2024Activity reportProject-TeamSODA
RNSR: 202224249S- Research center Inria Saclay Centre
- Team name: Computational and mathematical methods to understand health and society with data
- Domain:Digital Health, Biology and Earth
- Theme:Computational Neuroscience and Medicine
Keywords
Computer Science and Digital Science
- A3.3. Data and knowledge analysis
- A3.4. Machine learning and statistics
- A9.1. Knowledge
- A9.2. Machine learning
Other Research Topics and Application Domains
- B2.3. Epidemiology
- B9.1. Education
- B9.5.6. Data science
- B9.6.1. Psychology
- B9.6.3. Economy, Finance
1 Team members, visitors, external collaborators
Research Scientists
- Gael Varoquaux [Team leader, INRIA, Senior Researcher]
- Judith Abecassis [INRIA, ISFP]
- Marine Le Morvan [INRIA, Researcher]
- Jill Jenn Vie [INRIA, Researcher]
Post-Doctoral Fellows
- Riccardo Cappuzzo [INRIA, Post-Doctoral Fellow, until Sep 2024]
- Lihu Chen [Inria, from Jun 2024 until Oct 2024]
- Myung Kim [INRIA, Post-Doctoral Fellow]
- Jingang Qu [INRIA, Post-Doctoral Fellow, from Feb 2024]
- Clémence Reda [UNIV Rostock, Post-Doctoral Fellow]
PhD Students
- Julie Alberge [INRIA]
- Marie Generali-Lince [INRIA, from Oct 2024]
- Samuel Girard [INRIA]
- Leo Grinsztajn [INRIA, until Sep 2024]
- Felix Lefebvre [INRIA]
- Sebastien Melo [INRIA, from Oct 2024]
- Alexandre Perez [INRIA]
- Jovan Stojanovic [INRIA]
Technical Staff
- David Arturo Amor Quiroz [INRIA, Engineer, until Jan 2024]
- Hiba Bederina [INRIA, Engineer, from Jun 2024, Research engineer]
- Franck Charras [INRIA, Engineer, until Jan 2024]
- Jerome Dockes [INRIA, Engineer, until Aug 2024]
- Jeremie Du Boisberranger [INRIA, Engineer, until Jan 2024]
- François Goupil [INRIA, Engineer, until Jan 2024]
- Olivier Grisel [INRIA, Engineer, until Jan 2024]
- Tristan Haugomat [INRIA, Engineer, from May 2024]
- Guillaume Lemaitre [INRIA, Engineer, until Jan 2024]
- Vincent Maladiere [INRIA, Engineer, until Jan 2024]
Interns and Apprentices
- Achraf Chaouch [INRIA, Intern, from May 2024 until Aug 2024]
- Marie Generali-Lince [INRIA, Intern, from Apr 2024 until Sep 2024]
- Sebastien Melo [INRIA, Intern, from Apr 2024 until Aug 2024]
Administrative Assistants
- Marie Enee [INRIA]
- Ekaterina George [INRIA]
External Collaborators
- Audrey Berges [AP/HP, from Mar 2024]
- Lihu Chen [IMPERIAL COLLEGE LDN, from Oct 2024]
- Matthieu Doutreligne [HAS]
- Lea Hoisnard [AP/HP, from Nov 2024]
- Theo Jolivet [AP/HP]
- Yann Lechelle [SENS DIGITAL]
- Elise Liu [AP/HP, from Mar 2024]
- Koh Takeuchi [Kyoto University]
- Camille Troillard [Probabl]
2 Overall objectives
2.1 Context
2.1.1 Application context: richer data in health and social sciences
Opportunistic data accumulations, often observational, bare great promises for social and health sciences. But the data are too big and complex for standard statistical methodologies in these sciences.
Health databases
Increasingly rich health data is accumulated during routine clinical practice as well as for research. Its large coverage brings new promises for public health and personalized medicine, but it does not fit easily in standard biostatistical practice because it is not acquired and formatted for a specific medical question.
Social, educational, and behavioral sciences
Better data sheds new light on human behavior and psychology, for instance with on-line learning platforms. Machine learning can be used both as a model for human intelligence and as a tool to leverage these data, for instance improving education.
