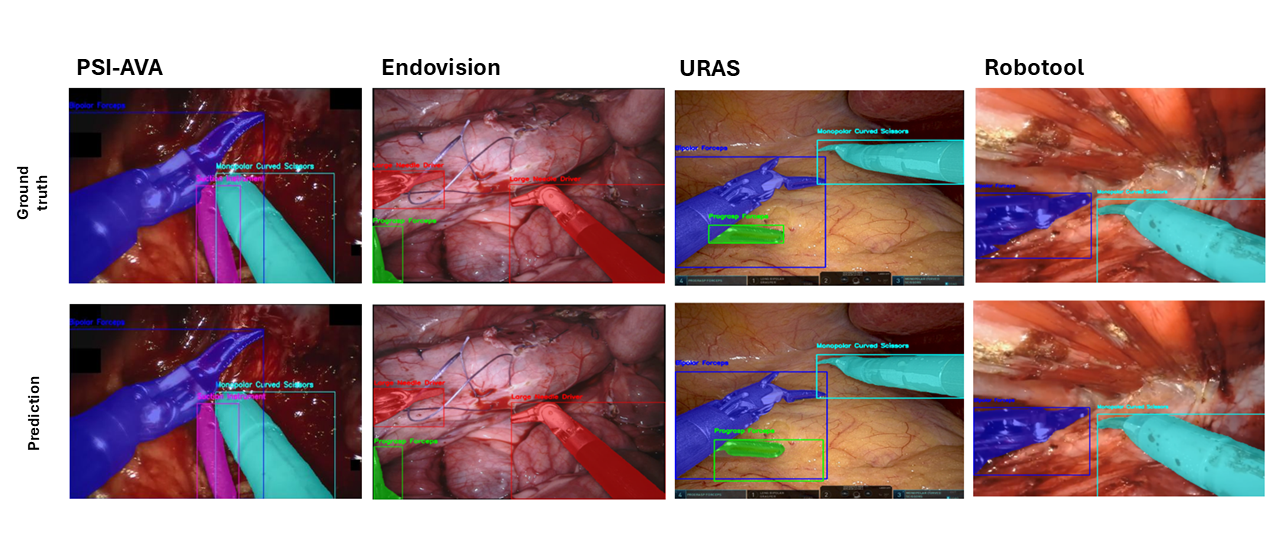
YOLOv8 performance in recognizing robotic and laparoscopic instruments in robot-assisted surgeries: multi-instrument results across various datasets
8.1.5 AI for 3D-2D Registration of CT and Fluoroscopy: A Step Forward in TAVI
This work was funded by 3IA Cote d'Azur.
Keywords:
Participants: Federica Facente [Correspondant], Hervé Delingette, Nicholas Ayache, Pierre Berthet-Rayne.
TAVI is a minimally invasive procedure for treating severe aortic valve stenosis, where accurate valve replacement is essential for optimal outcomes. During the preoperative phase, a CT scan is acquired, and the procedure is planned. The intervention is fluoroscopy-guided, but poor soft tissue visibility complicates guidance and valve positioning. To overcome this, we aim to generate augmented fluoroscopies through 3D-to-2D CT-fluoroscopy registration using a cascade CNN (Convolutional Neural Network), combining coarse and fine stages for accurate, real-time alignment (see Fig. 8).
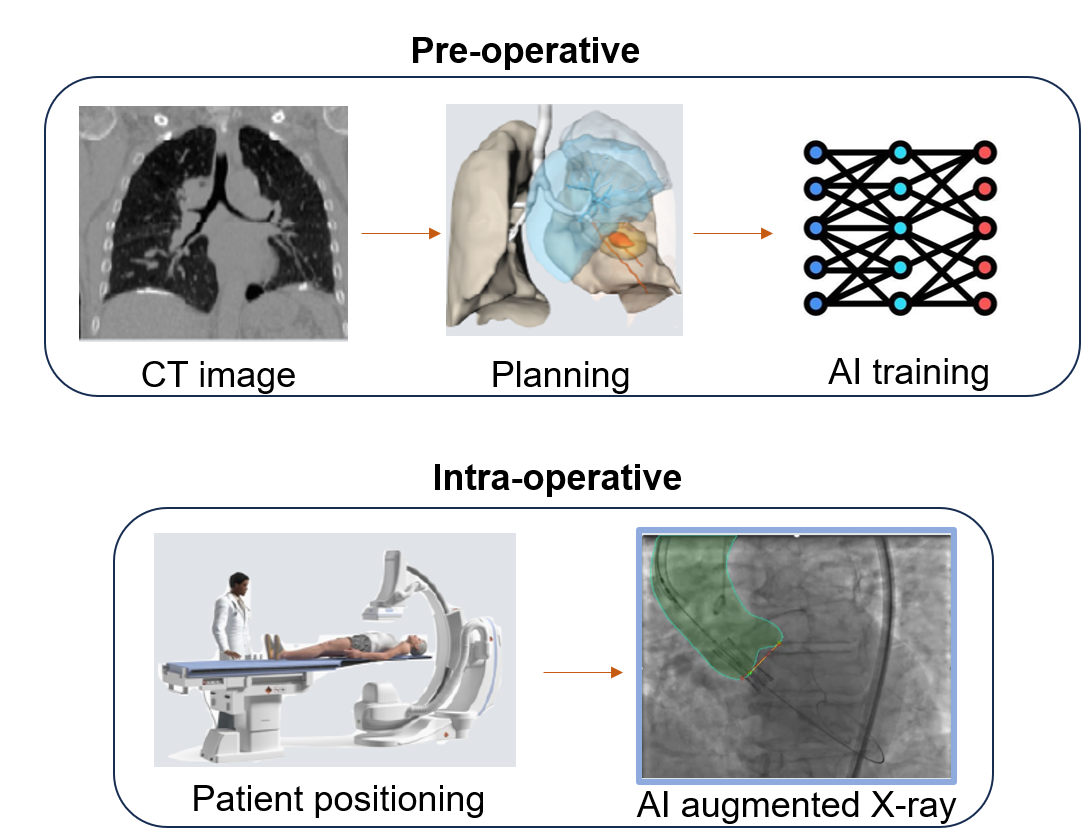
Illustration of the procedure with preoperative CT planning with AI training and the integration of AI-augmented intraoperative fluoroscopy for real-time guidance and improved valve positioning
8.1.6 Development of predictive models in patients with Peripheral Artery Disease
This work was partially funded by the Horizon-Europe project VASCUL-AID (ID 101080947).
Keywords:
Participants: Sébastien Goffart [Correspondant], Hervé Delingette, Juliette Raffort-Lareyre.
Peripheral artery disease (PAD) affects 230 million people due to artery narrowing from atherosclerosis. Despite advanced treatments, disease progression and post-operative complications remain common. To improve pre-surgical planning and outcomes, we developed a PAD amputation risk model using clinical data from 2,366 patients 19. We address the limitations of the Cox Proportional Hazards model by benchmarking survival machine learning models, reassessing key predictors, and creating a patient-level risk tool allowing patients risk stratification (see Fig. 9). Next, we aim to identify imaging patterns predictive of patient outcomes and develop predictive models integrating clinical and imaging data.
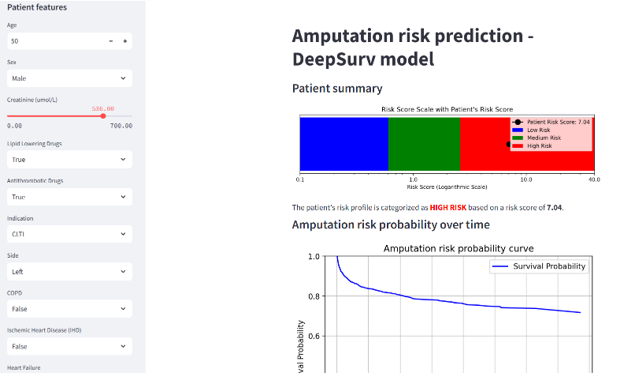
Screenshot of the online tool predicting an amputation risk. The risk score is calculated using 9 pre-operative clinical and biological features and provides patients‘ specific risk score with a classification into 3 risk groups (low, medium or high).
8.1.7 Differentiable Soft Morphological Filters for Medical Image Segmentation
This work was funded by 3IA Côte d'Azur.
Keywords:
Participants: Lisa Guzzi [Correspondant], Maria A. Zuluaga, Fabein Lareyre, Gilles Di Lorenzo, Sébastien Goffart, Juliette Raffort-Lareyre, Hervé Delingette.
The aim of this work is to extend binary morphological operations (see Fig. 10) to probability maps to enable their integration into neural networks 33. Our contributions are as follows:
- A novel definition of probabilistic morphological operators, formulated as the expectation of their binary counterparts.
- A method to translate any binary operation based on Boolean expressions into a single multi-linear or proxy polynomial. These soft morphological filters are differentiable and require no hyperparameter tuning.
- A demonstration of the integration of these filters in segmentation networks, either used inside a loss function or as the final layer of a U-Net, tested in 2 medical imaging applications.
This framework for differentiable soft morphological filters shows that their integration in deep learning architectures improves topological and connectivity performance in medical image segmentation.
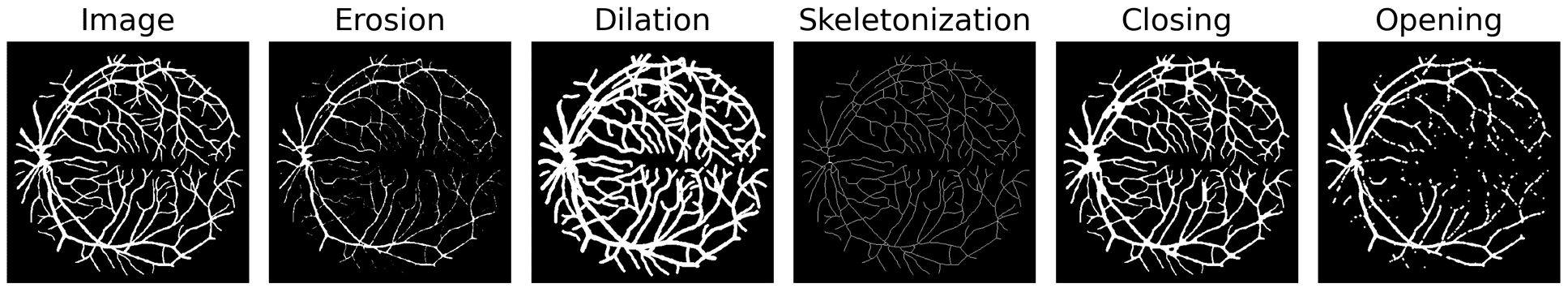
Example of differentiable morphological operations on a binary input of retinal blood vessels.
8.1.8 Generative Medical Image Anonymization Based on Latent Code Projection and Optimization
This project has been supported by the French government, through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002.
Keywords:
Participants: Huiyu Li [Correspondant], Nicholas Ayache, Hervé Delingette.
- We address the medical image anonymization problem with a two-stage solution: latent code projection and optimization (Fig. 11).
- We design a streamlined encoder and propose a co-training scheme to enhance the projection process.
- We optimize the latent code using two deep loss functions: identity loss and utility losss.
- We achieve a favorable trade-off between identity removal and utility preservation 51.
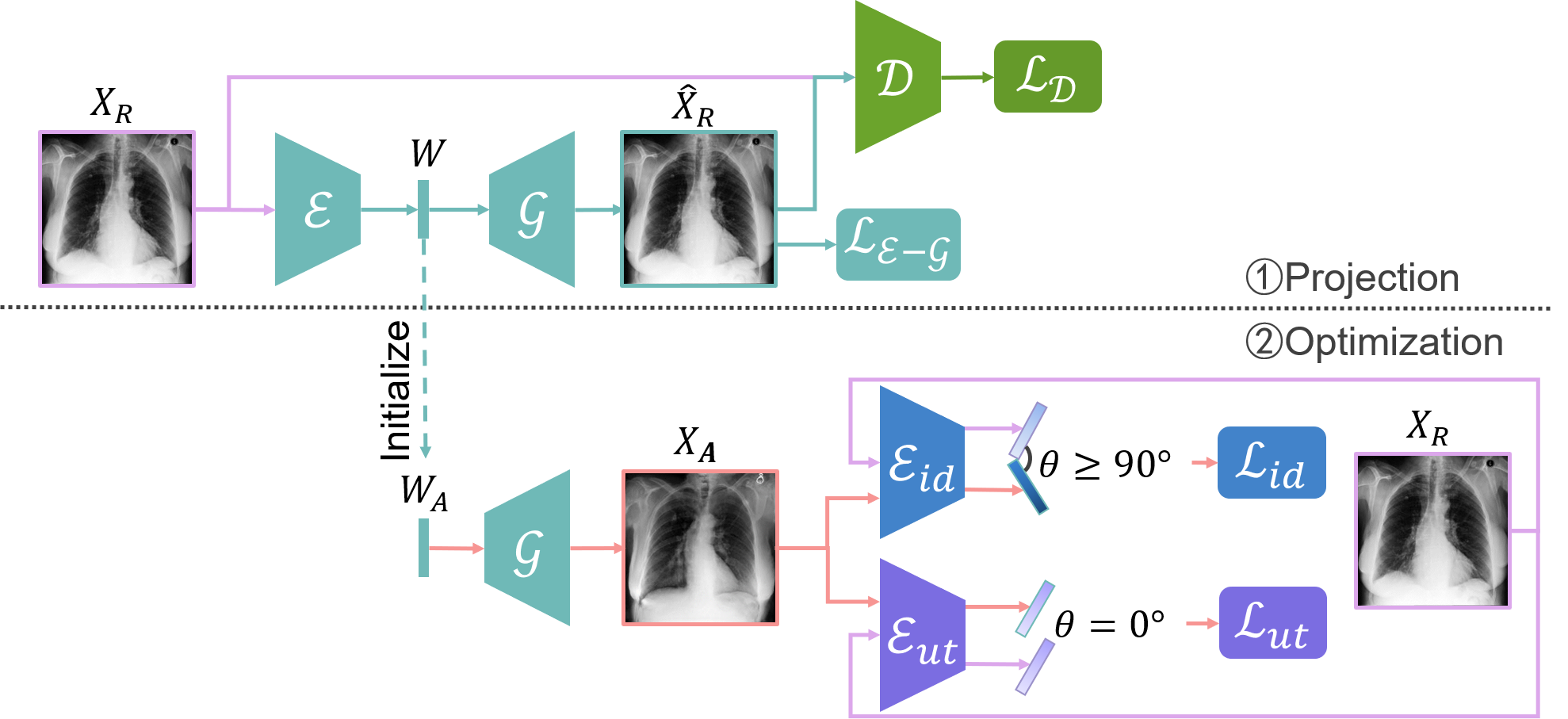
Overview of the proposed method, consisting of two key stages: (1) AE-GAN network for latent code projection, and (2) Latent code optimization using identity loss and utility losss.
8.1.9 Improving the robustness of cancer detection models in medical images studies
The PhD project is funded by the project ANR TRAIN ANR-22-FAI1-0003.
Keywords:
Participants: Huyen Trang Nguyen [Correspondant], Marco Lorenzi, Olivier Humbert.
The goal of the PhD project consists of studying novel approaches to quantify and correct data heterogeneity in medical imaging studies, to lead to robust and representative models for automatic diagnosis of pathological conditions. Specific focus will be given to the definition of novel robust approaches for tumour modeling and segmentation based on the analysis of CT and FDG-PET imaging data (Fig. 12). This project is in collaboration with the Antoine Lacassagne Cancer Center of Nice, France.
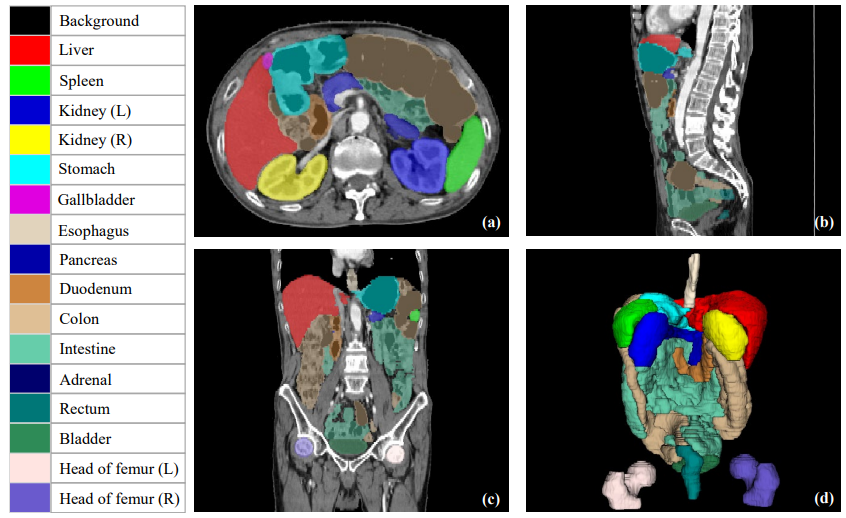
Organ segmentations example. The subfigures (a), (b), (c) show axial, coronal and sagittal planes respectively, while subfigure (d) presents 3D visualization of organ segmentations. The PhD project aims to identity and rectify inaccurate segmentations to enhance quality control for segmentation tools.
8.1.10 Thyrosonics
This project has received funding through BoostUrCAreer from the European Union's Horizon 2020 research and innovation program under grant agreement 847581. It has been co-funded by the Region Provence-Alpes-Côte d'Azur and IDEX UCA
Keywords:
Participants: Hari Sreedhar [Correspondant], Hervé Delingette, Guillaume Lajoinie, Charles Raffaelli.
- This project investigates thyroid ultrasound machine learning applications in the context of inter-expert variability, annotation difficulties, and the physics of nonlinear propagation (see Fig. 13).
- The thesis 52 resulting from the doctoral work of this project was successfully defended on October 25th, focusing on a study of inter-expert variability in thyroid nodule ultrasound evaluation, active learning techniques to facilitate training of AI algorithms for thyroid ultrasound, and an AI-assisted strategy for the estimation of an acoustic parameter using a physics-based strategy assessing nonlinear propagation.
- Elements of the work pertaining to inter-expert variability were presented at the 2024 Atelier Thyroïde de Sète (May 18).
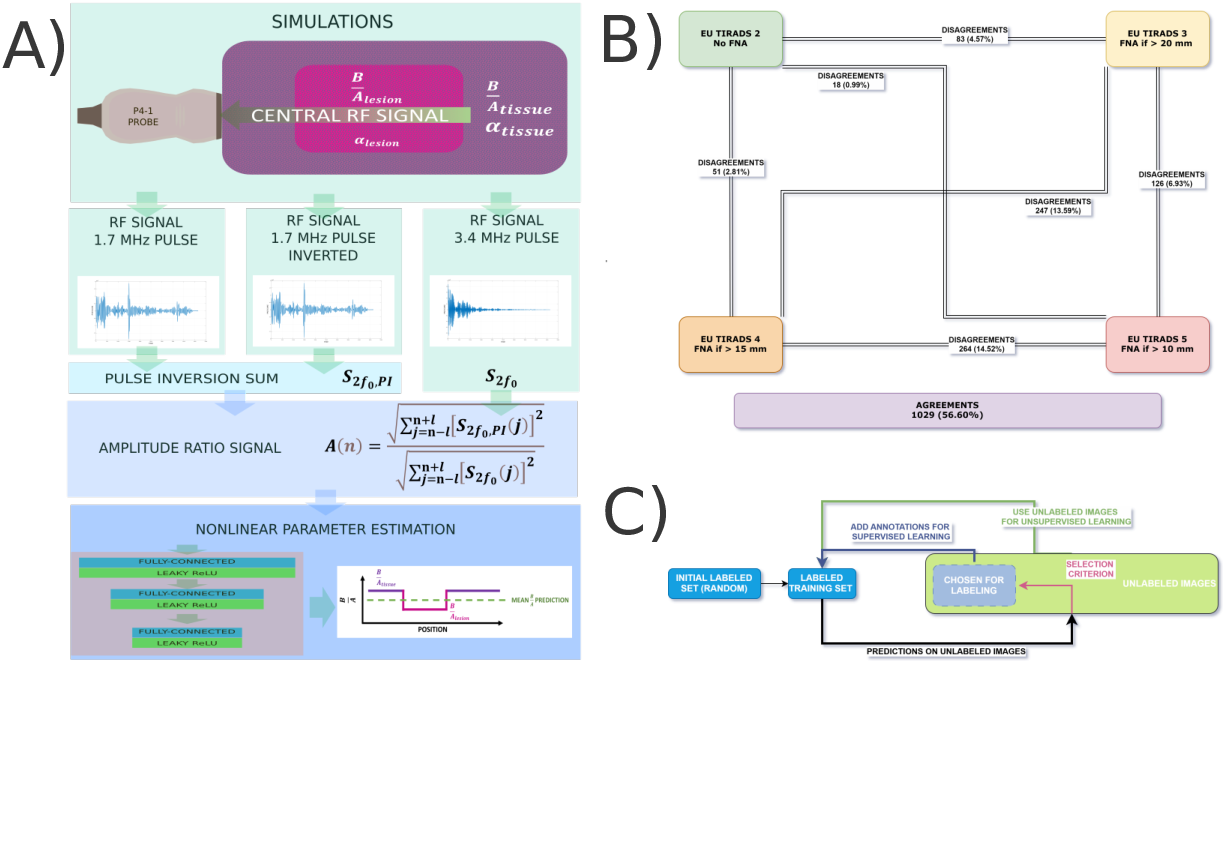
A) Strategy for acoustic nonlinear parameter estimation in soft tissue-like simulated media. B). Illustration of disagreements among French thyroid ultrasound experts in EU-TIRADS scoring. C) Cycle of pool-based active learning strategies.