Likewise, activity traces can provide empirical evidence for economical or political science, but their complexity requires new statistical practices.
AI in society
AI increasingly impacts multiple aspects of society. As such, it calls for rigorous evaluation, whether it is a benchmark of its ability, or a broader assessment of its impacts.
2.1.2 Related data-science challenges
Data management: preparing tabular data for analytics
Assembling, curating, and transforming data for data analysis is very labor intensive. These data-preparation steps are often considered the number one bottleneck to data-science. They mostly rely on data-management techniques. A typical problem is to establish correspondences between entries that denote the same entities but appear in different forms (entity linking, including deduplication and record linkage). Another time-consuming process is to join and aggregate data across multiple tables with repetitions at different levels (as with panel data in econometrics and epidemiology) to form a unique set of “features” to describe each individual. This process is related to database denormalization and might require schema alignment when performed across multiple data sources with imperfect correspondence in columns.
Progress in machine learning increasingly helps automating data preparation and processing data with less curation.
From machine learning to statistically-valid answers
Machine learning can be a tool to answer complex domain questions by providing non-parametric estimators. Yet, it still requires much work, eg to go beyond point estimators, to derive non-parametric procedures that account for a variety of bias (censoring, sampling biases, non-causal associations), or to provide theoretical and practical tools to assess validity of estimates and conclusion in weakly-parametric settings.
A question that is increasingly important in all applications of machine learning is that of auditing the model used in practice. This question arises in fundamental-research settings (medical research, political science...) for statistical validity, and in applications to assess societal biases, or safety of AI systems.
3 Research program
3.1 Table representation learning
Soda develops develop deep-learning methodology for relational databases, from tabular datasets to full relational databases. The stakes are i) to build machine-learning models that apply readily to the raw data so as to minimize manual cleaning, data formatting and integration, and ii) to extract reusable representations that reduce sample complexity on new databases by transforming the data in well-distributed vectors and bringing background information. The success of embarking such background knowledge in foundation models such as large language models motivates a quest for table foundation models.
3.2 Mathematical aspects of statistical learning for data science
While complex models used in machine learning can be used as non-parametric estimators for a variety of statistical tasks or for decision making, the statistical procedures and validity criterion need to be reinvented. Soda contributes statistical tools and results for a variety of problems important to data science in health and social science (epidemiology, econometrics, education). Statistical topics of interest comprise:
- Missing values and survival analysis
- Causal inference
- Model validation and auditing
- Uncertainty quantification
3.3 Machine learning for health and social sciences
Soda targets applications in health and social sciences, as these can markedly benefit from advanced processing of richer datasets, can have a large societal impact, but fall out of mainstream machine-learning research, which focus on processing natural images, language, and voice. Rather, data surveying humans needs another focus: it is most of the time tabular, sparse, with a time dimension, and missing values. In term of application fields, we focus on the social sciences that rely on quantitative predictions or analysis across individuals, such as policy evaluation. Indeed, the same formal problems, addressed in the two research axes above, arise across various social sciences: epidemiology, education research, and economics. The challenge is to develop efficient and trustworthy machine learning methodology for these high-stakes applications.
3.4 Turn-key machine-learning tools for socio-economic impact
Societal and economical impact of machine learning requires easy-to-use practical tools that can be leveraged in non-specialized organizations such as hospitals or policy-making institutions.
Soda works on scikit-learn, one of the most popular machine-learning tool world-wide, as well as skrub, a younger project that specializes machine learning for tables. Our goal is to transfer outside of the lab the understanding of machine learning and data science accumulated by the various research efforts.
Soda also works on other important software tools to foster growth and health of the Python data ecosystem in which scikit-learn is embedded.
4 Application domains
4.1 Precision medicine, public health, and epidemiology
Data management is the focus of the field of medical informatics as it is notably challenging in healthcare settings, due to the multiplicity of sources and the richness of the data that encompasses many modalities. We apply the our machine techniques for statistical analysis, including causal inference, in medicine to facilitate clinical research and public-health evidence. The central questions are that of personalized medicine –prediction at the individual level, for diagnosis, prognosis, or drug recommendation– and of public health –evaluation of treatments and policy, estimation of risk factors. The data on which we work are patient history and claims databases: mid-dimensional data with longitudinal coverage (as opposed to “omics” or imaging data, which is high dimensional and much less frequently available in clinical settings).