8.2 Imaging & Phenomics, Biostatistics
8.2.1 Assessing Ionic Current Blockades and Electromechanical Biomarkers' Interrelations Through a Novel Multi-Channel Causal Variational Autoencoder
This work was supported by the European Union's Horizon 2020 Research and Innovation Program SimCardioTest project which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No. 101016496.
Keywords:
Participants: Safaa Al-Ali [Correspondant], Maria Teresa Mora, Beatriz Trenor, Maxime Sermesant, Irene Balelli.
Certain drugs can disrupt normal cardiac activity, increasing the risk of life-threatening arrhythmias like Torsade de Pointes (TdP). In 29, a novel Multi-Channel Causal Variational Autoencoder (MC
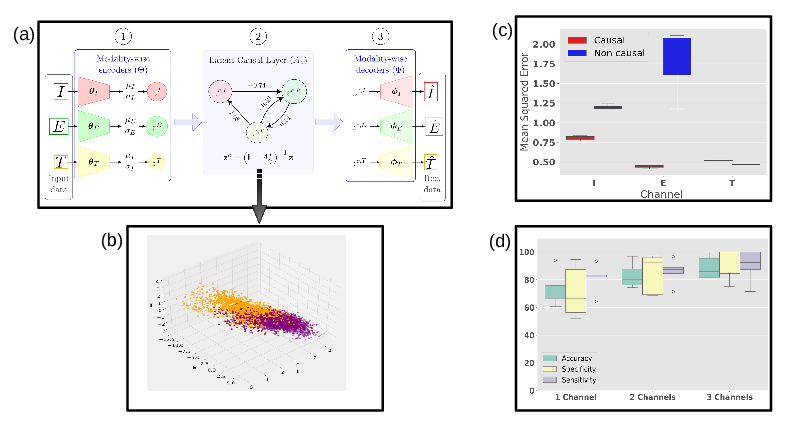
(a) MC
8.2.2 A Multi-omic Integration Approach to Understand the Etiology of Fragile X Syndrome
This PhD is funded by the Neuromod Institute.
Keywords:
Participants: Khatir Wassila [Correspondant], Balelli Irene, Lorenzi Marco, Gwizdek Carole.
This project seeks to investigate the etiology of Fragile X Syndrome (FXS) (Fig. 15 A) by leveraging omic data from public databases, in combination with newly generated data through a collaboration with the IPMC. Over the course of this year, we:
- Collected transcriptomic and translatomic data from 19 studies and proposed to use a multi-channel variational autoencoder (MCVAE) to jointly analyze these two omic layers. This model performs cross-modal reconstruction to create a shared latent space, capturing the underlying relationships between the two modalities. We established a baseline for normal transcription-translation relationships by training the MCVAE on a subset of fmr1 +/+ samples and then tested it on fmr1 -/- samples. Abnormalities were defined as reconstruction error size effect exceeding a specific threshold.
- Worked on proteomic analysis to study the effect of pharmacological rescue of the overactivated Rac1 and sex dimorphism on starvation in FXS models (Fig. 15 B).
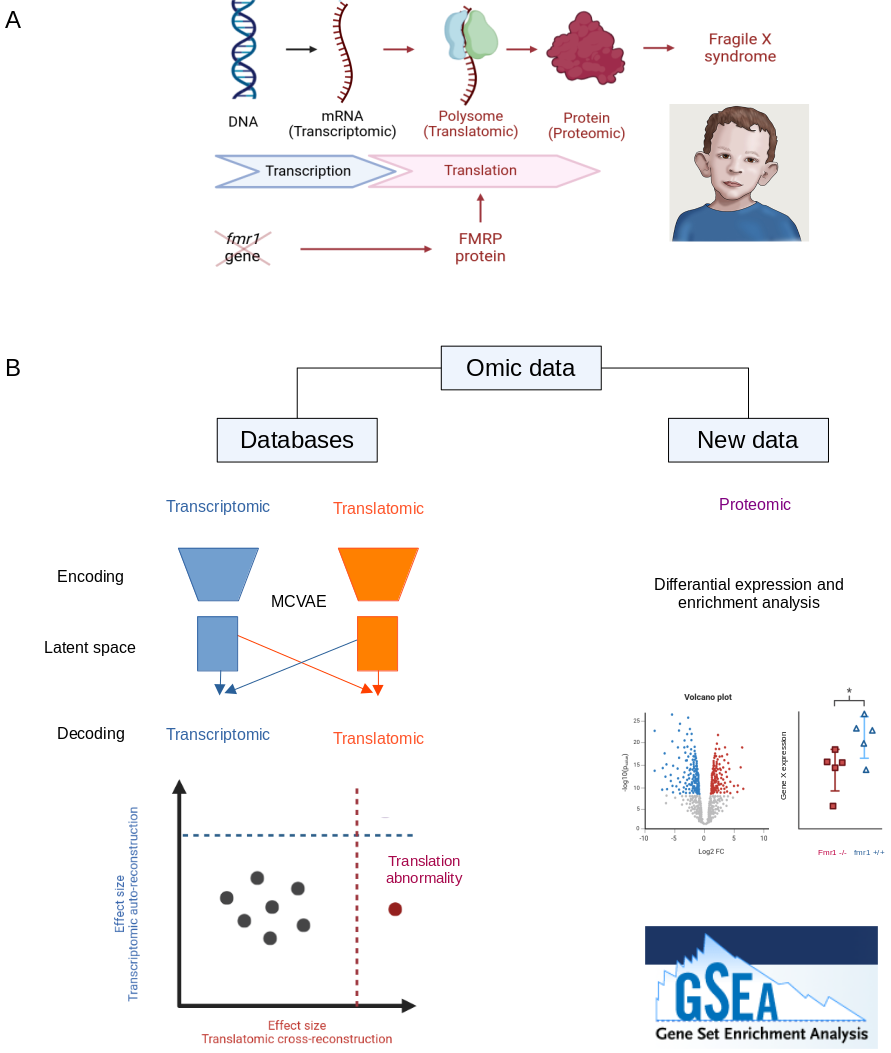
(A) FXS results from a lack of expression of the fmr1 gene, which encodes the FMRP protein, a regulator of translation. (B) To investigate the etiology of FXS, we employed an MCVAE approach to identify abnormalities in the transition between transcriptomic and translatomic levels. In parallel, we generated proteomic data and performed differential expression and enrichment analyses.
8.2.3 Disease Progression Modelling and Stratification for detecting sub-trajectories in the natural history of pathologies
The work has been supported by Michael J. Fox Foundation for Parkinson's Research (MJFF), and by the French government, through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA- 0002, by the TRAIN project ANR-22-FAI1-0003-02, and by the ANR JCJC project Fed-BioMed 19-CE45-0006-01.
Keywords:
Participants: Alessandro Viani [Correspondant], Marco Lorenzi, Emile D'Angremont, Boris Gutman.
- Development of the Disease Progression Modeling and Stratification (DPMoSt) method, designed to analyze biomarker sensitivity in subpopulations 45. The source code is available on GitHub.
- Application of DPMoSt to Parkinson's disease on both ENIGMA and PPMI datasets.
- Application of DPMoSt to the ALzheimer's disease on ADNI dataset. Notably, this analysis revealed an association between APOE4 and accelerated cognitive decline (see Fig. 16).
- Presentation of the results at the Longitudinal Disease Tracking and Modeling with Medical Images and Data workshop, part of the MICCAI conference.
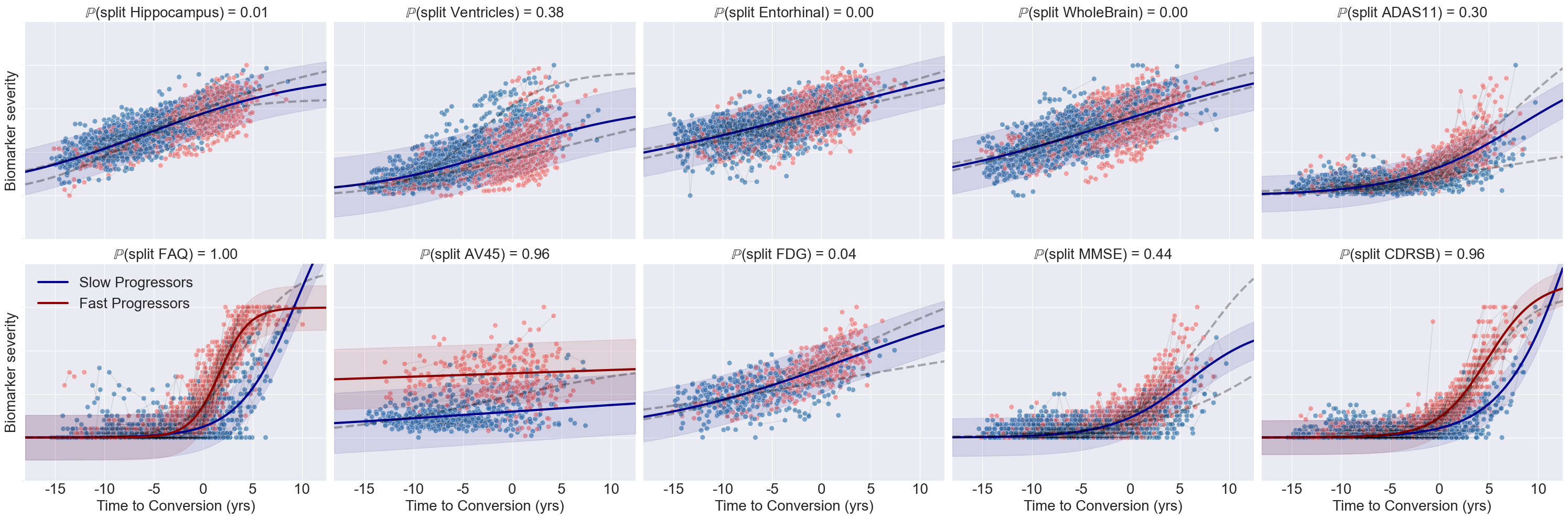
DPMoSt-estimated biomarker trajectories derived from the ADNI dataset. The y-axis represents biomarker severity, while the x-axis denotes re-parameterized time. Dots indicate individual longitudinal measurements for each subject. Solid lines show the estimated trajectories, with shaded bands representing standard deviations. Colors distinguish the two subtypes identified by the model. Each plot title displays the probability of biomarker specificity for the corresponding subtype.
8.3 Computational Anatomy & Geometric Statistics
8.3.1 Riemannian Metrics on Correlation Matrices and Quotient Geodesics
This work was supported by ERC grant #786854 G-Statistics from the European Research Council under the European Union's Horizon 2020 research and innovation program, and by the French government through the 3IA Côte d'Azur Investments ANR-19-P3IA-0002 managed by the National Research Agency.
Keywords:
Participants: Olivier Bisson [Correspondant], Xavier Pennec.
Correlation matrices are widely used to describe brain connectivity in anatomical and functional neuroimaging. The contributions of this work can be summarized as follows (see Fig. 17):
- Implement various Riemannian metrics for the space of full-rank correlation matrices in the Python package geomstats.
- Derive an expression for the differential of the stretch tensor in any dimension, expressed as a solution to a Sylvester equation.
- Use this formula to link the Riemannian geometry of the quotient homogeneous space
- Investigate an expression for the quotient geodesics of the Frobenius metric on covariance matrices and its extension to full-rank correlation matrices 15.
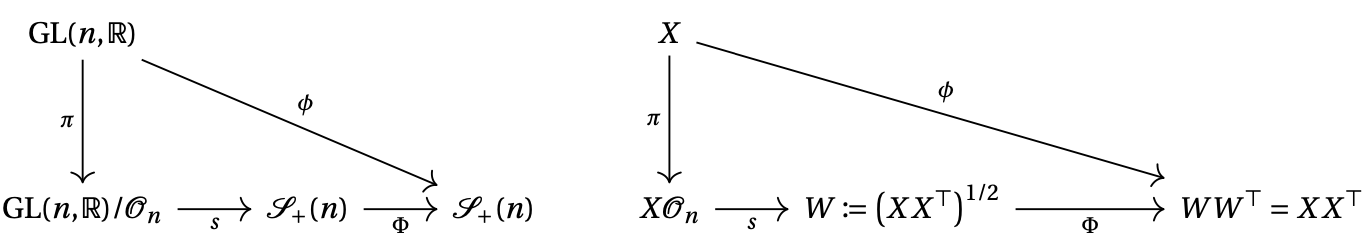
Commutative diagram of the principal bundle and its symmetric section.
8.3.2 Numerical optimization on stratified sets
This work was supported by the ERC grant #786854 G-Statistics from the European Research Council under the European Union's Horizon 2020 research and innovation program and by the French government through the 3IA Côte d'Azur Investments ANR-19-P3IA-0002 managed by the National Research Agency.
Keywords:
Participants: Guillaume Olikier [Correspondant], Irène Waldspurger.
This work considers nonconvex optimization problems for which even computing an approximate local minimizer is intractable. For such problems, algorithms are only expected to find a stationary point, i.e., a point that satisfies a necessary condition for local optimality. The paper 64 establishes that the projected gradient descent algorithm enjoys the strongest stationarity properties that can be expected. The paper 36 studies two definitions of a retraction, and shows that the weaker, although possibly generating discontinuous curves (Fig. 18), should be preferred in numerical optimization.
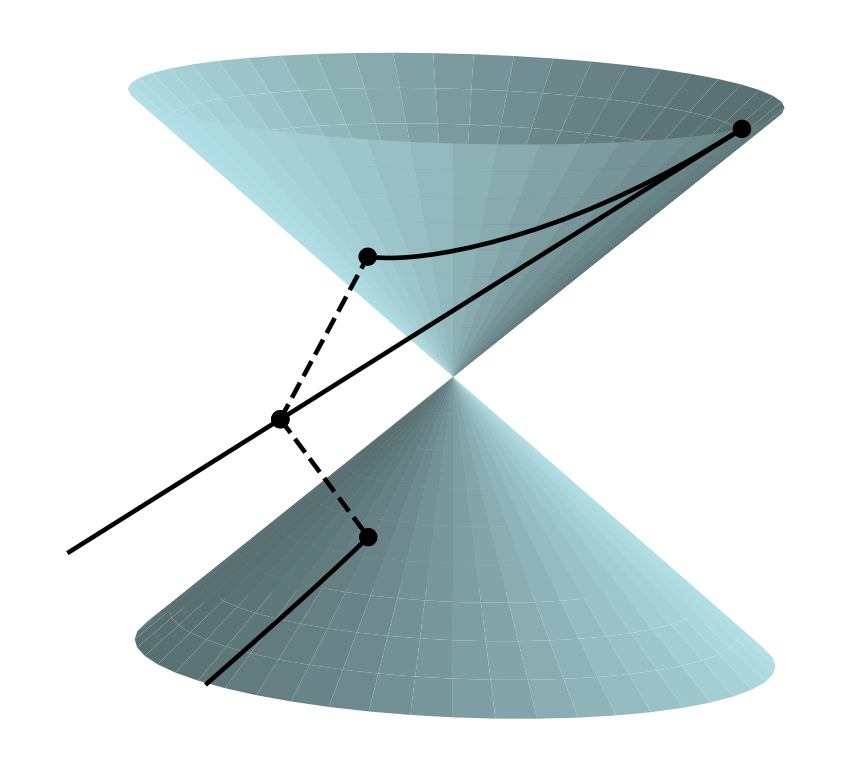
The double cone is a representation of the set of 2-by-2 real symmetric singular matrices. For every positive real number
8.3.3 The curse of isotropy: from principal components to principal subspaces
This work was supported by the ERC grant #786854 G-Statistics from the European Research Council under the European Union Horizon 2020 research and innovation program and by the French government through the 3IA Côte d'Azur Investments ANR-19-P3IA-0002 managed by the National Research Agency.
Keywords:
Participants: Tom Szwagier [Correspondant], Xavier Pennec.
Principal components of a covariance matrix with equal eigenvalues are defined up to an arbitrary rotation within the eigenspace they span and they cannot be interpreted alone. As a consequence, the eigenvectors associated to almost-equal eigenvalues suffer from a large intersample variability (see Fig. 19). We call this effect the “curse of isotropy” and we analyze it in 67. We notably aim at measuring how many samples we need to identify faithfully eigenmodes of neighboring eigenvalues from a statistical point of view.

Illustration of the curse of isotropy. When two population-covariance eigenvalues are equal, the associated sample-covariance eigenvectors have an isotropic variability, which is a pitfall for interpretation.
8.4 Computational Cardiology & Image-Based Cardiac Interventions
8.4.1 Identification of sub-groups with high risk of stroke by the shape of the left atrium
This work was supported by the PEPR Santé Numérique, ChroniCardio project.
Keywords:
Participants: Nicolas Drettakis [Correspondant], Maxime Sermesant.
- Clustering on 61 features directly extracted from a 3D representation of the left atrium following the work of Josquin Harrison. Reflection on how to determine the best clustering on a set of parameters using a score based on the idea of minMax (Fig. 20).
- Clustering directly on the meshes representing the left atriums. First we tried to use distance matrices after aligning the meshes, then we tried to use the Laplace-beltrami operator to represent the mesh by his first n eigen values, finally we decided to use neural network for meshes like autoencoders to build a representation of every mesh.
- The end of the internship was used to prepare a PhD which will first focus on extending Josquin's pipeline to new data and then try to apply our methods to these new meshes.
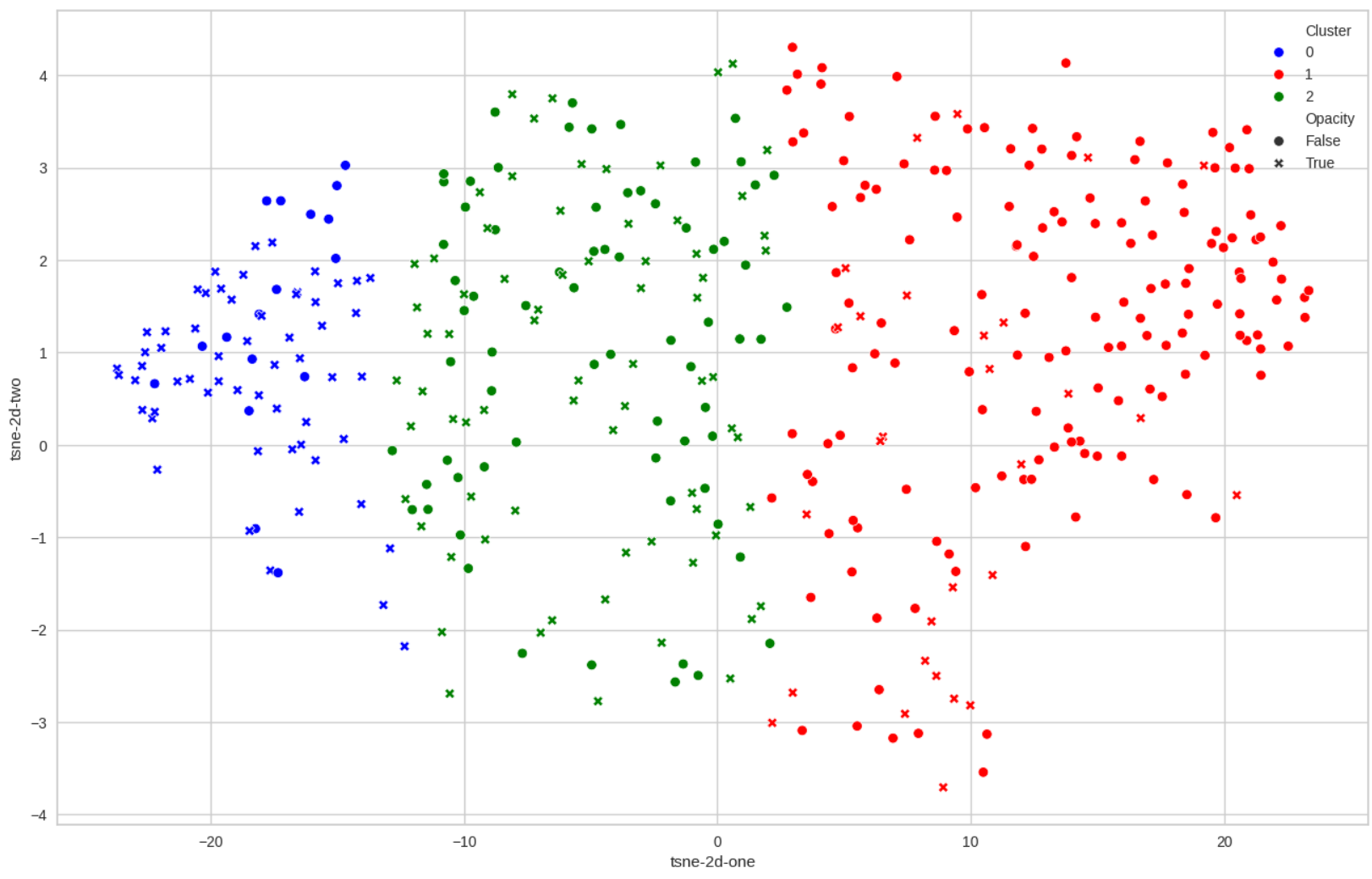
The clustering which obtained the best score. Blue is the high level of risk, green the mid one and red the low one. Opacity represents a defect of opacity which is known to be linked with the risk of stroke.
8.4.2 A Reduced-Order 0D Model for Cardiac Mechanics: Integration of Ultrafast Ultrasound-Derived Stiffness for Patient-Specific Analysis
This doctoral research is carried out under a Contrat doctoral de droit public by Inria, fully funded through the PEPR Santé Numérique program via ANR Financement d'Agences de financement de la recherche. It is conducted as part of the ChroniCardio project, with the funding period spanning from September 1, 2024, to August 31, 2027.
Keywords:
Participants: Camilla Ferrario [Correspondant], Maxime Sermesant, Jairo Rodriguez Padilla, Maelys Venet, Olivier Villemain.
-
Multiscale Cardiac Modeling: Developed a 0D reduced-order model coupled to a closed-loop circulatory system, integrating cardiac mechanics with systemic and pulmonary circulation. The model employs a Windkessel framework, using an RCR representation for arteries and an RC representation for veins, where R denotes resistance and C capacitance in the electro-hydraulic analogy. This computationally efficient approach models left and right ventricles as spherical chambers characterized by wall thickness (
- Biomechanical and Clinical Insights: Integrated myocardial stiffness (MS), a material property derived from ultrafast ultrasound, to quantify active and passive contributions to cardiac contractility. By linking stiffness to clinical data, the model facilitates personalized assessments of cardiac function in health and disease.
-
Sensitivity and Parameter Analysis: Conducted a systematic sensitivity analysis on key parameters, such as
- Clinical Validation and Application: Validated the model using ultrafast ultrasound data from 20 healthy individuals, establishing baseline dynamics for comparison. Future applications aim to extend this work to pathological conditions, including hypertrophic cardiomyopathy (HCM), to better understand disease progression and inform treatment strategies.
- Bridging Scales: The research emphasizes multiscale integration, linking cellular dynamics (e.g., calcium regulation) to whole-heart behavior. This approach enables comprehensive insights into cardiac performance and potential responses to therapeutic interventions.
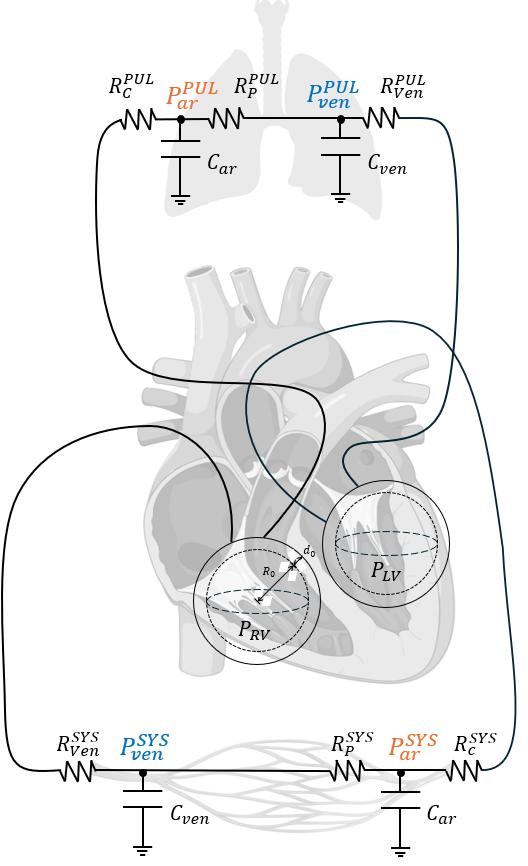
Schematic of the reduced-order 0D circulatory model. Left (LV) and right (RV) ventricles are modeled as spheres, with systemic and pulmonary circulations represented using RCR (arteries) and RC (veins) Windkessel frameworks. The model integrates myocardial stiffness components for patient-specific analysis.
8.4.3 From images to geometric features of the Left Atrium
This Work has been funded by PARIS - ERACoSysMed grant number 15087, by G-Statistics - ERC grant number 786854 and has been supported by the French government through the National Research Agency (ANR) Investments in the 3IA Côte d'Azur (ANR-19-P3IA-000). The authors are grateful to the OPAL infrastructure from Université Côte d'Azur for providing computing resources and support.
Keywords:
Participants: Josquin Harrison [Correspondant], James Benn, Anaelle Zanella, Hubert Cochet, Maxime Sermesant.
In this project 50, we study the link between the anatomy of the left atrium and the risk of stroke. We do this by building a pipeline to extract geometric features of the left atrium from CT-scans (see Fig. 22). We then use these features to derive a risk score. In parallel, we developed a novel technique improving the use of surface processing via neural networks (published in 34). This new method was then used within the main project to increase the robustness and speed of the pipeline.
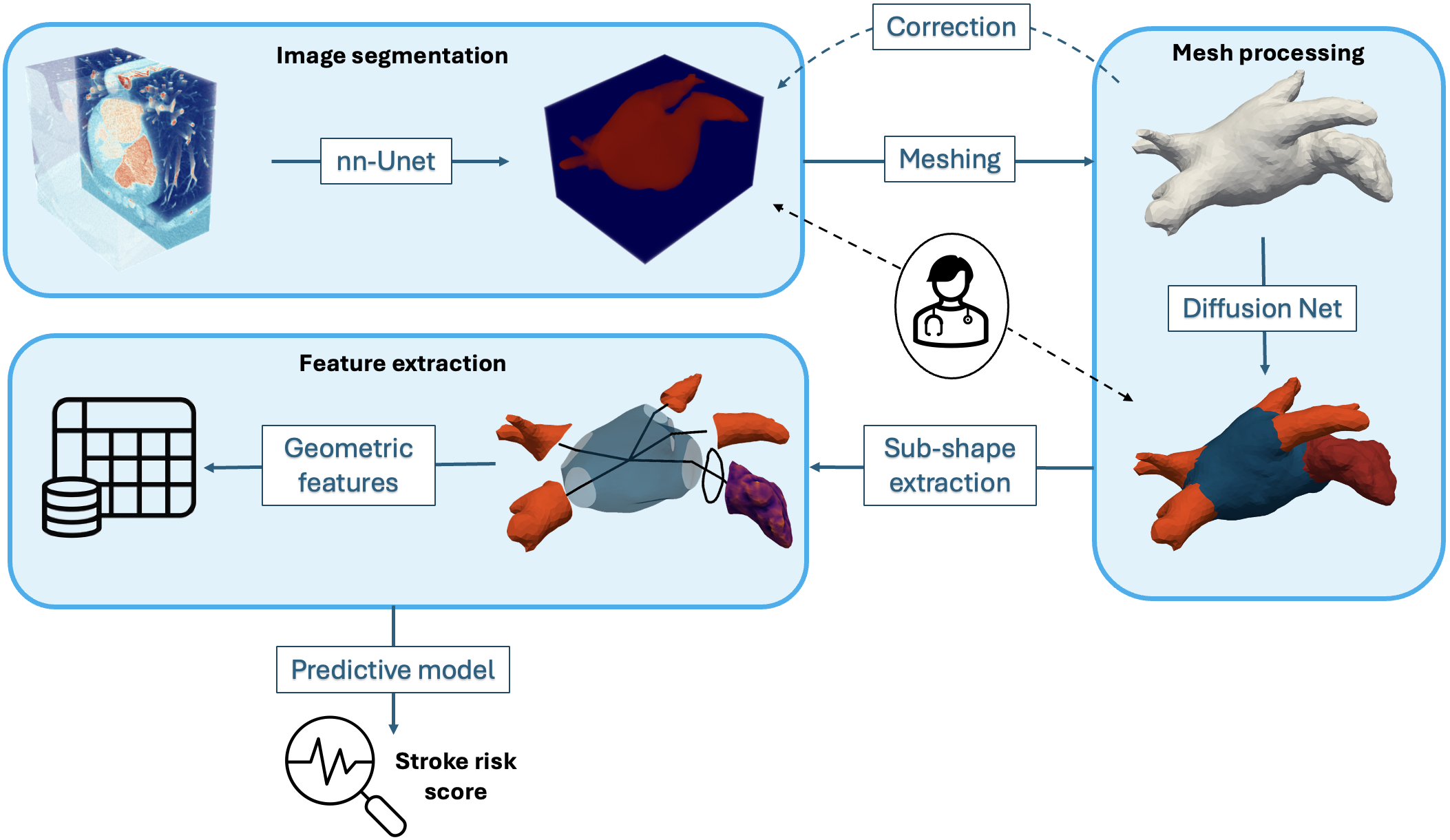
An end-to-end pipeline to represent the left atrium as a set of geometric features from CT-scans.
8.4.4 Deep Learning Meets Numerical Modelling AI and Biophysics for Computational Cardiology
This work has been supported by the French government through the National Research Agency (ANR) DeepNum project, contract number ANR-21-CE23-0017.
Keywords:
Participants: Maëlis Morier [Correspondant], Maxime Sermesant, Patrick Gallinari.
The aim of this project is to develop a faster tool to understand the propagation of cardiac action potential, in order to prevent arrhythmia. Using AI and mathematical tools, the objectives are (Fig. 23):
- to accelerate standard simulations;
- to use AI to determine the at-risk areas;
- to automatically parameterize Partial Differential Equations.
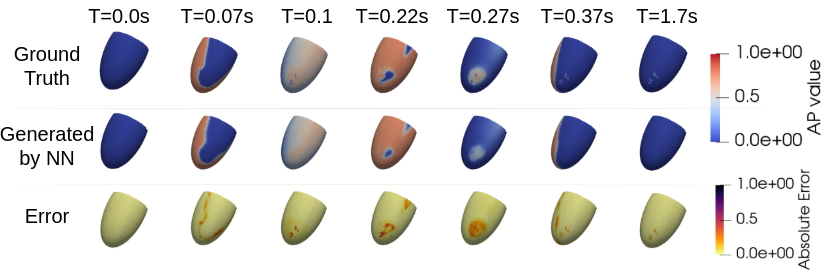
Comparison of simulations generated using the Finite Element Method (FEM) (top, ground truth) and a Graph Neural Network (GNN) (middle, generated by NN). The absolute error (bottom) demonstrates accurate propagation of the action potential, with minor discrepancies in the scar region. The FEM simulation required approximately 40 minutes, whereas the GNN inference completed in 5 minutes.
8.4.5 Multi-Modality Risk Stratification of Sudden Cardiac Death based on imaging, clinical data, and ECG
This work has been supported by the French government through France 2030 (MediTwin), the National Research Agency (ANR) Investments in the Future with 3IA Côted'Azur (ANR-19-P3IA-000), LIRYC (ANR-10-IAHU-04) and ChroniCardio (ANR-22-PESN-0015).
Keywords:
Participants: Evariste Njomgue [Correspondant], Buntheng Ly, Maxime Sermesant, Hubert Cochet.
Sudden cardiac death (SCD) remains a leading cause of mortality worldwide, accounting for a significant proportion of cardiovascular deaths. Despite advancements in medical therapies and device-based interventions, predicting individuals at risk of SCD remains a complex challenge. Current clinical risk stratification models often rely on single-modality metrics, such as left ventricular ejection fraction (LVEF), which, while valuable, lack the sensitivity and specificity to identify a large proportion of at-risk individuals. The objective of this study is to integrate state-of-the-art biomarkers to enhance the precision and effectiveness of SCD risk stratification:
- Sequential fusion (see Fig.24) for multi-modal ventricular arrhythmia (VA) classification: Inspired by constraints-based models, we proposed the Sequential Fusion model. Constraints-based model refers to a type of modeling approach that incorporates constraints to guide the learning process or restrict the possible solutions 70.
- Handling “scar age” missing values: scar age is statistically significant to separate both populations (VA+/VA-). In addition, precise estimation of scar age is essential for assessing clinical outcomes and optimizing treatment strategies in post-myocardial infarction management.
- Post ventricular tachycardia (VT) ablation survival analysis: Survival analysis after VT ablation is pivotal in understanding the long-term efficacy and safety of the procedure.
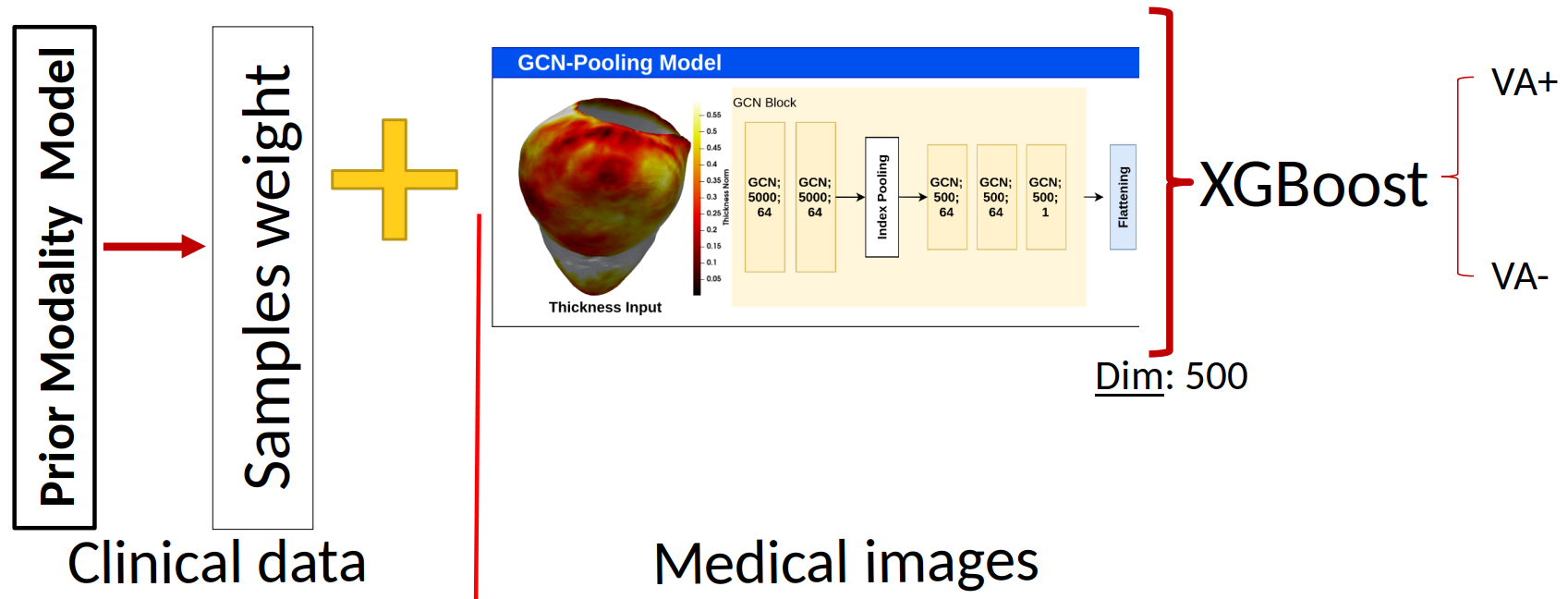
Sequential Fusion Model
8.4.6 Fast electromechanical models for translational research
This work has received funding from the European Union Horizon 2020 research and innovation programme under grant agreement No 101016496 (SimCardioTest), and from the ANR's PEPR project ChroniCardio, contract number 2023000289.
Keywords:
Participants: Jesus Jairo Rodríguez Padilla [Correspondant], Javier Villar-Valero, Buntheng Ly, Rafael Silva, Beatriz Tenor, Mihaela Pop, Maxime Sermesant.
In this work 37 we propose to leverage on computer simulations to study the inducibility of ventricular tachycardia (VT) by replicating the stimulation protocols typically employed in the clinics, but testing different locations of the stimulus site (Top frame in Fig. 25). This will allow us to gain insight into the effect of stimulus location on the electrophysiological response in the presence of scar tissue.