We collaborate actively with AP-HP and Ministère de la Santé. APHP provides access to its very rich and complex data mart, with dozens of tables following millions of individuals, both a challenge and an opportunity, and we work with various medical specialists (neurology, diabetology, public health) on specific clinical questions related to prognostic, treatment evaluation, and risk factors. With Ministère de la Santé, we process the claims data from the national insurance database to establish trajectories of individuals as a function of their future health risks. The short-term goal is to find which medical conditions can be predicted and with what reliability. The longer-term goal is to define prevention strategies.
4.2 Educational data mining
In educational data mining, we are interested in developing mathematical methods of learning to personalize education through adaptive assessment (developing algorithms that select questions for measuring efficiently the latent knowledge of examinees or for optimizing learning), recommending learning resources, generating exercises automatically. It is a challenging problem as it is hard to quantify learning, unlike in traditional reinforcement learning scenarios, and it is hard to measure the effect of courses on learning. This is why it is traditionally modeled as a partially-observable Markov decision process (POMDP). We are interested in modeling the evolution of uncertainty over the latent knowledge of examinees over time, for example using Bayesian approches, or model-based reinforcement learning.
Soda is actively collaborating with the national platform Pix.fr for certifying the digital competencies of all French citizens. Jill-Jênn Vie is one of the original core developers and they jointly received a Paris Region PhD grant in 2023 allowing them to co-supervise the PhD of Samuel Girard about optimizing human learning. In 2023, Jill-Jênn Vie joined the scientific committee of the French Ministry of Education (CSEN, conseil scientifique de l'Éducation nationale), leading to collaborations with Franck Ramus and ongoing discussions with Camille Terrier, Marc Gurgand, Hugo Gimbert via the scientific committee of MonProjetSup, a state startup about a study path recommender system.
4.3 Data management
Data preparation for analytics is intrinsically related to data management. For instance, linked open data provides consistent views on data across silos, but integrating these data into a statistical model to answer a given question still requires a lot of user efforts. Database operation increasingly relies on machine learning. While Soda is in no way expert in database research, the analytic tools that we build for relational data are increasingly used for data management. We are collaborating with Paolo Papotti (Eurecom) on this topic.
4.4 Broader data science
The tools, practical and theoretical, that we develop are central to many applications of data science. For instance, we often discuss with banks and insurances, which use machine learning but face statistical problems that we tackle: censoring or other sampling biases, forecasting, uncertainty quantification. Marketing and business intelligence also face the same exact problems. Even more generally, data preparation from relational databases is a challenge is most data-science applications. We interact with data scientists in a broad set of applications via the user base of the software tools that we develop (eg scikit-learn) and the various courses and lectures that we give around these tools to industry audiences.
We have started a collaboration in economics (Margherita Comola, Paris School of Economics) on using machine learning to understanding communication strategies of politicians from social-network data.
4.5 Behavioral sciences
A methodological challenge in health and educational sciences common to behavioral science is that the quantities of interest are difficult to measure, e.g. intelligence or progress of a student. Supervised machine learning can infer proxies from indirect signs, such as psychological traits from brain imaging, diagnosis from clinical traces, or socio-economical status from demographics. This notion of proxies is central in policy evaluation, serving as indirect signals in causal inference, to provide secondary outcomes for treatment effect estimation or to control confounders not directly observed.
An ongoing project with Pass Culture (via Inria-Ministry of Culture convention) is to adapt the recommender system of the app to encourage diversity, i.e. not only optimize click-through rate, but making students discover new things. This is done by modeling this problem as contextual bandits, and a diversity term acts as regularizer in the objective function.