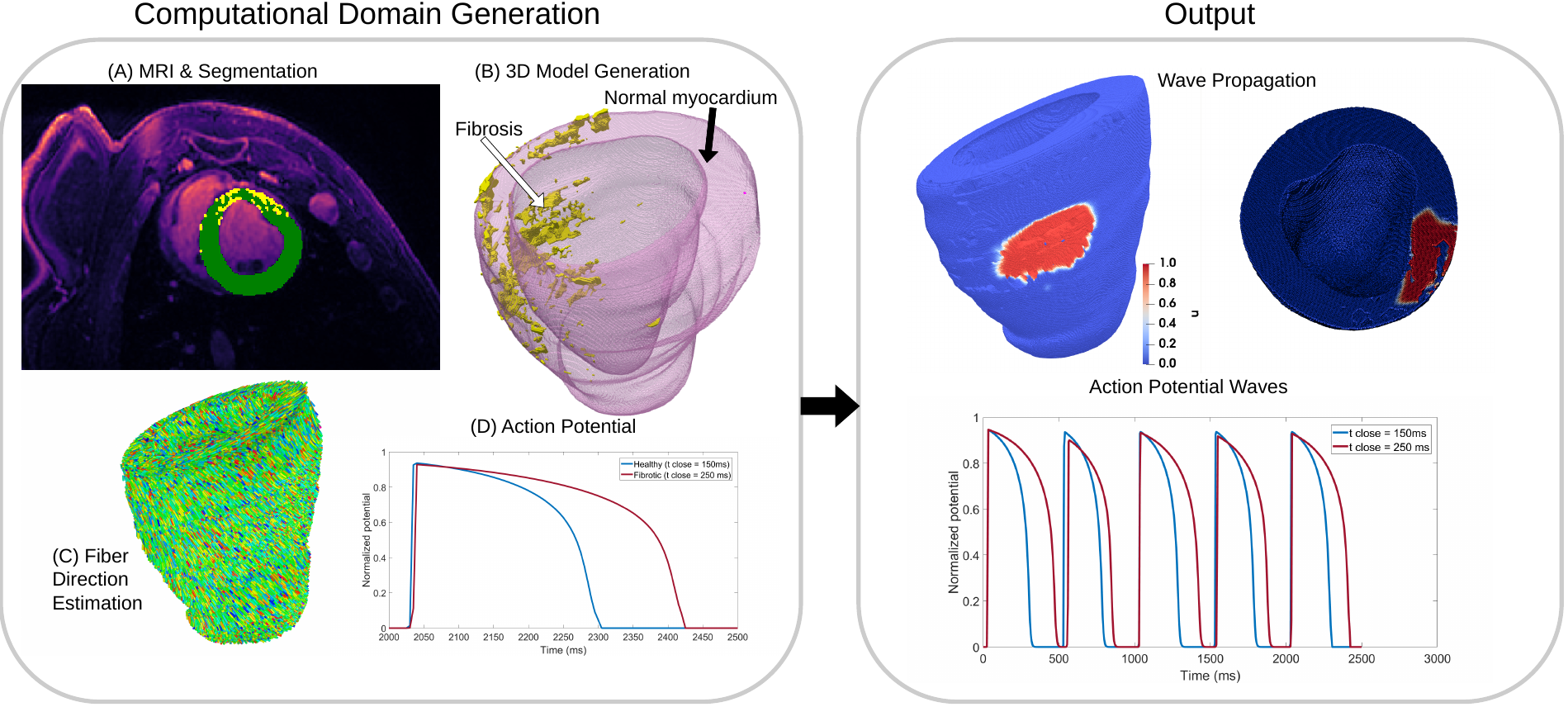
Additionally, we explored the effect of the doxorubicin drug, widely used to treat cancer, on cardiac wave propagation 43. This drug has been shown to produce, as a side effect, diffuse fibrosis in the myocardium. We combine computer simulations and pre-clinical data to study the effect of this induced diffused fibrosis on action potential propagation. Figure 25, bottom frame, shows the pipeline that we employ in this study.
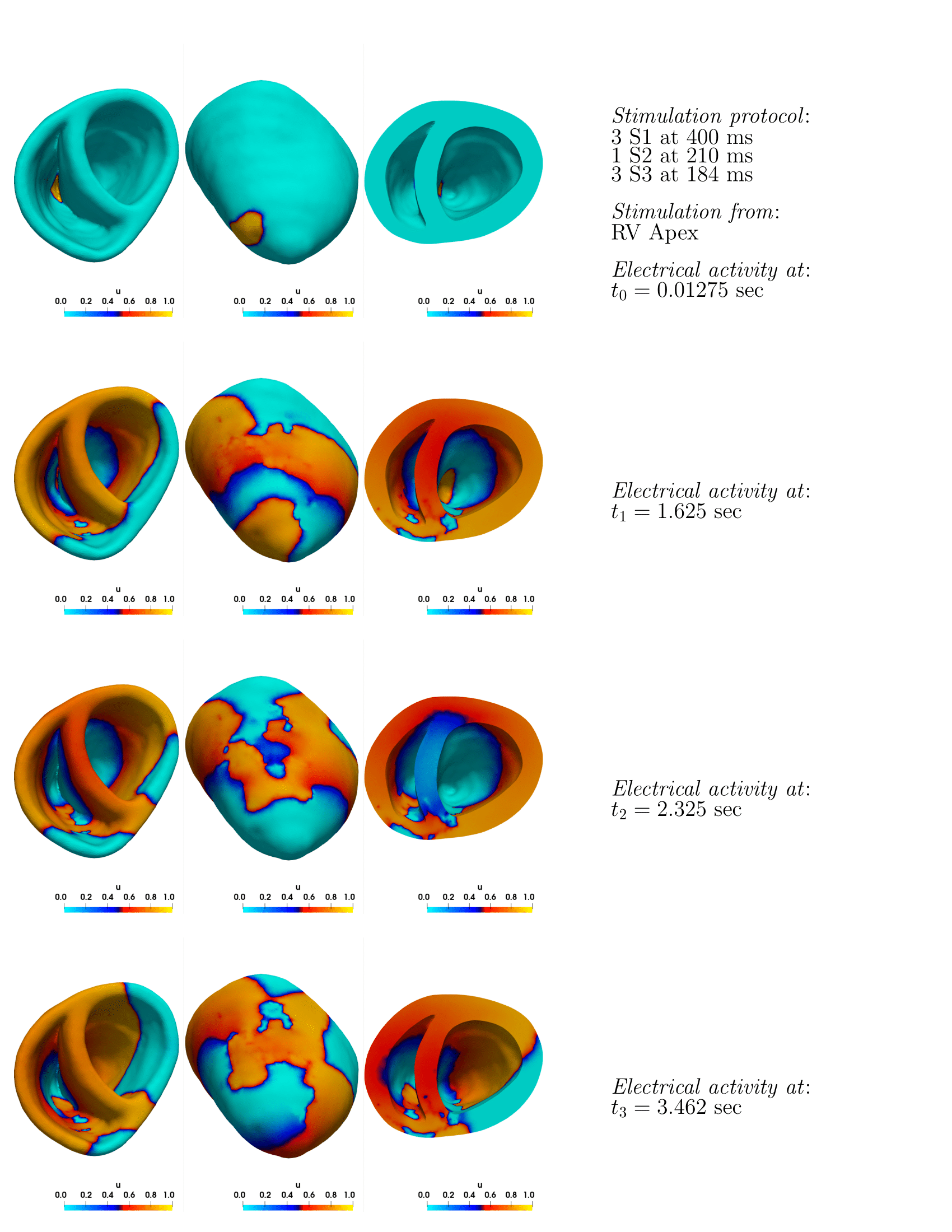
Top frame: ventricular tachycardia project; Bottom frame: chemotherapy induced cardiotoxicity project.
8.4.7 Automatic Extraction of Activation Recovery Intervals from Intracardiac Electrograms
This work was supported by the SimCardioTest project which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No. 101016496, by the French government, through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002 and by Canadian CIHR project grant (PJT) 1532121, which also provided the preclinical data.
Keywords:
Participants: Rafael Silva [Correspondant], Jairo Rodriguez-Padilla, Mihaela Pop, Maxime Sermesant.
This study proposes a methodology to automatically extract activation and recovery times from Intracardiac Electrogram Recordings (iEGMs) 38, 68, often manually annotated by clinicians. It uses surface ECG fiducials and wavelet decomposition to detect activation/recovery times and analyzes Activation Recovery Interval (ARI) data from infarcted porcine hearts. Results reveal ARI differences across tissue types and rhythms, highlighting the need for personalized repolarization studies (Fig. 26).
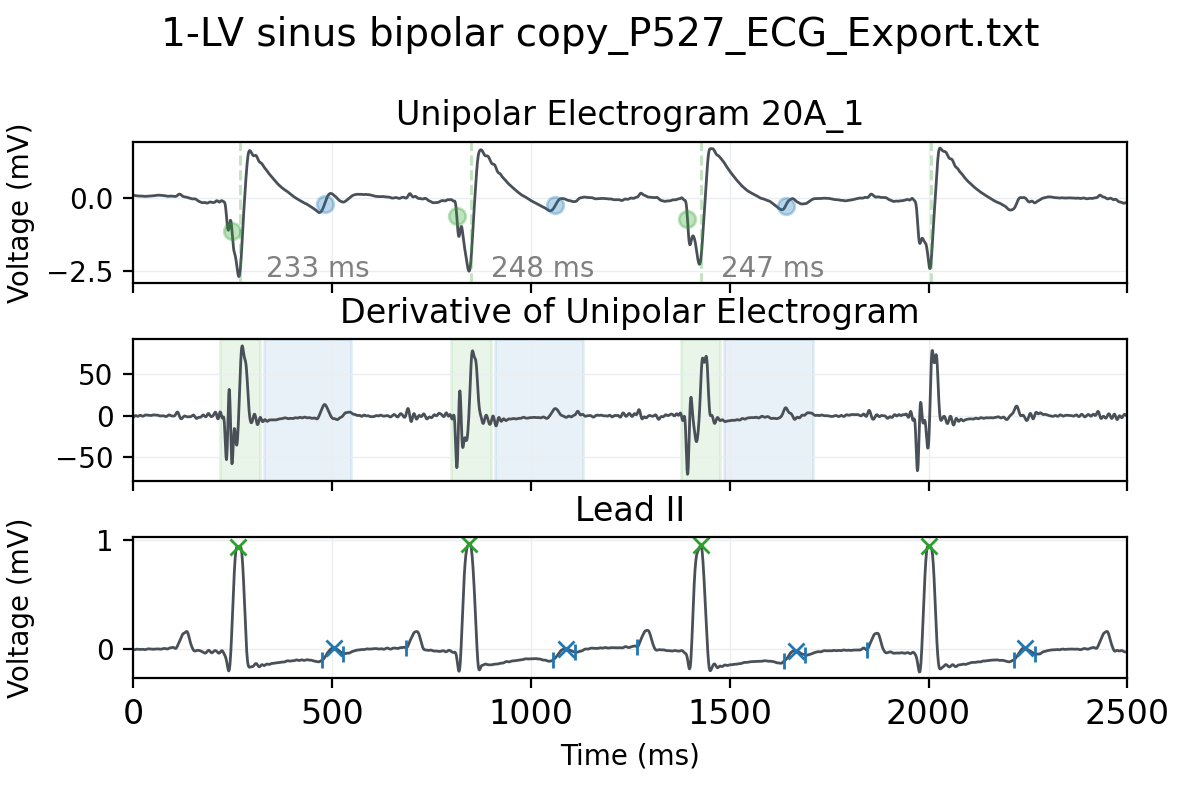
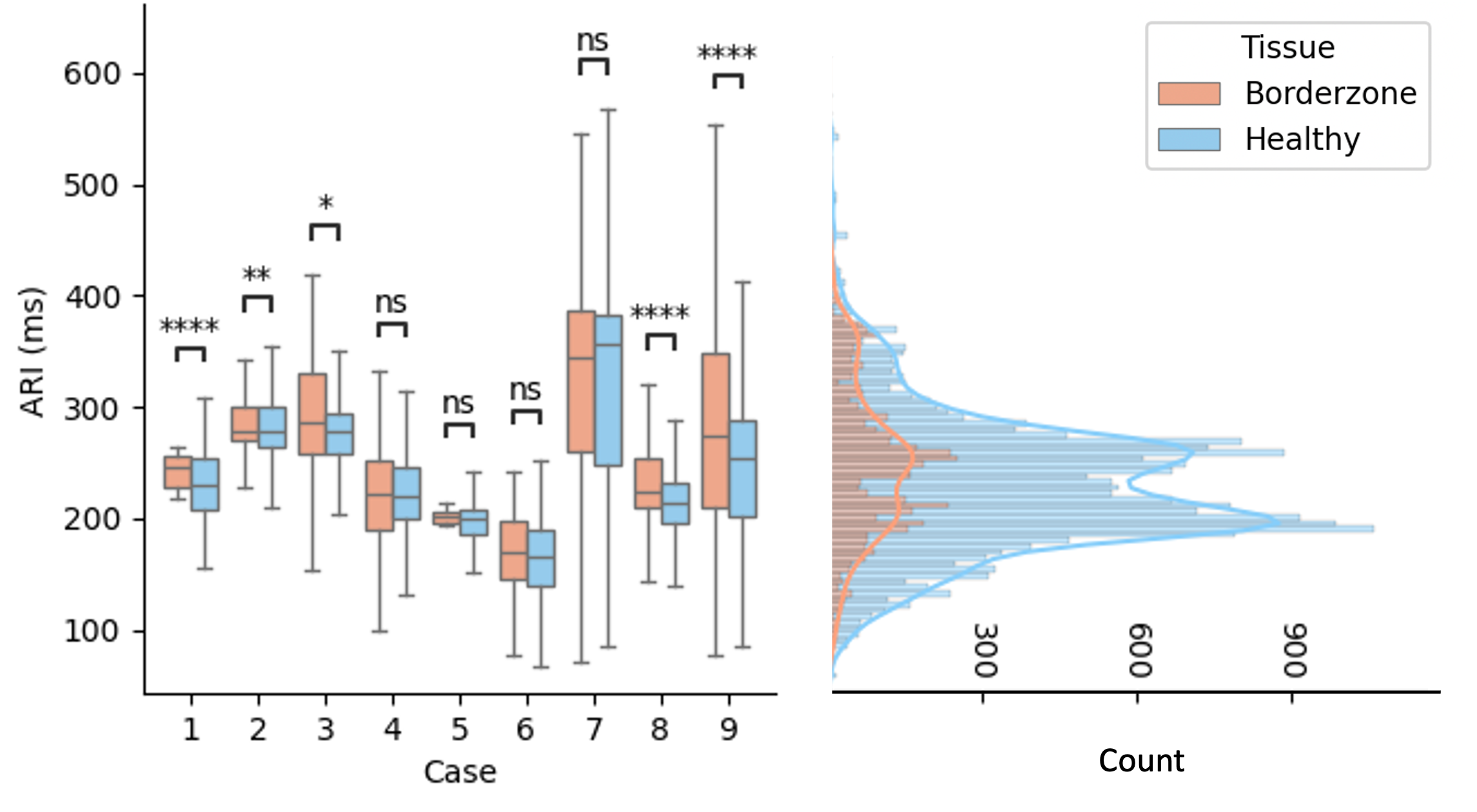
Top frame: (A) UE signal showing activation (green) and recovery (blue) times, and the corresponding ARIs. (B) first derivative of the UE signal, along with the detection windows for activation (green) and recovery (blue) times. (C) lead II and R-peaks and T-wave limits. Bottom frame: (right) Distribution of tissue-stratified ARI values per case with the results from the Mann-Whitney U test and (left) the combined distribution. Statistical significance (p-value):
8.4.8 Digitization of ECG Images (The George B. Moody PhysioNet Challenge 2024)
This work was supported by the SimCardioTest project which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No. 101016496, by the French government, through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002.
Keywords:
Participants: Rafael Silva [Correspondant], Yingyu Yang, Maëlis Morier, Safaa Al-Ali, Maxime Sermesant.
The George B. Moody PhysioNet Challenge 2024 focused on extracting and classifying ECG time series from images. Team “Epione” proposed YOUR-Lead: YOLO and U-Net for Reconstruction of ECG Lead Signals 39 (see Fig. 27). Contributions include an image generation pipeline for realistic ECG scans, a YOLO model for lead identification, a U-Net for trace enhancement, and a reconstruction algorithm to convert images into ECG signals.
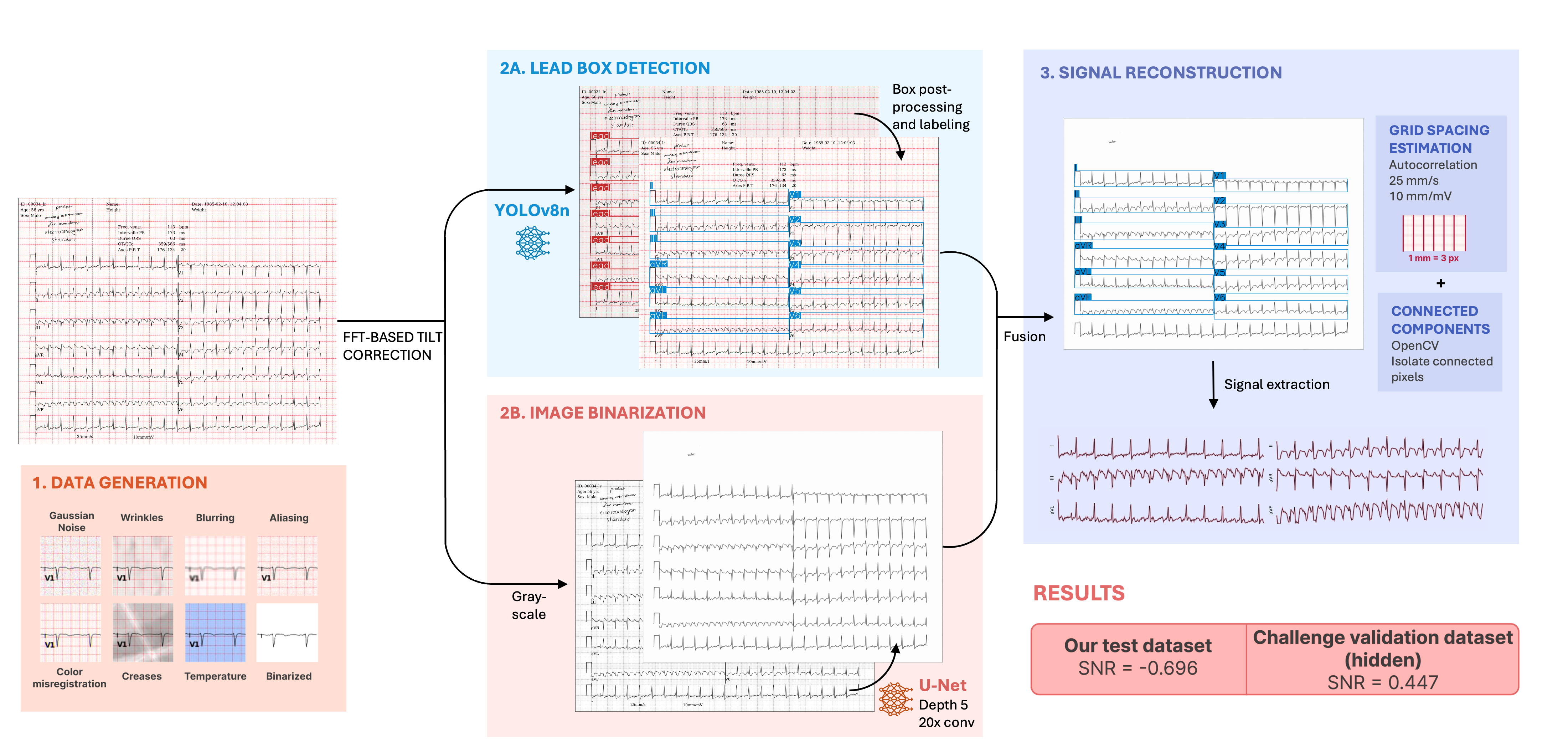
YOUR-Lead pipeline: ECG paper images are generated with varying characteristics to mimic real-life challenges. A U-Net model binarizes the images, extracting relevant parts, while a fine-tuned YOLO model detects signal box leads. The cleaned image and detected leads are then used to extract 1D ECG signals.
8.4.9 Artificial Intelligence for Automated Cardiac Defibrillation in Embedded Systems
This work was supported by the French government, through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002.
Keywords:
Participants: Rafael Silva [Correspondant], Caroline Stehlé, Maxime Sermesant.
With the collaboration of 3IA Techpool and Inn'Pulse, this work develops a Deep Learning system for real-time detection of shockable rhythms using single-lead ECGs on embedded systems (Fig. 28) 69. It optimizes neural networks via hyperparameter tuning, data preprocessing, and augmentation, improving robustness. The system balances detection speed, reliability, and hardware constraints, meeting defibrillator standards. Models are tested on real datasets and benchmarked for deployment feasibility.
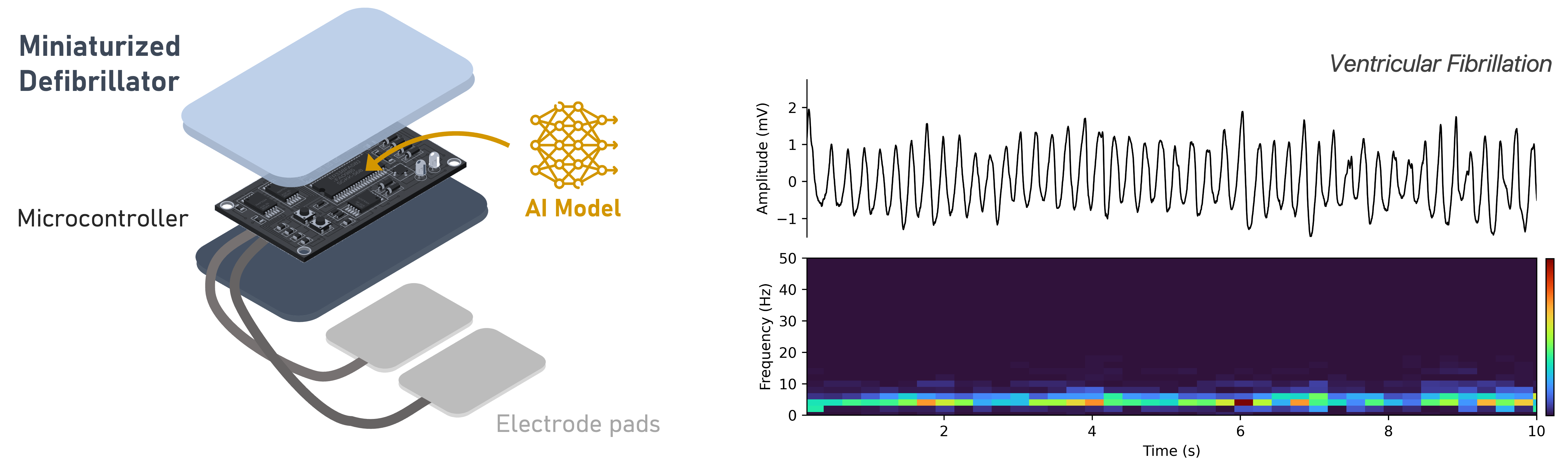
(Left) AI-powered embedded system for automated cardiac defibrillation, featuring a miniaturized defibrillator and an integrated AI model for detecting ventricular fibrillation. (Right) The ECG signal can be represented in various forms, such as time series and spectrogram.
8.4.10 Multi-modal Cardiac function analysis using echocardiography and electrocardiogram
This work was funded by 3IA Côte d'Azur (ANR-19-P3IA-0002) and Inria.
Keywords:
Participants: Yingyu Yang [Correspondant], Pamela Moceri, Marie Rocher, Maxime Sermesant.
- We extend our previous work on echocardiography analysis, which utilized segmentation and motion tracking to extract interpretable features such as ejection fraction and left heart longitudinal strain, to multi-center datasets 28 (refer to Fig. 29). This extension demonstrates the generalizability of our proposed method and enhances the detection of myocardial infarction (MI) patients using multi-view echocardiography data.
- Furthermore, we investigate the integration of uncertainty in uni-modal MI detection and propose a trustworthy decision fusion approach based on uni-modal uncertainty. By combining echocardiography and electrocardiogram data, we significantly improve multi-modal decision accuracy by 4% 44.
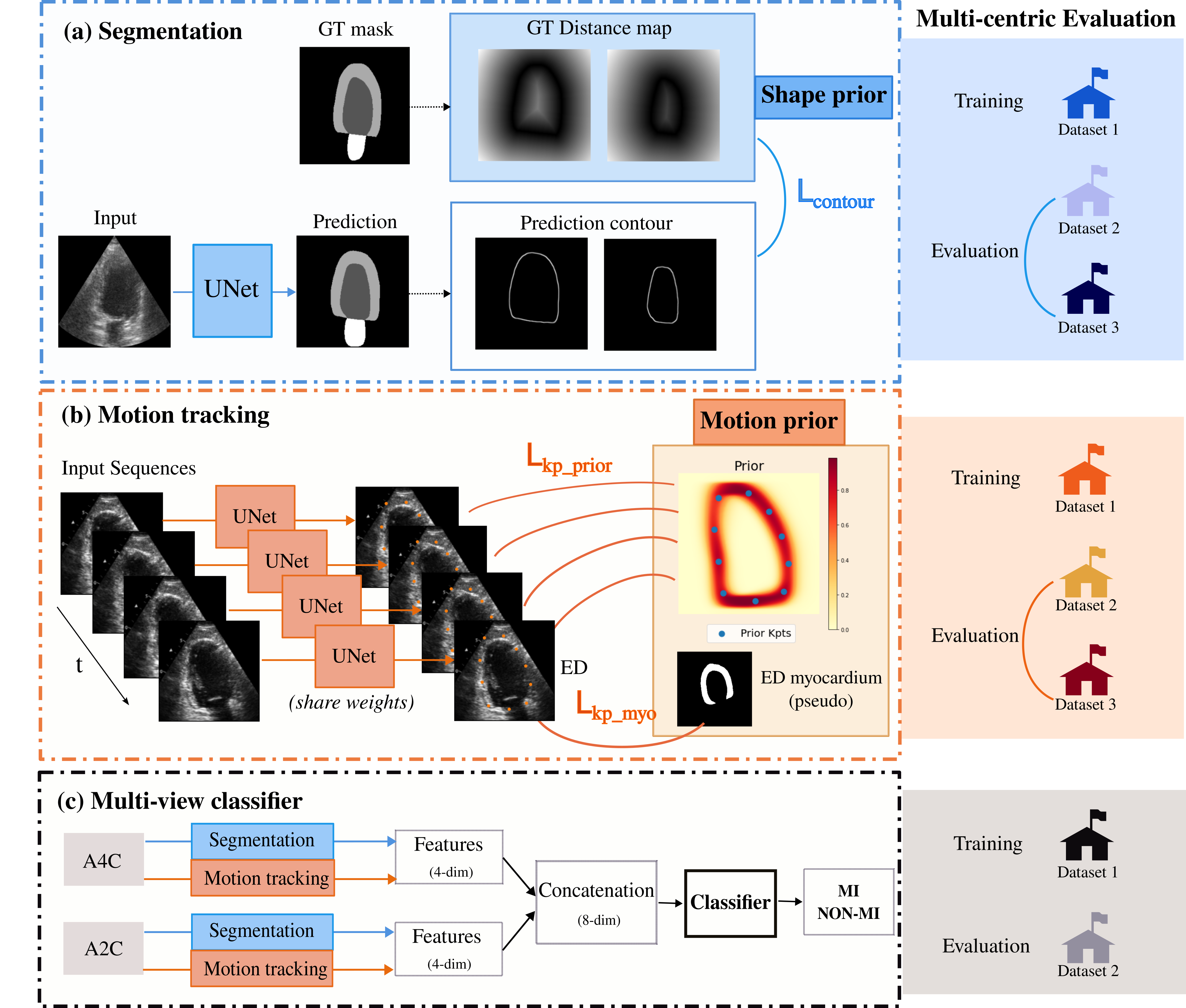
Echocardiography Analysis with Deep Learning using Priors with Multi-centric Evaluation of Generalization. (a) Segmentation prior. (b) Motion tracking prior. (c) Multi-view classifier for MI detection.
8.5 Multi-centric data and Federated Learning
8.5.1 Real-world Deployment of Federated Learning in Biomedical Research Consortia with Fed-BioMed
This project received financial support by the French government through the Agence Nationale de la Recherche (ANR), project Fed-BioMed ref. num. ANR-19-CE45-0006.
Keywords:
Participants: Francesco Cremonesi [Correspondant], Sergen Cansiz, Yannick Bouillard, Lucie Chambon, Marc Vesin, Marco Lorenzi.
Fed-BioMed is an open-source initiative aimed at enabling real-world deployment of federated learning in biomedical research. In 2024, Fed-BioMed has been actively developed and improved, and is currently in release v5.4. Most notably, security and encryption have been significantly improved in terms of speed, stability, and computational costs, and Low-Overhead Masking (LOM) 41 has replaced Joye-Libert as the default algorithm for secure aggregation. Fed-BioMed has been deployed as part of the first demonstrator of the European Cancer Imaging (EUCAIM) infrastructure, achieving full integration with the project's proof-of-concept infrastructure (see Fig. 30). Additionally, Fed-BioMed has been selected as the core technology for DTRIP4H, a Horizon project aimed at supporting the creation of digital twin federated infrastructure for healthcare research.
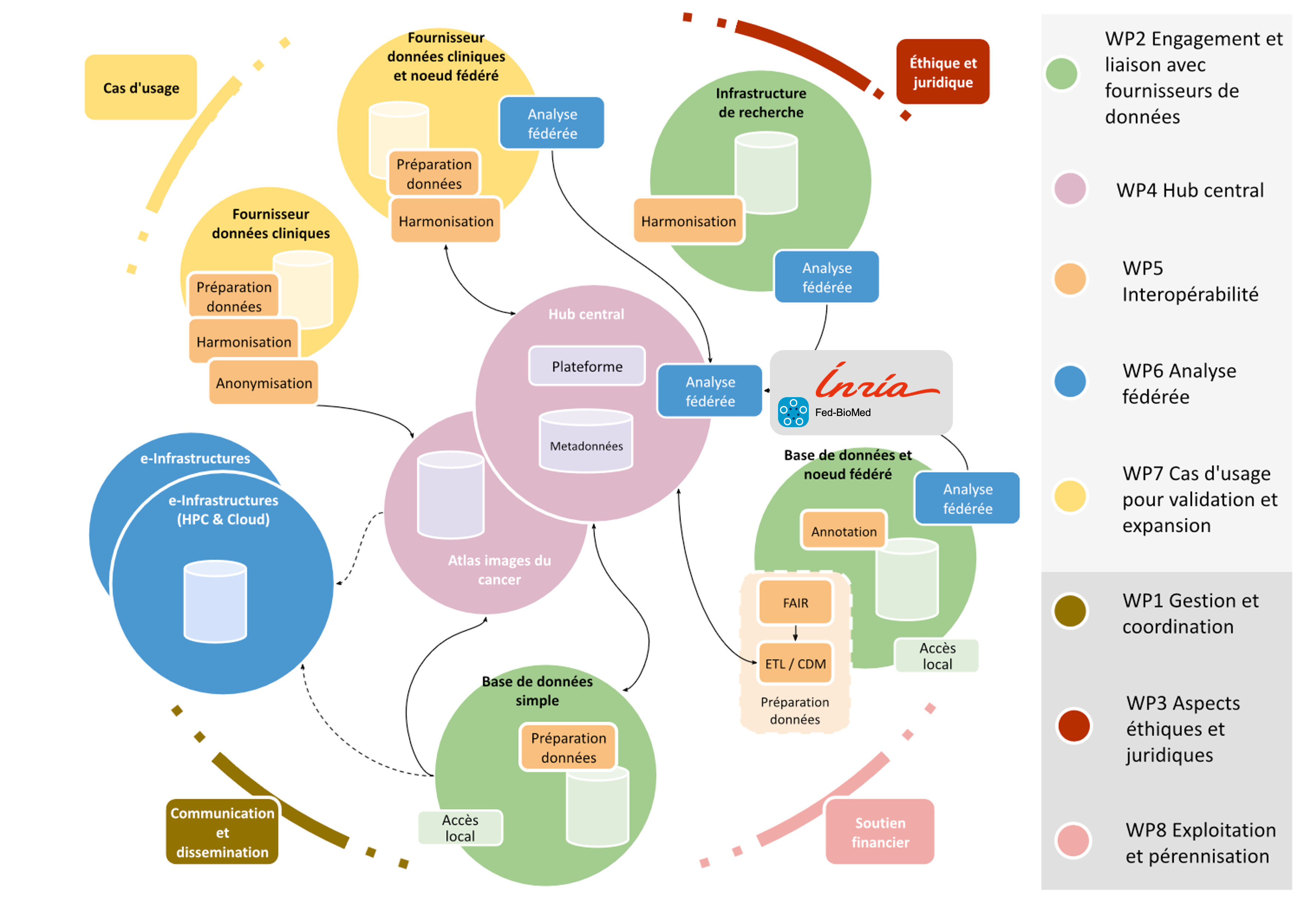
Planned architecture for the European Cancer Imaging research infrastructure (EUCAIM), including Fed-BioMed as the core technology for enabling federated learning and privacy-preserving multi-centric data analysis.
8.5.2 Cross-Domain Image Reconstruction and Segmentation by Representation Alignment in Deep Generative Models
This work was funded by the Franco-German project ANR TRAIN.
Keywords:
Participants: Giuseppe Antonio Orlando [Correspondant], Marco Lorenzi, Maria Zuluaga.
Modern machine learning applications face the challenge of domain variability in data collected from different sources. This project addresses this challenge by developing a generative model that aligns distinct domains by identifying a common latent space representation. Our contributions are as follows (see also Fig. 31):
- We formulate the problem of cross domain alignment by optimizing the encoding of each domain to minimize their discrepancy in the latent space.
- We demonstrate this approach for the problem of multi centric prostate MRI segmentation.
- We show that our model is able to extract and reproduce domain specific artifacts, allowing to obtain robust cross-domain segmentation results.
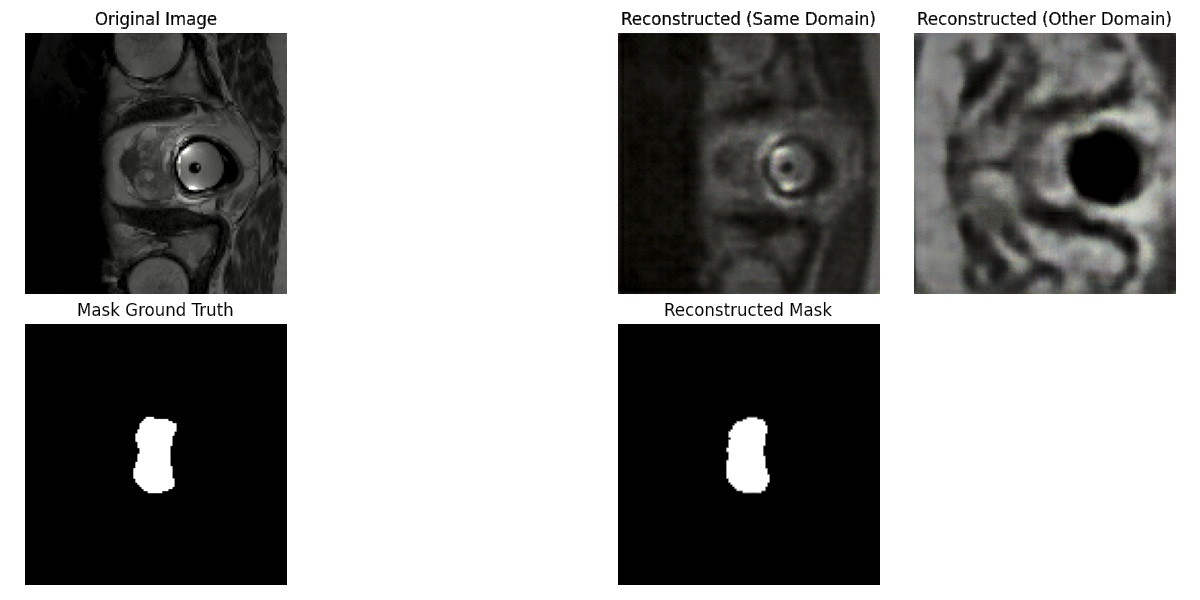
Reconstructed prostate MRI scan. Columns represent 1) Original image and segmentation mask, 2) Same domain (center) image reconstruction and common reconstructed segmentation, 3) Cross domain (center) reconstruction.
8.5.3 StratifyAging: Interoperatibility of Clinical Studies and Routine Care for the Advent of a Stratified Medicine of Aging Using Clinical, Imaging, and Omics Data
This project received financial support by the PEPR Santé Numerique.
Keywords:
Participants: Ghiles Reguig [Correspondant], Marco Lorenzi.
The aim of this project is in the development of robust analysis tools for neuroimaging data, that can find application in multi-centric studies such as the French CATI. We tested methods using the frequency representation given by the Fourier transform without success. We are currently working on aggregating models trained on data coming from different sites using a "multihead" approach where the final model is built using the site specific models.
- We tested methods using the frequency representation given by the Fourier transform.
- We are currently working on aggregating models trained on data acquired on different sites using a "multihead" approach, where the final model is built using site specific models (Fig. 32). The sites were simulated by adding specific biases to the data (blurring, bias, motion, ghosting, gamma contrast). The model was trained to remove the simulation.
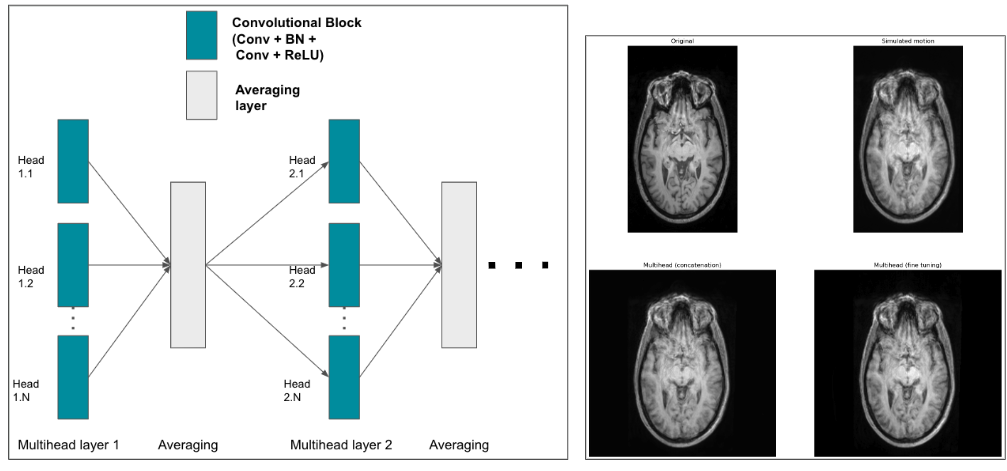
Proposed encoder multihead architecture (left) and examples of predictions given by two multihead models (right). On the left, we show the structure of a multihead encoding block which aggregates the outputs of several convolutional blocks by averaging them. The decoding part remains the same as a classical nnunet. On the right, an MRI is shown (top left), along with its version with a motion simulation (top right) and the predictions given by two multihead models (a multihead model that aggregates already trained nnunets and only trains the decoder part on the bottom left; a multihead model that was finetuned from the same trained nnunets on the bottom right).
8.5.4 Multi-centric AI-based Framework for Prostate Cancer Patients Management on Active Surveillance Through Robust Federated Learning
This work has been supported by the French government, through the 3IA Côte d'Azur Investments.
Keywords:
Participants: Lucia Innocenti [Correspondant], Francesco Cremonesi, Michela Antonelli, Sebastien Ourselin, Marco Lorenzi.
Collaborative Learning (CL) provides a powerful setting for exploiting collective expertise and data resources from a federation of clients without the need to share data or sensitive information (Fig. 33).
Our study 58 provides a comprehensive examination of CL methods, by benchmarking them in terms of performance and cost-effectiveness for an extended benchmark of biomedical data analysis tasks.
We compared two CL approaches: Federated Learning (FL) and Consensus-Based Methods (CBM). In this specific scenario, CBM and FL are comparable in terms of performance. At the same time, CBM is more cost-effective, requiring fewer resources to be trained and naturally addressing some FL open problems like client dropout.
These insights will benefit researchers and practitioners seeking to optimize collaborative learning in healthcare, improving patient care and data security.
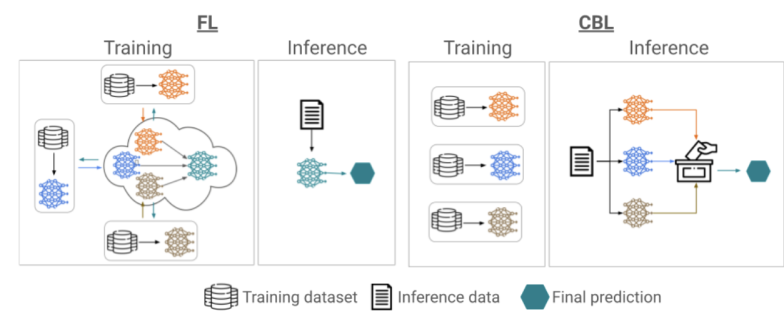
Training and inference phases for federated learning (FL, on the left) and consensus-based learning (CBL, on the right). In FL training is performed collaboratively to produce a common global model across clients. The global model is subsequently used for inference on new data instances. CBL instead requires clients to train a model on the respective local data independently. Inference on new data instances is performed collaboratively through consensus.
8.5.5 Data Structuration and Security in Large-scale Collaborative Healthcare Data Analysis
This work has been supported by the French government, through the 3IA Côte d'Azur Investments.