5 Social and environmental responsibility
5.1 Footprint of research activities
The main footprint of Soda's activity is the carbon footprint of our travels (surpassing our compute cost, as we seldom run very intensive computation). For this reason, we try to be careful with our long-distance travel and try to take the plane as little as possible. Not flying at all is not possible, as it would cut us off from the world-wide research community sometimes mediated by crucial conferences in North America. However, we favor online seminars, or on-premise talks accessible by train.
Because of a race to scale, artificial intelligence is starting to have a large environmental footprint. As this is the result of collective action, as opposed to a single research group, we are trying to bring this problem to the attention of the community 42. Whenever possible, we also work on algorithms with small computational costs. For instance using tree-based models instance of neural networks can sometimes bring sizable computational and statistical benefits 35. Such work requires solving fundamental challenges, as trees are not differentiable, and is sometimes difficult to get accepted because it not fashionable.
5.2 Impact of research results
While data science can improve health and education, working with personal data or providing decision tools that affect individuals comes with responsibilities.
We make sure that work at Soda do not risk having direct negative impact. All research real-life health data (hospital-level or nation-wise) is started only after approval by the corresponding ethical board. Soda does not put any tools in production: none of the works of soda directly leads to automated decisions. Consequently none of our work has directly impacted individuals. Soda works on pseudonymized data, and we leave the –pseudonymized– electronic health data on servers inside the protected environment of the hospital where they have been acquired and are used. Going further, Soda runs research on privacy-preserving synthetic data generation, to provide open datasets for research and development without privacy concerns.
Soda is also active on assess and discussing the broader impacts and risks associated to AI, participating in national 34 and international 36 efforts to create consensus.
6 Highlights of the year
6.1 Awards
-
Gaël Varoquaux
Clarivate highly-cited researcher
7 New software, platforms, open data
7.1 New software
7.1.1 Scikit-learn
-
Keywords:
Clustering, Classification, Regression, Machine learning
-
Scientific Description:
Scikit-learn is a Python module integrating classic machine learning algorithms in the tightly-knit scientific Python world. It aims to provide simple and efficient solutions to learning problems, accessible to everybody and reusable in various contexts: machine-learning as a versatile tool for science and engineering.
-
Functional Description:
Scikit-learn can be used as a middleware for prediction tasks. For example, many web startups adapt Scikitlearn to predict buying behavior of users, provide product recommendations, detect trends or abusive behavior (fraud, spam). Scikit-learn is used to extract the structure of complex data (text, images) and classify such data with techniques relevant to the state of the art.
Easy to use, efficient and accessible to non datascience experts, Scikit-learn is an increasingly popular machine learning library in Python. In a data exploration step, the user can enter a few lines on an interactive (but non-graphical) interface and immediately sees the results of his request. Scikitlearn is a prediction engine . Scikit-learn is developed in open source, and available under the BSD license.
- URL:
- Publications:
-
Contact:
Gael Varoquaux
-
Participants:
Thomas Moreau, Jerome Dockes, Alexandre Gramfort, Bertrand Thirion, Gael Varoquaux, Loic Esteve, Olivier Grisel, Guillaume Lemaitre, Jeremie Du Boisberranger, Julien Jerphanion
-
Partners:
Axa, BNP Parisbas Cardif, Dataiku, Nvidia, Chanel, Probabl
7.1.2 joblib
-
Keywords:
Parallel computing, Cache
-
Functional Description:
Facilitate parallel computing and caching in Python.
- URL:
-
Contact:
Gael Varoquaux
-
Participant:
Thomas Moreau
-
Partner:
Probabl
7.1.3 skrub
-
Keyword:
Data analysis
-
Functional Description:
Joins, aggregates, and vectorizes tables to enable statistical learning, including with badly formated entries
- URL:
-
Contact:
Gael Varoquaux
-
Participants:
Jerome Dockes, Riccardo Cappuzzo
-
Partner:
Probabl
8 New results
8.1 Table representation learning
Participants: Gael Varoquaux.
Tabular deep learning
Neural networks traditionally underperform tree-based learners on tabular data. However, Holzmuller et al3 show that an array of modifications (initializations, learning-rate scheduler, feature standardization...), enables classic architectures (such as the multi-layer perceptron) to catch up. This work suggests that defaults must be adapted to the data modality, and tables call for new defaults.