Keywords:
Participants: Riccardo Taiello [Correspondent], Melek Önen, Olivier Humbert, Marco Lorenzi.
Our work aims to study the impact of privacy-preserving methods in Federated Learning (FL) for biomedical applications. In 23, we extended our previous work, *Privacy-Preserving Image Registration (PPIR)*, by introducing new cost functions (see Fig. 34). In 42, we proposed the Eagle and Owl secure aggregation (SA) protocols for FL, which address stragglers and reduce resource overhead. Using Fed-BioMed 41, we implemented the Joye-Libert and Low Overhead Masking SA protocols, achieving minimal accuracy loss and negligible resource overhead.
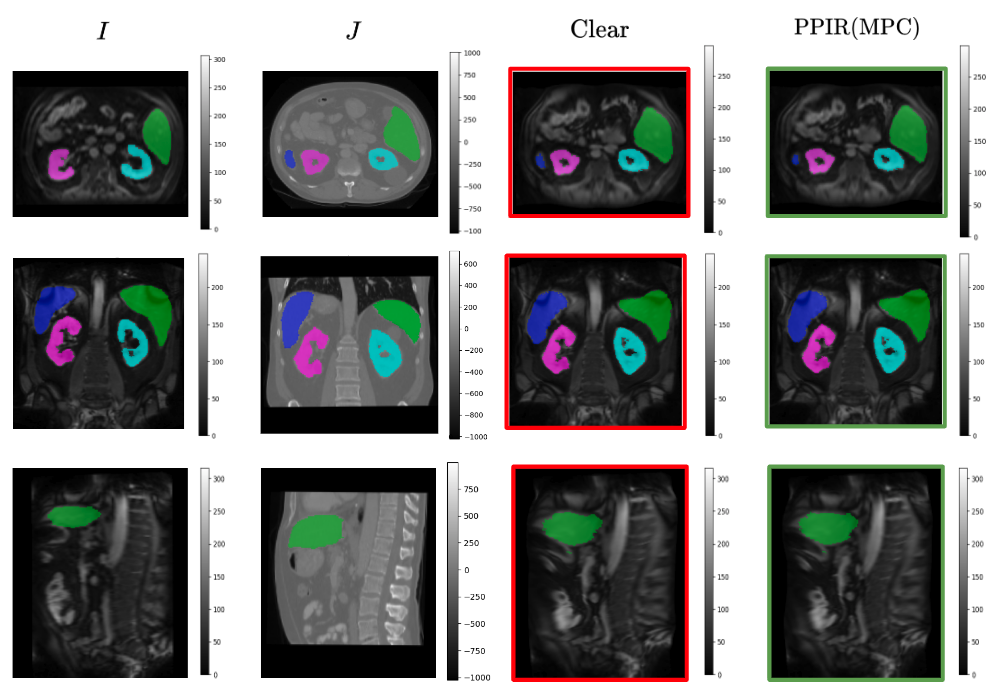
The images are presented in a 3x4 grid, with the first row representing the axial axis, the second row the coronal axis, and the third row the sagittal axis. The first and second columns show, respectively, MRI and CT images. The third column shows the MRI transformed using Clear, while the fourth column shows the MRI transformed using PPIR.
9 Bilateral contracts and grants with industry
9.1 Bilateral contracts with industry
9.1.1 Spin-off company inHEART
Participants: Maxime Sermesant.
inHEART is a spin-off of the Epione team and IHU Liryc funded in 2017. inHEART provides a service to generate detailed anatomical and structural meshes from medical images, that can be used during ablation interventions. inHEART received 2 awards, one from Aquitaine region and one i-LAB from the BPI. It raised 3.2 million euros in 2020. It currently employs 27 people. It is FDA and CE certified.
9.1.2 Start-up Inn'Pulse
Participants: Rafael Silva, Caroline Stehle, Maxime Sermesant.
Inn'Pulse is a start-up developing an ultra-portable automatic cardiac defibrillator. We are designing AI algorithms for better signal processing in collaboration with the 3IA TechPool.
9.1.3 Koelis
Participants: Nicholas Ayache, Hervé Delingette.
The company Koelis participates in the thesis work of Zhijie Fang and Manasi Kattel on the multimodal registration of MR and ultrasound images. The objective is to improve the accuracy of the targeted biopsies inside the prostate.
10 Partnerships and cooperations
10.1 International collaborations
- Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology. Benjamin Billot is currently collaborating with Prof. Polina Golland and Dr. Neel Dey on developing equivariant networks for motion tracking in fetal MRI time series. This work is also in collaboration with Prof. Ellen Grant from the Boston Children's Hospital.
- Harvard Medical School and Massachusetts General Hospital. Benjamin Billot is a collaborator of Dr. Juan Eugenio Iglesias and Prof. Bruce Fischl to maintain some parts of the FreeSurfer package, and to develop new tools for neuro-imaging based on domain randomization strategies.
- King's College London (KCL), London, UK Marco Lorenzi is a collaborator of the School of Biomedical Engineering & Imaging Sciences at King's College London. This collaboration focuses on the development of novel collaborative learning methods to improve prostate cancer detection in multicentric studies.
- Laboratory of Physics of Fluids, University of Twente, NL Hervé Delingette is collaborating with Assistant Professor Guillaume Lajoinie, on the topics of Deep Learning for ultrasound imaging in the framework of the BoostUrCareer Cofund program and the thesis of Hari Sreedhar.
- University College London (UCL), London, UK Marco Lorenzi is a collaborator of the COMputational Biology in Imaging and geNEtics (COMBINE) group within the Centre for Medical Image Computing (CMIC) of UCL. His collaboration is on the topic of spatio-temporal analysis of medical images and imaging-genetics, with special focus on brain imaging analysis and biomarker development.
- Laboratory of Neuroimaging of Aging (LANVIE), Faculty of Medicine, Geneva University Hospitals, HUG Marco Lorenzi collaborates with the LANVIE laboratory led by Prof. Giovanni B. Frisoni. The collaboration consists in developing and translating novel approaches for disease progression modeling in neurodegenerative disorders, such as Alzheimer's disease.
- Illinois Institute of Technology (IIT, IL, USA) Marco Lorenzi is currently a collaborator of IIT for the investigation of the complex relationship between brain atrophy and genetics in Alzheimer's disease, in particular for demonstrating the effectiveness of multivariate statistical models in providing a meaningful description of the relationship between genotype and brain phenotype.
10.1.1 Participation in other International Programs
Project MDPARK, funded by Michael J. Fox Fundation
Participants: Marco Lorenzi.
- Title: Multimodal Dynamic Modeling and Prediction of Parkinsonian Symptom Progression
-
Partner Institution(s):
- Illinois Institute of Technology, USA
- Amsterdam University Medical Center, The Netherlands
- University of Southern California, USA
- Northwestern University School of Medicine, USA
- Date/Duration: 2022-2026
- Additionnal info/keywords: Connectomics, Disease models, Network analysis, Neuroimaging
10.2 International research visitors
Mihaela Pop
-
Status:
Researcher
-
Institution of origin:
Sunnybrook Research Institute
-
Country:
Toronto, Canada
-
Dates:
Jan.-Apr. 2024
-
Context of the visit:
Mihaela Pop contribuyted to the SImCardioTest and Chronicardio projects, bringing her expertise in experimental data, cardiac modeling and cardio-oncology.
-
Mobility program/type of mobility:
Research stay
Javier Villar Valero
-
Status:
PhD
-
Institution of origin:
Polytechnic University of Valencia
-
Country:
Valencia, Spain
-
Dates:
Apr.-Jul. 2024
-
Context of the visit:
Javier Villar Valero came to the lab in order to work on a joint project within SimCardioTest, evaluating in silico the impact of chemotherapy on the cardiac function, leveraging experimental data from Mihaela Pop.
-
Mobility program/type of mobility:
Research stay
Sasha Panfilov
-
Status:
Emeritus Professor
-
Institution of origin:
Ghent University
-
Country:
Ghent, Belgium
-
Dates:
Sep. 2024
-
Context of the visit:
Sasha Panfilov came to the team in order to interact on the mathematical modeling of cardiac electrophysiology, discussing potential improvements to the reaction-diffusion models used in the lab.
-
Mobility program/type of mobility:
Research stay
Anna Custo
-
Status:
Research Associate
-
Institution of origin:
University of Geneva
-
Country:
Switzerland
-
Dates:
Sep. 2024
-
Context of the visit:
Anna Custo visited the team to extend the functionalities of the tool SimulAD for the simulation of the effect of anti-amyloid treatments in Alzheimer's disease.
-
Mobility program/type of mobility:
Research stay
10.3 European initiatives
10.3.1 Horizon Europe
EUCAIM
Participants: Marco Lorenzi.
- Title: EUCAIM
- Type: H2020
- Program: Research and Innovation
- Duration: 2023-2027
- Inria Contact: Marco Lorenzi
- Coordinator: University of Valencia, EIBR
-
Summary:
EUropean Federation for CAncer IMages (EUCAIM) is the cornerstone of the European Commission-initiated European Cancer Imaging Initiative, a flagship of Europe’s Beating Cancer Plan (EBCP), which aims to foster innovation and deployment of digital technologies in cancer treatment and care to achieve more precise and faster clinical decision making, diagnostics, treatment and predictive medicine for cancer patients.
EUCAIM will establish and deploy a pan-European digital federated infrastructure of FAIR pan-cancer anonymized images from Real-World Data, preserving the data sovereignty of providers. Furthermore, it will provide an Experimentation Platform for the development and benchmarking of AI tools toward Precision Medicine in cancer diagnosis and treatment. EUCAIM will address the fragmentation of the existing cancer image repositories by building a distributed atlas of cancer images, including both common and rare types of cancer, from existing initiatives, including related Research Infrastructures’ networks and successful Horizon 2020 projects. EUCAIM will target clinicians, researchers, and innovators, providing the means to build reproducible clinical decision-making systems supporting diagnosis, treatment, and predictive medicine. This Infrastructure will benefit citizens through improved healthcare procedures, and will stimulate the European market through the innovation of new tools and services.
10.3.2 H2020 projects
ERC G-Statistics
Participants: Xavier Pennec, Guillaume Olikier, Tom Szwagier.
- Title: Geometric Statistics
- Type: ERC
- Program: H2020
- Duration: 2018-2024
- Inria Contact: Xavier Pennec
- Coordinator: Inria
-
Summary:
G-Statistics aims at exploring the foundations of statistics on non-linear spaces with applications in the Life Siences. Invariance under gauge transformation groups provides the natural structure explaining the laws of physics. In life sciences, new mathematical tools are needed to estimate approximate invariance and establish general but approximate laws. Rephrasing Poincaré: a geometry cannot be more true than another, it may just be more convenient, and statisticians must find the most convenient one for their data. At the crossing of geometry and statistics, G-Statistics aims at grounding the mathematical foundations of geometric statistics and to exemplify their impact on selected applications in the life sciences. So far, mainly Riemannian manifolds and negatively curved metric spaces have been studied. Other geometric structures like quotient spaces, stratified spaces or affine connection spaces naturally arise in applications. G-Statistics will explore ways to unify statistical estimation theories, explaining how the statistical estimations diverges from the Euclidean case in the presence of curvature, singularities, stratification. Beyond classical manifolds, particular emphasis will be put on flags of subspaces in manifolds as they appear to be natural mathematical object to encode hierarchically embedded approximation spaces. In order to establish geometric statistics as an effective discipline, G-Statistics will propose new mathematical structures and characterizations of their properties. It will also implement novel generic algorithms and illustrate the impact of some of their efficient specializations on selected applications in life sciences. Surveying the manifolds of anatomical shapes and forecasting their evolution from databases of medical images is a key problem in computational anatomy requiring dimension reduction in non-linear spaces and Lie groups. By inventing radically new principled estimations methods, we aim at illustrating the power of the methodology and strengthening the “unreasonable effectiveness of mathematics” for life sciences.
SimCardioTest
Participants: Maxime Sermesant, Irene Balelli, Jairo Rodriguez, Safaa Al Ali.
-
Title:
Simulation of Cardiac Devices & Drugs for in-silico Testing and Certification
- Link: SimCardioTest project on cordis.europa.eu
-
Duration:
From January 1, 2021 to June 30, 2025
-
Partners:
- Universidad Pompeu Fabra (UPF), Spain
- Inria, France
- Virtual Physiological Human Institute for Integrative Biomedical Research VZW (VPH Institute), Belgium
- Sorin CRM SAS (LIVANOVA), France
- Exacture, France
- Simula Research Laboratory AS, Norway
- INSILICOTRIALS Technologies BV, Netherlands
- Universite de Bordeaux (UBx), France
- Boston Scientific Scimed IVC, United States
- Iniversitat Politecnica de Valencia (UPV), Spain
- INSILICOTRIALS Technologies S.P.A. (InSilicoTrials), Italy
-
Inria contact:
Maxime Sermesant
-
Coordinator:
Inria
-
Summary:
Despite massive investment in healthcare, huge R&D cost increase and regulatory pathway complexity hamper tremendously commercialization of new devices & medicines, putting patient populations at risk of not receiving adequate therapy. At the same time, outside healthcare, computer modeling and simulation (CM&S) is precisely recognized to increase speed & agility while reducing costs of development. CM&S can create scientific evidence based on controlled investigations including variability, uncertainty quantification, and satisfying demands for safety, efficacy & improved access.
Cardiac modeling has dramatically gained maturity over the last decades, with personalization to clinical data enabling validation. We selected a number of cardiac devices and medicines where CM&S is mature enough and that represent the most common cardiac pathologies, to demonstrate a standardized and rigorous approach for in-silico clinical trials.
SimCardioTest will bring a disruptive innovation by creating an integrated and secure platform standardizing & bridging model simulations, in-silico trials, and certification support. This environment will go beyond the state-of-the-art in computational multi-physics & multi-scale personalized cardiac models. Diseased conditions and gender/age differences will be considered to overcome clinical trials limitations such as under-representation of groups (e.g. women, children, low socio-economic status). Advanced big data, visual analytics & artificial intelligence tools will extract the most relevant information.
It is critical that Europe demonstrates its capacity to leverage in-silico technology in order to be competitive in healthcare innovation. SimCardioTest exploitation aims at delivering a major economic impact on the European pharmaceutical and cardiac devices industry. It will accelerate development, certification and commercialization, and will produce a strong societal impact contributing to personalised healthcare.
inEurHeart
Participants: Maxime Sermesant.
- Title: inEurHeart: AI, Digital Twin & Clinical Trial for a Disruption in Catheter Ablation
- Link: inEurHeart website
-
Partners:
- Inria
- Rotterdam University, Netherlands
- Inserm, France
- CHU Bordeaux, France
- inHEART, France
- Université de Bordeaux (UBx), France
- Duration: 2022-2024
- Inria Contact: Maxime Sermesant
- Coordinator: Inria
-
Summary:
inEurHeart is an innovation project in Artificial Intelligence, Digital Twin & a Clinical Trial for a Disruption in Catheter Ablation for Ventricular Tachycardia, making ablation therapy accessible to most patients. This project is a collaborative project between 5 organizations in France and Netherlands funded by EIT Health – the European Institute of Innovation and Technology, co-funded by the European Union. This project will exemplify how the academic-industrial relationships can be fostered and can lead to drastic changes in clinical practice. EIT Health provides a unique opportunity to transfer Artificial Intelligence tools to enable the scale-up phase, and to validate the technology through a randomized clinical trial.
10.3.3 Other european programs/initiatives
HELIOS
Participants: Francesco Cremonesi.
-
Title:
Haemoglobinopathies in European Liaison of Medicine and Science
-
Type:
COST Action
-
Duration:
2023-2027
-
Inria Contact:
Francesco Cremonesi
-
Coordinator:
The Cyprus Foundation For Muscular Dystrophy Research, Nicosia, Cyprus
-
Summary:
The goal of HELIOS is to build a network of excellence for integrating, harmonizing, and spreading the existing knowledge and for developing innovative services and tools, thus improving knowledge accessibility and healthcare equally across Europe and beyond. HELIOS comprises five working groups that will coordinate existing and emerging haemoglobinopathy-related activities in Europe, ranging from clinical and molecular research to data analysis and bioinformatics, aiming to advance health care systems, contribute to informed policymaking and improve survival and quality of life for existing and future patients. Francesco Cremonesi is the leader of WG 3 which is focused on Data management, sharing, and analysis. During the first year, the Working Group has conducted a thorough review of the available hemoglobinopathy data with a focus on research centers willing to set up scientific collaborations.
TRAIN
Participants: Marco Lorenzi.
-
Title:
Trustworthy and Robust Artificial INtelligence
-
Type:
German-French joint call for proposals on Artificial Intelligence
-
Duration:
2023-2027
-
Inria Contact:
Marco Lorenzi
-
Coordinator:
Inria
-
Summary:
The goal of TRAIN is to explore two main barriers to the widespread deployment of AI: the lack of trustworthiness and the lack of robustness. TRAIN, comprising of Inria and EURECOM from France and RUB and Fraunhofer IPT from Germany, will design, develop and evaluate new AI solutions that address these two challenges. The development of such AI systems requires careful attention at every aspect of the model development pipeline, starting from construction and training of models all the way to evaluation and deployment. The proposed research agenda focuses on the more challenging federated learning (FL) setting as a key paradigm to enable trustworthiness in the training phase. We also investigate trustworthiness and robustness of the evaluation and deployment phase in both centralized and collaborative machine learning. The project identifies four main objectives: (i) robust learning with data heterogeneity and bias; (ii) quantifying robustness with automated frameworks; (iii) privacy-preserving federated learning; (iv) adversarial robustness. TRAIN solutions will be integrated into real-world frameworks in two main application domains: healthcare and smart industry.
10.4 National initiatives
10.4.1 PEPR Digital Health ChroniCardio
Participants: Jairo Rodriguez, Mihaela Pop, Maxime Sermesant.
-
Duration:
2023 - 2027
-
Partners:
- Inria
- Hospices Civils de Lyon
- Assistance Publique - Hôpitaux de Marseille
- CREATIS
-
Inria contact:
Maxime Sermesant
-
Coordinator:
Inria
-
Summary:
ChroniCardio is a new 4-year multi-institution project funded by the French Research Priority Programme on Digital Health to accelerate the integration of multi-scale data (clinical, imaging, genetic, ECG, etc.) and the development of advanced modeling tools to predict the long-term evolution of non-ischemic dilated and hypertrophic cardiomyopathies. This includes the risk of arrhythmia, heart failure and sudden cardiac death. Our consortium brings together research scientists and engineers from Inria (Sophia Antipolis, Lyon, Rennes, Bordeaux) and INSA / Lyon /University / CNRS (Lyon), and clinicians from Lyon and Marseille University Hospitals. The project is coordinated by Maxime Sermesant, from Inria Epione team.
10.4.2 PEPR Digital Health Rewind
Participants: Marco Lorenzi.
-
Duration:
2023 - 2027
-
Partners:
- Inria
- CNRS
- INSERM
- Université Grenoble Alpes
-
Inria contact:
Stephanie Allassonniére
-
Coordinator:
Inria
-
Summary:
The project Rewind will focus on the development of new mathematical and statistical approaches for the analysis of multimodal multiscale longitudinal data. These models will be designed, implemented as prototypes and then transferred to an easy-used-well-documented platform where researchers from diverse communities, in particular physicians, will be able to analyze their own data set.
10.4.3 PEPR Digital Health Secure, safe and fair machine learning for healthcare
Participants: Marco Lorenzi.
-
Duration:
2023 - 2027
-
Partners:
- Inria
- LAMSADE (CNRS, Dauphine-PSL)
- CEA
-
Inria contact:
Aurelien Bellet
-
Coordinator:
Inria, Dauphine-PSL
-
Summary:
The goal of this project is to overcome the challenges that prevent the effective use of personalized health data. To achieve this, we will develop new machine learning algorithms that are designed to handle the unique characteristics of multi-scale and heterogeneous individual health data, while providing formal privacy guarantees robustness against adversarial attacks and changes in data dynamics, and fairness for under-represented populations. By addressing these barriers, we hope to unlock the full potential of personalized health data for a wide range of applications.