Table foundation models
Much of the success of deep learning has been driven by the ability to reuse pretrained models –fitted on very large datasets, foundation models pushing this idea very far with models that provide background information useful for a wide variety of downstream tasks. A crucial part of these foundation model is the attention mechanism, stacked in a transformer architecture, that bring associative memory to the inputs by contextualizing them.
With the CARTE model 4, we adapted these ideas to tables. The strings –in the tables entries and column names– give the information that enables transfer from one table to another: data semantics. Here, key is to have an architecture that 1) models both strings and numerical values 2) applies to any set of tables while using the column names to route the information. For this purpose, CARTE uses a new dedicated attention mechanism that accounts for column names. It is pre-trained on a very large knowledge base. As a result, it outperform the best models (including tree-based models) in small sample settings (up to ).
This result is very significant as it opens the door to foundation models for tables. It is giving birth to a very active line of research.
8.2 Statistical aspects of machine learning
Participants: Marine Le Morvan, Gael Varoquaux.
Prediction with missing values
Asymptotic results shows that to predict well with missing values, it is neither necessary nor sufficient to impute well these missing values by their most-likely value. Le Morvan et al5 studied the finite-sample question empirically, in the missing at random settings where, in theory, imputation in most likely to give benefits. Results show that indeed, better recovery of missing values leads to better prediction, but with diminishing returns: a large improvement in recovery quality –which typically comes at a sizable computational cost– leads to a small improvement in prediction accuracy. Additionally, the more flexible the final learner, the weaker the link is. However, adding a missing-value indicator, an extra column that indicates which values have been imputed, is always beneficial.
Assessment of large language models
Large language models (LLMs), such as chatGPT, may produce answers that are plausible but not factually correct, the so-called “hallucinations”. A variety of approach try to assess how likely a statement is to be true, for instance by sampling multiple responses from the language model. However, the challenge is to threshold these assessments, or assign a probability of correctness.
Chen et al1 investigates the confidence of LLMs in their answers. The work shows that the probabilities computed are not only overconfident, but also that there is heterogeneity (grouping loss): on some groups of queries the overconfidence is more pronounced than on others. For instance, for an answer on a notable individual, the LLMs' confidence is reasonably calibrated if the individual is from the United States, but severely overconfident for individuals from South East Asia (fig:llmconfidencenationality). Characterizing the corresponding groups opens the door to correcting the corresponding bias, a “reconfidencing” procedure.
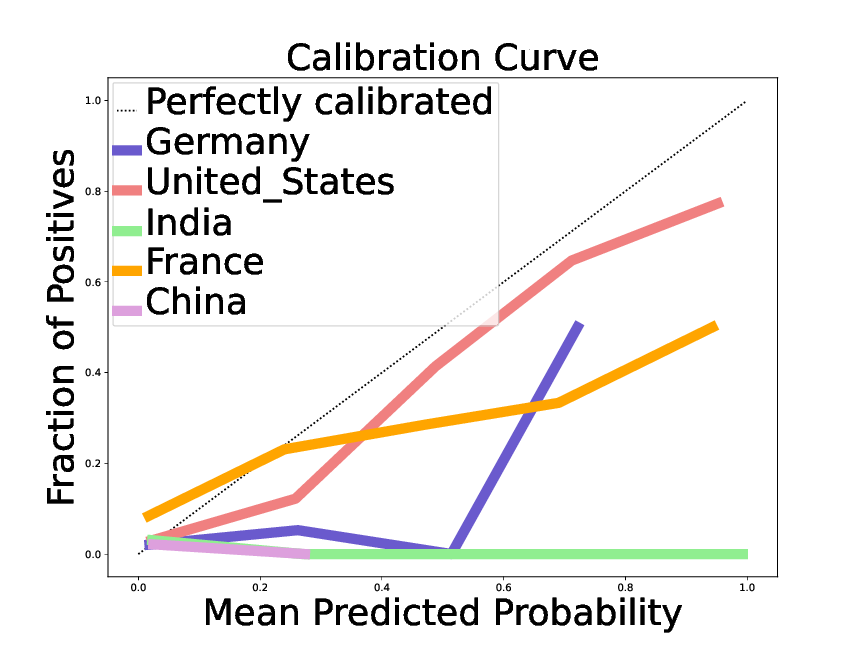
A reliability plot, giving the fraction of true statements as a function of the predicted probability for dates of birth, in replies on individuals of different nationality.
A reliability plot, giving the fraction of true statements as a function of the predicted probability for dates of birth, in replies on individuals of different nationality.
8.3 Machine learning for health and social sciences
Participants: Gael Varoquaux, Judith Abécassis, Jill-Jênn Vie.
Causal machine learning on large scale observational data
Causal approaches offer a compromise between purely predictive machine learning models that have no causal interpretation and randomized experiments that are costly and difficult to organize. Causal machine learning proposes a framework of strong assumptions to assess the causal effect of a treatment on an outcome. Those assumptions focus on the inclusion of confounding variables in the model and a non-zero probability to get the treatment for any unit in the dataset.
Dumas et al2 apply this strategy to systematically analyze the impact of chronic diseases and medications at the time of breast cancer (BC) diagnosis on cancer survival using the French Social Security data (SNDS). This would be infeasible and inefficient to do it on actual BC patients, for cost and ethical considerations, but the scale and the exhaustivity of the SNDS data enables such a study, at least to narrow down potential therapeutic candidates. In the analysis of a cohort of 235,368 French women and 288 medications with sufficient subcohort size to draw statistical conclusions, several medications have a statistically significant positive or negative effect on BC survival. Those results should not be directly interpreted as candidates for additional treatment after the BC diagnosis as chronic conditions pre-existing the diagnosis are considered here, but offer some insights about potential drug interactions or mechanism that affect the onset of BC, in particular through the immune system. This large-scale systematic study also provides a proof-of-concept of the relevance and the precision of medical knowledge that could be extracted from large claim or EHR datasets.
Reinforcement learning for adaptive recommendation of learning resources
Massive Open Online Courses (MOOCs) have greatly contributed to making education more accessible. However, many MOOCs maintain a rigid, one-size-fits-all structure that fails to address the diverse needs and backgrounds of individual learners. Learning path personalization aims to address this limitation, by tailoring sequences of educational content to optimize individual student learning outcomes. Existing approaches, however, often require either massive student interaction data or extensive expert annotation, limiting their broad application.
Vassoyan et al. 6 have framed learning path personalization as a partially observable Markov decision process. This is actually the first RL environment for dynamic cognitive diagnosis, where we assume that students learn when we show them documents within their frontier of knowledge (i.e. zone of proximal development) and our goal is to optimize their learning outcomes. By propagating information on a bipartite graph of keywords and documents, we could learn a policy (using REINFORCE algorithm) for selecting the best learning resource for learning a topic. By using word embeddings on the content of documents, we could alleviate item cold-start. We conducted experiments with simulated students on a real corpus of MOOCs. We went deeper in reducing the data needed to provide relevant recommendations of documents: dozens of episodes instead of thousands of episodes. We also showed that our method can generalize to unseen corpuses of documents. This is a collaboration with Anan Schütt & Elisabeth André from U. Augsburg, Arun Narayanan from U. Pittsburgh, and Nicolas Vayatis from Centre Borelli.
8.4 Turn-key machine-learning tools for socio-economic impact
Participants: Gael Varoquaux.
Releases of scikit-learn
With 3 major releases in 2024 (1.4 in Jan, 1.5 in May, and 1.6 in December), Scikit-learn is always improving, adding features for better and easier machine learning in Python. We list below a few highlights that are certainly not exhaustive but illustrate the continuous progress made.