10.4.4 PEPR Digital Health Stratify Aging
Participants: Marco Lorenzi.
-
Duration:
2023 - 2027
-
Partners:
- CEA
- Inria
- CNRS
- INSERM
-
Inria contact:
Marco Lorenzi
-
Coordinator:
CEA
-
Summary:
The goal of StratifyAging is to focus on using high-quality, curated data from clinical research to advance the field of patient stratification through the use of hypothesis-driven approaches or AI algorithms. By harmonizing and aggregating data from various studies, it will be possible to reach a large enough sample size to effectively stratify patients. This process will also lead to the development of standard protocols that can be applied in routine care. As a result, a virtuous cycle may be created, in which standardized data from routine care is collected and analyzed through the Health Data Hub and used to perform population monitoring and develop normative charts and decision support tools.
10.4.5 MediTwin
Participants: Maxime Sermesant, Hervé Delingette, Xavier Pennec.
-
Duration:
2024 - 2028
-
Partners:
- Dassault Systemes
- Inria
- IHUs
- Start-ups
-
Inria contact:
Maxime Sermesant
-
Coordinator:
Dassault Systemes
-
Summary:
The MEDITWIN project will offer personalized virtual twins of organs, metabolism and cancer, for better diagnosis and treatment. In particular, MEDITWIN will enable doctors to simulate future scenarios for a patient. MEDITWIN will enable the industrialization, clinical validation and standardization of these innovations, so that these technologies can be deployed in a standardized way and benefit as many people as possible. The best standards of care will be incorporated into virtualized experiences made accessible worldwide, setting a new benchmark for quality in healthcare and providing a decisive learning ground for progress in medical science. The benefits of virtual twins will be assessed for medical teams, patients, and the healthcare system, notably in terms of improving the efficiency of care, quality of multidisciplinary decision-making, and effectiveness and safety of medical practices and interventions.
10.4.6 DAICAP
Participants: Hervé Delingette.
-
Duration:
2020 - 2025
-
Partners:
- AP-HP
- Inria
- Incepto
- CHU Bordeaux, CHU Lille, CHU Strasbourg, Hopitaux Civils de Lyon
-
Inria contact:
Hervé Delingette
-
Coordinator:
AP-HP
-
Summary:
The DAICAP project aims at creating a large multi-centric study (8 clinical centers on 5 partner university hospitals) combining multiparametric MR images of the prostate and histology for the detection and characterization of prostate cancer.
Inria participates to the data collection, quality control of restrospective and prospective data from 1250 patients. It also performs the training and evaluation of AI algorithms for prostate lesion detection. The infrastructure of the Health Data Hub is used to centralize the data and to fine-tune the AI models.
The DAICAP project was selected by the Health Data Hub, the Grand Défi « Amélioration des diagnostics médicaux par l’Intelligence Artificielle », and Bpifrance in July 2020.
10.4.7 AICOO
Participants: Hervé Delingette.
-
Duration:
2024 - 2028
-
Partners:
- Incepto
- Inria
- AP-HP
- France Imagerie Territoires
- EDL
- Easydoct
-
Inria contact:
Hervé Delingette
-
Coordinator:
Incepto
-
Summary:
The AICOO project has been selected among the winners of the French national « Innovation in medical imaging » call for projects. It aims to transform patient care at the early stage of cancer detection by developing an oncology coordination platform. The platform includes AI solutions for the early detection of Prostate Cancer and the extraction of advanced biomarkers.
In this project, Inria develops advanced machine learning solutions for the automatic characterization of prostate lesion malignancy using multiparametric Magnetic Resonance Imaging.
10.4.8 RHU ReBONE
Participants: Hervé Delingette, Alix de Langlais.
-
Duration:
2024 - 2029
-
Partners:
- Partenaires académiques : Université Côte d’Azur, Inserm, Inria, CNRS, Institut Pasteur,Université de Bretagne Occidentale, Université Paris Cité, Université Aix-Marseille, Mines de Paris
- Partenaires industriels : Abys Medical, Newclip Technics, Addidream, Aguila Expertise, Digital Medical Hub
- Partenaires Cliniques : CHU Nice, AP-HP
-
Inria contact:
Hervé Delingette
-
Coordinator:
CHU Nice
-
Summary:
The ReBone project has been selected among the winners of the French national « Recherche Hospitalo-Universitaire en santé » (RHU) call for projects. It aims to develop novel personalized, automated, collaborative and validated preoperative planning tools to simulate, prepare and then perform a secure and patient-specific surgical intervention in osteoarticular surgery.
In this project, Inria works on the collection and quality control of medical image datasets associated with 3 use cases. We also develop new automated methods to delineate fractured bony structures in CT images and participate to their validations in terms of clinical and industrial use.
10.4.9 RHU TALENT
Participants: Maxime Sermesant, Irene Balelli, Marco Lorenzi.
-
Duration:
2024 - 2029
-
Partners:
- Partenaires académiques : U Bordeaux, IHU Liryc, Inria
- Partenaires industriels : Cardiologs, inHEART, Incepto, AMPS
- Partenaires Cliniques : CHU Bordeaux, CHU Dijon
-
Inria contact:
Maxime Sermesant
-
Coordinator:
Université de Bordeaux
-
Summary:
The consortium of academic centres and private companies brought together in the TALENT project aims to revolutionize the prediction of stroke risk by developing digital tools capable of detecting this increased risk. The work will focus on the analysis of widely available data, including chest CT scans and/or electrocardiograms, and simpler clinical data such as age or the presence of diabetes. Inria is in charge of image and shape analysis, causal discovery and multimodal learning.
10.4.10 IHU RespirERA
Participants: Hervé Delingette, Nicholas Ayache.
-
Duration:
2024 - 2034
-
Partners:
- Founders of the institute : Inria, Inserm, CHU Nice, Université Côte d’Azur
-
Inria contact:
Hervé Delingette
-
Coordinator:
CHU Nice
-
Summary:
The RespirERA institute has been funded following the third wave of the French national « Institut Hospitalo-Universitaire » (IHU) call for proposals. The institute aims to improve the care in the field of respiratory diseases. The objectives are to reduce the incidence of lung diseases linked to pollution and age and the impact of the exposome (all exposures to environmental factors), extend the life expectancy of patients, delay dependency and progression to respiratory failure and avoid hospitalizations.
Inria is coordinating a workpackage in this new institute focusing on AI solutions for lung cancer screening, and biomarker extraction from heterogenous data for the diagnosis of respiratory diseases. Hervé Delingette and Nicholas Ayache are the members of the executive team.
10.4.11 Other national initiatives
Consulting for Industry
- Nicholas Ayache has joined the Scientific Advisory Board of Caranx Medical in Oct 2021.
- Maxime Sermesant is a scientific advisor for the company inHEART (Bordeaux).
Institute 3IA Côte d'Azur
-
The 3IA Côte d'Azur is one of the four "Interdisciplinary Institutes of Artificial Intelligence" that were created in France in 2019. Its ambition is to create an innovative ecosystem that is influential at the local, national and international levels, and a focal point of excellence for research, education and the world of AI.
-
Epione is heavily involved in this institute since 5 permanent researchers (N. Ayache, H. Delingette, M. Lorenzi, M. Sermesant and X.Pennec) are chair holders in this institute, and N. Ayache serves as scientific director. The 5 Epione chairs were renewed in 2023 by an international jury. H. Delingette and N. Ayache are members of its scientific committee.
Funded projects
- Marco Lorenzi is co-PI of the project FEDERATED-PET (2022-2026), with Prof. Olivier Humbert (CAL, Nice). The project is funded by the Institut National du Cancer (INCa), and aims at developing the first French federated learning infrastructure in a network of hospitals from the Unicancer consortium. Marco Lorenzi is also principal investigator of the project TRAIN, funded by the ANR, and co-PI of the project StratifyAging of the PEPR Santé Numerique. He is also partner of the Horizon Europe Project EUCAIM.
Collaboration with national hospitals
- The Epione project team collaborates with the following 3 French IHU (University Hospital Institute): the IHU-Strasbourg (Pr J. Marescaux and L. Soler) on image-guided surgery, the IHU-Bordeaux (Pr M. Haïssaguere and Pr P. Jaïs) on cardiac imaging and modeling and the IHU-Pitié Salpétrière (Dr. O. Colliot and Pr. B. Stankoff) on neuroimaging.
- The IHU RespirERA was selected in May 2023 among the 12 new institutes in France. This IHU is based in Nice, and focuses on respiratory diseases. Inria is one of its 4 founding institutions together with the University Hospital of Nice, the Université Côte d'Azur and INSERM. Hervé Delingette and Nicholas Ayache are the members of the executive team and are leading a workpackage focusing on AI algorithms for data analysis.
- Several research projects of Epione are part of the joint laboratory Bernouilli between Inria and Assistance Publique des Hôpitaux de Paris (AP-HP), in particular, the DAICAP, and PAIMRI projects with Pr Raphaele Renard-Penna (Hospital La Pitié Salpêtrière), on prostate cancer detection and characterization.
- We also have long term collaborations with the CHU Nice, the Centre Antoine Lacassagne of Nice, and the Hospital Lenval of Nice.
10.5 Regional initiatives
10.5.1 PLICIA
Participants: Hervé Delingette.
-
Duration:
2024 - 2027
-
Partners:
- Quantificare
- Inria
- Hosteur
- I3S, CNRS
-
Inria contact:
Hervé Delingette
-
Coordinator:
Quantificare
-
Summary:
The PLICIA project is a I-Démo regional collaborative project which aims to create a software platform for managing clinical photographies and integrating AI solutions. More precisely, it focuses on the improvement of medical data management by building and providing a secure research and development infrastructure in AI.
In this project, we work on the anonymization of sensitive medical photographies by using generative AI models.
11 Dissemination
Participants: Nicholas Ayache, Irene Balelli, Benjamin Billot, Hervé Delingette, Marco Lorenzi, Xavier Pennec, Maxime Sermesant.
11.1 Promoting scientific activities
11.1.1 Scientific events: organisation
Member of the organizing committees
- I. Balelli is member of the organizing committee of the Complex days (to be held in Nice, February 2025) and of the spring school GeMSS/Statlearn (to be held in Sophia Antipolis, April 2025).
- H. Delingette was a member of the organizing committee of the MIDL 2024 conference that was held in Paris from July 3rd until July 5th and gathered around 500 persons.
- X. Pennec was one of the organizers of the conference "Geometric Sciences in Action: from geometric statistics to shape analysis", held at CIRM, Luminy, 27-31 May, 2024. His ERC G-Statistics financed half of the 80 participants.
- M. Sermesant was a member of the organizing committee of the STACOM MICCAI Workshop, held in Marrakech, October 2024.
11.1.2 Scientific events: selection
Chair of conference program committees
- B. Billot is Area Chair for the conference MIDL 2025 (Medical Imaging with Deep Learning) that will take place in Salt Lake City, USA.
- M. Lorenzi is Area Chair for the conference NeurIPS 2025 (Annual Conference on Neural Information Processing Systems) that will take place in San Diego, USA.
Reviewer
- I. Balelli was PC Member for the ECAI 2024 conference (Spain) and was the recipient of an Outstanding PC Member Award.
11.1.3 Journal
Member of the editorial boards
- N. Ayache is the co-founder and the Co-Editor in Chief with J. Duncan (Professor at Yale) of Medical Image Analysis journal. This scientific journal was created in 1996 and is published by Elsevier.
- N. Ayache is a member of the editorial board of the following journal: Journal of Computer Assisted Surgery (Wiley).
- H. Delingette is a member of the editorial board of the journal Medical Image Analysis (Elsevier).
- M. Lorenzi is member of the editorial board of the journal Medical Image Analysis (Elsevier). He is also member of the Board of Statisticians of the Journal of Alzheimer's Disease (IOS Press).
- X. Pennec is a member of the editorial board of the journal Medical Image Analysis (MedIA, Elsevier), of the International Journal of Computer Vision (IJCV, Springer), and of the Journal of Mathematical Imaging and Vision (JMIV, Springer).
Reviewer - reviewing activities
- I. Balelli was a reviewer for the following journals: Vaccine, Medical Image Analysis, Computers in Biology and Medicine, Neuroimage, SMAI J. of Computational Mathematics.
- B. Billot was reviewer for the following journals: Nature Biomedical Engineering, Scientific Data, Medical Image Analysis, IEEE Transactions on Medical Imaging.
- F. Cremonesi was a reviewer for the following journals: Journal of the American Medical Informatics Association (Oxford Academic), Medical Image Analysis (Elsevier), Patterns (Cell press).
- H. Delingette was a reviewer for the following journals: Medical Image Analysis, IEEE Transaction in Medical Imaging.
- M. Lorenzi was a reviewer for the following journals: Journal of Alzheimer's Disease, Medical Image Analysis, IEEE Transactions on Medical Imaging.
- X. Pennec was a reviewer for the following journals: Medical Image Analysis, SIAM Journal on Mathematics of Data Science (SIMODS), Medical Image Analysis, Journal of the Royal Statistical Society Series B, Information Geometry.
- M. Sermesant was a reviewer for the following journals: Journal of Machine Learning Research, Journal of the American College of Cardiology, IEEE Transactions on Medical Imaging, IEEE Transactions on Biomedical Engineering, Medical Image Analysis and Computers in Biology and Medecine.
11.1.4 Invited talks
- N. Ayache gave the following invited lectures: Invited Lecture for the History of Inserm in Digital Twin Modeling, organized by Inserm, Paris, 2024 (available on Youtube); Invited Lecture on the Future of Digital Twins, Académie de médecine, Paris, 2024; Invited talk at an expert panel on the impact of AI on Sciences, organized by 3IA Prairie, Paris, 2024.
- I. Balelli gave invited talks at IABM 2024 (Grenoble, March 24, 2024), i2m seminar (Marseille, March 11, 2024) and HeKa seminar (online, June 3, 2024).
- F. Cremonesi presented Fed-BioMed and the EUCAIM project at Rencontres Bernoulli Lab 2024 in Paris, on March 7th 2024.
- H. Delingette was an invited speaker at the French Congress of Histologists and Embryologists in Nice on March 22nd , at the French Radiological Congress (Journées Françaises de Radiologie) on Oct 7th in Paris.
- M. Lorenzi was keynote speaker at the the Congress "Medicine 4.0", organized by the Académie Nationale de Chirurgie and Medicen. He was also keynote speaker to the workshop POND (Progression over neurodegenerative disorders), held at University College London.
- X. Pennec gave invited talks at the workshop "Interactions of Statistics and Geometry (ISAG) II" in Singapour, October 14-26, the workshop "Plant Computational Biology", Lyon, November 4-8, the workshop "Challenges in Neuroimaging Data Analysis", IMSI, Chicago, August 26-30, the "Shape Workshop: Math in Maine", Andover, Maine, USA, August 18-24, the Special Session on "Computational Techniques to Study the Geometry of the Shape Space" at the Joint Mathematics Meetings AMS, San Francisco (CA, USA), January 6.
- M. Sermesant was an invited speaker at the IABM conference (AI in biomedical imaging), Grenoble, March and gave an invited talk at the French Academy of Sciences following the innovatino prize, Paris, May, gave an invited talk at JFICV (French conference on cardiovascular imaging) on digital twins. He also gave an invited talk at FIRE (interventional radiology).
11.1.5 Research administration
- N. Ayache has been a member of the scientific council of the Health Data Hub for 3 years until Sept 2024. He has been the scientific director and chair of the Scientific Committee of the 3IA Cote-d'Azur since its creation in 2019. He has been a member of the scientific council of the Mécénat Santé program of the AXA group since June 2024. He is on the scientific advisory board of the start-up companies inHeart (digital heart) and Caranx Medical (Medical Robotics).
- I. Balelli is a member of the scientific advisory board of the GIS (scientific interest group) FC3R since Jul. 2023, and of the Scientific committee of the Academy 2 (Complex Systems) since Nov. 2023. I. Balelli is in charge of the pedagogical orgization of the AI for Health track of the Data Science & AI Master, Univ. Côte d'Azur, France.
- H. Delingette is one of the 2 scientific directors of the IdEx program UCA JEDI under the direction of the IdEX vice-president of the Université Côte d'Azur. He is an administrator and a member of the scientific committee of the Groupement de Coopération Sanitaires (GCS) CARES involving the Université Côte d'Azur and the 3 local hospitals (CHU Nice, Centre Antoine Lacassagne, Fondation Lenval). He is also a member of the research and innovation committee organized by the employer union UPE06.
- H. Delingette is a representive of Inria at the Federation Hospitalo-Universitaire Oncoage led by the CHU Nice. He is also the contact person at the Inria center of Université Côte d'Azur for research data management. H. Delingette is the Inria representative at the executive committee of the DATAZUR structure, helping Université Côte d'Azur researchers handle their research data.
- M. Lorenzi is member of the Member of the Comité de Centre of the Centre Inria d'Université Côte d'Azur. He was Selection Committee Member for the France-Japan call for proposals on Edge Artificial Intelligence. He is External Advisory Board of the HealthData@EU Pilot project.
- X. Pennec is co-director of the Ecole doctorale STIC of Université Côte d'Azur. He is a member of the committee of EDSTIC, of the Doctoral follow-up Committee (CSD) at Inria Sophia Antipolis. He was also a member of the executive committee of the Academy 4 (Living systems Complexity and diversity) of the IDEX JEDI at University Côte d'Azur.
- X. Pennec was a member of the CRCN/ISFP recruitment committee of Inria Nancy (March / April).
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
- Master: H. Delingette and X. Pennec, Medical Image Analysis based on generative, geometric and biophysical models, 21h course (28.5 ETD), Master 2 MVA, ENS Saclay, France.
- Master: H. Delingette and X. Pennec, Medical Image Processing, 30h course, Master Data-Science and Artificial Intelligence, Université Côte d'Azur, France.
- Master: J. Rodríguez Padilla, Cardiac digital twin, 6h, Domaine Professionnel TMS: Technologies Médicales et de Santé, at École d'Ingénieurs ISEN Yncréa Ouest, Caen, France.
- Master: F. Cremonesi and Y. Bouillard, Federated Learning, 15h ETD, École d'ingénieur ISIS, Castres, France
- Master: H. Delingette, AI in Digital Pathology, 3h course, Master of Science Biobanks & Complex Data Management, Univ. Côte d'Azur, France.
- Master: H. Delingette, AI in Oncology 2h course, Master Cancérologie et Recherche Translationnelle, Univ. Côte d'Azur, France.
- Master: M. Lorenzi and V. Alessandro, Bayesian Learning, 30h course, Master Data Science, Univ. Côte d'Azur, France.
- Master: M. Lorenzi, Model Selection and Resampling Methods, 30h course, Master Data Science, Univ. Côte d'Azur, France.
- Institutional Diploma: K. Manasi, 22h, Data Analysis and Visualization, Centrale Méditerranée, Nice, France.
- Master: I. Balelli, Research awareness, 3h EURECOM, Sophia Antipolis.
- Licence: I. Balelli, Advanced statistical modeling, 22.5h ETD, Univ. Côte d'Azur, France.
- Licence: I. Balelli, Statistical modelling for complex data and Big Data, 37.5h ETD, Univ. Côte d'Azur, France.
- Licence: R. Silva, Electronique Analogique, 18H, and Électronique avec ARDUINO, 18H, Polytech, France.
- License: O. Bisson, L1 Math (Approfondissement 2), 40h + 10h de khôlles ETD, and L2 ECUE MATH (Compléments d'Algèbre Linéaire), 15h ETD, Univ. Côte d'Azur, France.
- University Diploma IA & Healthcare: R. Silva, 6h, Université Côte d’Azur, France.
- University Diploma IA & Healthcare: H. Delingette, 2h, Université Côte d’Azur, France.
11.2.2 Supervision: defended PhDs
- Josquin Harrison, Medical Imaging and learning for the prediction of strokes in the case of atrial fibrillation, Université Côte d'Azur. Started in 2020. Directed by Maxime Sermesant 50.
- Huiyu Li, Anonymization and protection of medical data based on deep neural networks. Université Côte d'Azur. Started in 2020. Co-directed by Hervé Delingette and Nicholas Ayache 51.
- Hari Sreedhar, Learning-based detection and classification of thyroid nodules from ultrasound images, Université Côte d'Azur. Started in 2020. Co-directed by Hervé Delingette and Charles Raffaelli (CHU-Nice) 52.
- Riccardo Taiello, Data Structuration and Security in Large-Scale Collaborative Healthcare Data Analysis, Université Côte d'Azur. Started in 2021. Co-supervised by Melek Önen (EURECOM) and Olivier Humbert (Centre Antoine Lacassagne) 53.
- Théodore Soulier, Prediction of re-myelination from Multi-sequence MRI for patients with Multiple Sclerosis. Collaboration with Bruno Stankov (Pitié-Salpêtrière Hospital, Paris) and Olivier Colliot (3IA Prairie, Paris) 40.
11.2.3 Supervision: ongoing PhDs
- Olivier Bisson, Géométrie, Stratification et application des matrices de corrélation structurées. Directed by X. Pennec. 3IA PhD started in October 2023.
- Alix de Langlais, Automatic generation of three-dimensional models of extremity fractures of proximal humerus for preoperative planning and intraoperative assistance in mixed reality, Inria-INSERM funded PhD started in June 2024. Co-directed by Hervé Delingette, and Marc-Olivier Gauci (Orthopedics Surgeon, CHU Nice, IBV).
- Federica Facente, Learning Statistical and Biomedical Models for multimodal image analysis – application to Image Guided Surgical Robotics, 3IA PhD started in September 2023. Co-directed by Nicholas Ayache, Pierre Berthet-Rayne (CTO and co-founder of Caranx-Medical, 3IA affiliate chair holder) and Hervé Delingette.
- Camilla Ferrario, Electromechanical modelling of non-ischemic cardomyopathies, funded by PEPR Digital Health ChroniCardio, started in September 2024.
- Sebastien Goffart, Development of predictive models in patients with peripheral artery disease, Université Côte d'Azur. ANR grant handled by CHU Nice. Started in 2023. Co-directed by Hervé Delingette, and Juliette Raffort-Lareyre.
- Lisa Guzzi, Automatic segmentation of the vascular system to enhance AI-based decision support system for peripheral artery disease, Université Côte d'Azur, 3IA fellowship. Started in 2022. Directed by Hervé Delingette, Juliette Raffort-Lareyre
- Manasi Kattel, Deep learning methods for the analysis and registration of US images, Université Côte d'Azur. 3IA Côte d'Azur fellowship. Started in December 2023. Co-directed by Nicholas Ayache and Hervé Delingette.
- Wassila Khatir, Integromics analysis: a new approach to study the pathophysiology of X-Fragile Syndrome (FXS), Université Côte d'Azur. Neuromod fellowship. Started in March 2024. Co-directed by Irene Balelli, Marco Lorenzi and Carole Gwizdek (IPMC).
- Maelis Morier, Deep Learning Meets Numerical Modelling, AI and Biophysics for Computational Cardiology, started in 2023. Co-supervised by Maxime Sermesant and Patrick Gallinari (Sorbonne Universités).
- Huyen Trang Nguyen, Robust Biomarker Extraction in PET-CT Imaging Data for Immunotherapy in Lung Cancer. Franco-German ANR Train, co-directed with Olivier Humbert (Centre Antoine Lacassagne).
- Evariste Njomgue Fotso, Multimodal learning for sudden cardiac death risk prediction. Funded by MediTwin, started in 2023.
- Rafael Luis Soares Da Costa E Silva, Artificial Intelligence for Cardiac Monitoring: Portable Multimodal Cardiac Function Analysis. Started in 2023. Co-directed by Maxime Sermesant and Pamela Moceri (CHU Nice).
- Tom Szwagier, Rethinking statistical methods with Flags spaces, Université Côte d'Azur. Started in 2022. Directed by Xavier Pennec.
11.2.4 Juries
- I. Balelli was a member of the PhD jury of H. Liu (Inria and Sorbonne University, Paris, May 2024) and of V. Montalibet (Bordeaux University, Bordeaux, Sep. 2024).
- H. Delingette was the external reviewer for the PhD defense committee of Sidaty El Hadramy (Université de Strasbourg, Dec. 2024), of Alexandre Cafaro (Université Paris Saclay, Dec. 2024), a member of the PhD defense committee of Remy Winter (Université Côte d’Azur, Aug. 2024), of Hakim Benkirane (Université Paris-Saclay, Dec. 2024) , an opponent in the PhD defense committee of Nathan Blanken (University Twente, NL, Dec. 2024).
- M. Lorenzi was PhD reviewer of C. Hassan (Queensland University of Technology), A. El Moadine (IMT Atlantique). He was also PhD jury member for the thesis of G. Quintana (ENS Paris), O. Laousy (Centrale Supelec), and , R. Louiset (Telecom Paris). He was Reviewer for the Habilitation à diriger des recherches of K. Kallas (IMT Atlantique).
- X. Pennec was President of the PhD jury of Josquin Harrison (Univ. Côte d'Azur, June 2024) and of Raphael Mignot (Univ. Lorraine, Oct. 2024). He was opponent in the PhD jury of Mohsen Taheri Shalmani, (Stavanger Univ., NO, June 2024), and external expert for a tenure committe in the USA.
- M. Sermesant was the president of the PhD jury of Louis Ohl, Université Côte d'Azur, May 2024, external reviewer for the licenciate seminar of Giacomo Verardo, KTH, Sweden. He was a reviewer for the habilitation thesis of Mark Potse, Bordeaux University, and a member of the PhD jury of Hang Jung Ling, Creatis, Lyon, November.
11.3 Popularization
11.3.1 Specific official responsibilities in science outreach structures
- M. Lorenzi co-authored with M. Van Waerebeke the article "Apprendre à oublier : le nouveau défi de l'intelligence artificielle" published in The Conversation.
11.3.2 Live events
- R. Silva, M. Morier, S. Al-Ali, E. da Pozzo, N. Drettakis, B. Ly, E. Njomgue Fosto, J. Rodriguez and N. Cedilnik participated to the Fête de la Science in Villeneuve-Loubet.
- M. Kattel and F. Facente participated to the Fête de la Science in Nice.
12 Scientific production
12.1 Major publications
- 1 articleEfficient Patient-Specific Simulations of Ventricular Tachycardia Based on Computed Tomography-Defined Wall Thickness Heterogeneity.JACC: Clinical ElectrophysiologySeptember 2023HALDOI
- 2 articleDeep Clustering for Abdominal Organ Classification in US imaging.Journal of Medical Imaging1032023, 034502HALDOI
- 3 articleA General Theory for Federated Optimization with Asynchronous and Heterogeneous Clients Updates.Journal of Machine Learning Research24March 2023, 1-43HAL
- 4 miscClustered Sampling: Low-Variance and Improved Representativity for Clients Selection in Federated Learning.May 2021HAL
- 5 articleIntroduction to Riemannian Geometry and Geometric Statistics: from basic theory to implementation with Geomstats.Foundations and Trends in Machine Learning163February 2023, 329-493HALDOI
- 6 articleMorphologically-Aware Consensus Computation via Heuristics-based IterATive Optimization (MACCHIatO).Journal of Machine Learning for Biomedical Imaging2UNSURE 2022 Special IssueSeptember 2023, 361-389HALDOI
- 7 inproceedingsImproving Neural Network Surface Processing with Principal Curvatures.NeurIPS ProceedingsNeurips 2024 - 38th Annual Conference on Neural Information Processing Systems2024Vancouver, CanadaDecember 2024HAL
- 8 articleSimultaneous data assimilation and cardiac electrophysiology model correction using differentiable physics and deep learning.Interface Focus136December 2023HALDOI
- 9 articleLearning a Generative Motion Model from Image Sequences based on a Latent Motion Matrix.IEEE Transactions on Medical ImagingFebruary 2021HALDOI
- 10 articleApplications of artificial intelligence in cardiovascular imaging.Nature Reviews CardiologyMarch 2021HALDOI
- 11 articleMS-CLAM: Mixed Supervision for the classification and localization of tumors in Whole Slide Images.Medical Image Analysis852023, 102763HALDOI
- 12 articleBayesian Logistic Shape Model Inference: application to cochlear image segmentation.Medical Image AnalysisOctober 2021HAL
- 13 articleEchocardiography Analysis with Deep Learning using Priors: Multi-centric Evaluation of Generalisation.Journal of Machine Learning for Biomedical Imaging2November 2024November 2024, 2293-2325HALDOI
12.2 Publications of the year
International journals
- 14 articleDetection of ulcerative colitis lesions from weakly annotated colonoscopy videos using bounding boxes.Gastrointestinal Disorders61March 2024, 292-307HALDOI
- 15 articleDifferential of the Stretch Tensor for Any Dimension with Applications to Quotient Geodesics.Comptes Rendus. Mathématique362G12November 2024, 1847-1856HALDOIback to text
- 16 articleGraph Alignment Exploiting the Spatial Organization Improves the Similarity of Brain Networks.Human Brain Mapping451January 2024, e26554HALDOI
- 17 articleVascular liver segmentation: a narrative review on methods and new insights brought by artificial intelligence.Journal of International Medical Research529September 2024HALDOI
- 18 articleA Bayesian approach for clustering and exact finite-sample model selection in longitudinal data mixtures.Computational StatisticsMay 2024HALDOI
- 19 articleArtificial Intelligence to enhance future clinical trials in Vascular Surgery.Annals of Vascular Surgery111November 2024, 331-335HALDOIback to text
- 20 articleOn Tail Decay Rate Estimation of Loss Function Distributions.Journal of Machine Learning Research2525February 2024, 1−47HAL
- 21 articleBenchmarking feature selection and feature extraction methods to improve the performances of machine-learning algorithms for patient classification using metabolomics biomedical data.Computational and Structural Biotechnology Journal231December 2024, 1274-1287HALDOI
- 22 articleA Systematic Review of the Diagnostic Accuracy of Deep Learning Models for the Automatic Detection, Localization, and Characterization of Clinically Significant Prostate Cancer on Magnetic Resonance Imaging.European Urology Oncology2024HALDOI
- 23 articlePrivacy Preserving Image Registration.Medical Image Analysis94May 2024, 103129HALDOIback to text
- 24 articlePermutation-invariant log-Euclidean geometries on full-rank correlation matrices.SIAM Journal on Matrix Analysis and Applications452April 2024, 930-953HALDOI
- 25 articleWhARIO: whole-slide-image-based survival analysis for patients treated with immunotherapy.Journal of Medical Imaging1103May 2024HALDOI
- 26 articleMutual Information Guided Diffusion for Zero-Shot Cross-Modality Medical Image Translation.IEEE Transactions on Medical Imaging438March 2024, 2825-2838HALDOI
- 27 articleSecond order kinematic surface fitting in anatomical structures.Medical Image AnalysisFebruary 2025, 103488HALDOI
- 28 articleEchocardiography Analysis with Deep Learning using Priors: Multi-centric Evaluation of Generalisation.Journal of Machine Learning for Biomedical Imaging2November 2024November 2024, 2293-2325HALDOIback to text
International peer-reviewed conferences
- 29 inproceedingsAssessing Ionic Current Blockades and Electromechanical Biomarkers’ Interrelations Through a Novel Multi-Channel Causal Variational Autoencoder.Computing in Cardiology 20242024 Computing in Cardiology Conference51408Karlsruhe (Germany), GermanySeptember 2024HALDOIback to text
- 30 inproceedingsA Computer Model for In Silico Trials on Pacemaker Energy Efficiency.Computing in Cardiology 2024CinC 2024 - 51st international Computing in Cardiology conferenceKarlsruhe, GermanyDecember 2024HALDOI
- 31 inproceedingsSIFU: Sequential Informed Federated Unlearning for Efficient and Provable Client Unlearning in Federated Optimization.PMLR proceedingsAISTATS 2024 - International Conference on Artificial Intelligence and StatisticsPMLR-238International Conference on Artificial Intelligence and Statistics, 2-4 May 2024, Palau de Congressos, Valencia, SpainValencia, SpainMay 2024HAL
- 32 inproceedingsFederated Multi-centric Image Segmentation with Uneven Label Distribution.Lecture Notes in Computer ScienceMICCAI 2024 - 27th International Conference on Medical Image Computing and Computer Assisted InterventionLNCS-15010Marrakesh, MoroccoSpringer Nature SwitzerlandOctober 2024, 350-360HALDOI
- 33 inproceedingsDifferentiable Soft Morphological Filters for Medical Image Segmentation.Lecture notes in computer scienceMICCAI 2024 - Medical Image Computing and Computer Assisted InterventionLNCS-15008Medical Image Computing and Computer Assisted Intervention – MICCAI 2024 27th International Conference, Marrakesh, Morocco, October 6–10, 2024, Proceedings, Part VIIIMarrakesh, MoroccoOctober 2024HALback to text
- 34 inproceedingsImproving Neural Network Surface Processing with Principal Curvatures.NeurIPS ProceedingsNeurips 2024 - 38th Annual Conference on Neural Information Processing Systems2024Vancouver, CanadaDecember 2024HALback to text
- 35 inproceedingsGenerative medical image anonymization based on latent code projection and optimization.IEEE International Symposium on Biomedical Imaging (ISBI 2025)Houston (Texas), United StatesApril 2025HAL
- 36 inproceedingsRetractions on closed sets.IFAC-PapersOnLineMTNS 2024 - 26th International Symposium on Mathematical Theory of Networks and Systems5817Cambridge (GB), United Kingdom2024, 286-291HALDOIback to text
- 37 inproceedingsPersonalisation of Action Potentials Based on Activation Recovery Intervals in Post Infarcted Pigs: A Simulation Study.IEEE XploreCinC 2024 - Computing in Cardiology 202451Karlsruhe (Allemagne), GermanySeptember 2024HALback to text
- 38 inproceedingsAutomatic Analysis of Activation Recovery Interval in Heterogeneous Fibrotic Tissue from Chronically Infarcted Swine.Computing in CardiologyCinC 2024 - 51th International Computing in Cardiology 202451Karlsruhe, Germany2024HALDOIback to text
- 39 inproceedingsYOUR-Lead: YOLO and U-Net for Reconstruction of ECG Lead Signals.2024 Computing in Cardiology2024 Computing in Cardiology Conference51Karlsruhe (Germany), GermanySeptember 2024, 1-4HALDOIback to textback to text
- 40 inproceedingsGenerating PET-derived maps of myelin content from clinical MRI using curricular discriminator training in generative adversarial networks.SPIE Medical ImagingSan Diego, United StatesSPIEFebruary 2024HALDOIback to text
- 41 inproceedingsEnhancing Privacy in Federated Learning: Secure Aggregation for Real-World Healthcare Applications.Lecture notes in computer science5th MICCAI Workshop on Distributed, Collaborative and Federated Learning (in Conjunction with MICCAI 2024)LNCS-15274Medical Image Computing and Computer Assisted Intervention – MICCAI 2024 WorkshopsMarrachech, MoroccoSeptember 2024, 204–214HALDOIback to textback to text
- 42 inproceedingsLet Them Drop: Scalable and Efficient Federated Learning Solutions Agnostic to Stragglers.ACM Digital LibraryARES 2024 - 19th International Conference on Availability, Reliability and SecurityARES '24: Proceedings of the 19th International Conference on Availability, Reliability and Security13Vienna, AustriaACMJuly 2024, 1-12HALDOIback to text
- 43 inproceedingsExploring Chemotherapy-Induced Cardiotoxicity Combining A 3D Computational Model and Preclinical Cardiac Imaging Data.Lecture notes in computer scienceSTACOM 2024 - 15th workshop on Statistical Atlases and Computational Modeling of the HeartLNCS-Marrakesh, MoroccoNovember 2024HALback to text
- 44 inproceedingsUncertainty-Based Multi-modal Learning for Myocardial Infarction Diagnosis Using Echocardiography and Electrocardiograms.Lecture Notes in Computer ScienceASMUS 2024 - The 5th International Workshop of Advances in Simplifying Medical UltraSoundLNCS-15186Simplifying Medical Ultrasound : 5th International Workshop, ASMUS 2024, Held in Conjunction with MICCAI 2024, Marrakesh, Morocco, October 6, 2024, ProceedingsMarrakech, MoroccoSpringer Nature SwitzerlandOctober 2024, 177-186HALDOIback to text
Conferences without proceedings
- 45 inproceedingsDisease Progression Modelling and Stratification for detecting sub-trajectories in the natural history of pathologies: application to Parkinson's Disease trajectory modelling.LDTM 2024 - Longitudinal Disease Tracking and Modelling with Medical Images and Data (MICCCAI 2024)Marrachech, MoroccoAugust 2024HALback to text
Scientific books
- 46 bookTrustworthy AI in Medical Imaging.Elsevier2025HALDOIback to text
Scientific book chapters
- 47 inbookPreface (Trustworthy AI in medical imaging).Trustworthy AI in medical imagingAcademic PressNovember 2024HALback to text
- 48 inbookCausality: fundamental principles and tools.Trustworthy AI in Medical ImagingChapitre 14November 2024, 297-314HALback to text
- 49 inbookIntroduction to trustworthy AI for medical imaging: Lecture plan.Trustworthy AI in Medical ImagingAcademic Press; ElsevierNovember 2024, 536HALDOIback to text
Doctoral dissertations and habilitation theses
- 50 thesisMedical imaging, shapes and statistics for stroke prediction in atrial fibrillation.Université Côte d'AzurJune 2024HALback to textback to text
- 51 thesisData exfiltration and anonymization of medical images based on generative models.Université Côte d'AzurNovember 2024HALback to textback to text
- 52 thesisThyrosonics : learning-based detection and classification of thyroid nodules from ultrasound images.Université Côte d'AzurOctober 2024HALback to textback to text
- 53 thesisPrivacy-preserving machine learning for large-scale collaborative healthcare data analysis.Université Côte d'AzurSeptember 2024HALback to text
Reports & preprints
- 54 miscBRAIN NETWORK ALIGNMENT USING STRUCTURAL AND FUNCTIONAL CONNECTIVITY WITH ANATOMICAL CONSTRAINTS.January 2025HAL
- 55 miscMulti-Channel Causal Variational Autoencoder.August 2024HAL
- 56 miscThe Geometry of Right-Invariant Metrics.May 2024HAL
- 57 miscOn the Implementation of Geodesic Metric Spaces.June 2024HAL
- 58 miscKnowledge-based semantic enrichment of medical imaging data for automatic phenotyping and pattern discovery in metastatic lung cancer.February 2025HALback to text
- 59 miscA cautionary tale on the cost-effectiveness of collaborative AI in real-world medical applications.December 2024HAL
- 60 miscOptimizing Intraoperative AI: Evaluation of YOLOv8 for Real-Time Recognition of Robotic and Laparoscopic Instruments.December 2024HALDOIback to text
- 61 reportSoftMorph: Differentiable Probabilistic Morphological Operators for Image Analysis.TechrxivOctober 2024HALDOI
- 62 miscComputing Bouligand stationary points efficiently in low-rank optimization.September 2024HAL
- 63 miscLow-rank optimization methods based on projected-projected gradient descent that accumulate at Bouligand stationary points.July 2024HAL
- 64 miscProjected gradient descent accumulates at Bouligand stationary points.March 2024HALback to text
- 65 miscA geometric framework for asymptotic inference of principal subspaces in PCA.November 2024HAL
- 66 miscEffective formulas for the geometry of normal homogeneous spaces. Application to flag manifolds.December 2024HAL
- 67 miscThe curse of isotropy: from principal components to principal subspaces.August 2024HALDOIback to text
Other scientific publications
- 68 inproceedingsActivation Recovery Interval for Personalized Cardiac Assessment.CinC 2024 - (&th international Computing in CardiologyKarlsruhe, GermanyZenodo2024HALDOIback to text
- 69 inproceedingsArtificial Intelligence for Automated Cardiac Defibrillation in Embedded Systems.SophIA Summit 2024Biot, FranceZenodo2024HALDOIback to text
Scientific popularization
- 70 inproceedingsConstraint-Based Model in Multimodal Learning to Improve Ventricular Arrhythmia Prediction.STACOM 2024 - 15th Statistical Atlases and Computational Modeling of the Heart workshop (MICCAI 2024)Marrackech, MoroccoOctober 2024HALback to text