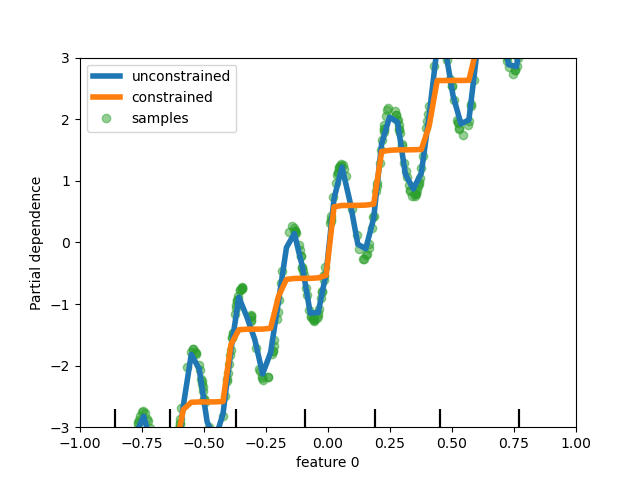
Monotonic constraints in trees clip out the bumps when fitting the addition of a sine wave and a constantly increasing slope
Monotonic constraints in trees clip out the bumps when fitting the addition of a sine wave and a constantly increasing slope
-
Controling classifier threshold
Given a fitted classifier, the FinedThresholdClassifier and TunedThresholdClassifier can adjust the threshold to maximize a given utility, either set theoretically with a cost matrix, or empirically to minimize the cost on a validation set.
-
FrozenEstimator
An already-fitted estimator can be given to FrozenEstimator so to have an object that is no longer modified at fit time. This is useful to inject in pipelines pre-trained models as it enables reusing standard model evaluation tools.
-
Categorical support
If an input is given as a dataframe (pandas or polars) with some columns typed as categories, HistGradientBoosting classifier and regressor will use a categorical splitter in the trees for these.
-
Polars output
using the set_output method of an estimator, transformers can output a polars dataframe, respecting the column names of the input if any.
-
Missing value support
Random Forest and Extra Trees now support missing-values natively, fitting them by special-casing them in the construction of the tree (a strategy sometimes called “Missing Incorporated Attribute").
-
Monotonic constraints in tree models
Different tree-based models (HistGradientBoosting regressor and classifier, random forests, extra trees) can now take constraints to force the prediction function to be monotonous along given features (fig:monotonictres).
skrub
The first release of skrub was late 2023. There have been 3 releases in 2024, leading to 0.4 in December 2024. Skrub is a package to facilitate machine learning on tables. The major features added in 2024 are:
-
TableReport
The TableReport gives an interactive display of dataframes, enabling inspection of the different columns and their distributions, that can be easily embedded, including in the programming environment used by data scientists.
-
TextEncoder
The TextEncoder uses a pretrained deep-learning language model to embed strings in a given column.
-
tabular_learner
The tabular_learner function builds a preprocessing pipeline that encodes messy data frames in a way that is well suited for a given a predictor.
joblib
joblib is a very simple computation engine in Python that is massively used worldwide, including as a dependency of packages such as scikit-learn for parallel computing.
Release 1.4 (May 2024). Many changes to follow evolutions of the ecosystem and improve behaviors (eg better error handling). Major changes are:
- Allowing to cache coroutines
- Optional unordered execution of parallel loop, to better use multiple CPUs
9 Bilateral contracts and grants with industry
Participants: Judith Abecassis, Gael Varoquaux, Jill-Jênn Vie.
9.1 Bilateral contracts with industry
Probabl
Probabl is an Inria spin-off in which Gaël Varoquaux has 10% of his time allocated. Probabl's mission is to develop and make sustainable an ecosystem of data-science commons. Probabl is the larger employer of scikit-learn maintainers. It builds a commercial offer around the scikit-learn ecosystem for the entreprise. Gaël Varoquaux is the point of contact at Soda.
Pass Culture
Within the Ministry of Culture-Inria convention, Samuel Girard and Jill-Jênn Vie have been involved in a partnership with Pass Culture (used by 3M students in France) to improve the diversity of their recommendations (12 months, started in June 2024). We hired an engineer, Hiba Bederina, from June 2024.
Collaboration with Ministère de la Santé
We have a 2-year long collaboration with Ministère de la Santé (HAS) on using the national healthcare data for prevention and policy evaluation. Gaël Varoquaux and Judith Abecassis are in charge at Soda.
9.2 Bilateral Grants with Industry
Collaboration with public interest group Pix
Jill-Jênn Vie got a Paris Region PhD 2023 funding with Pix (certification of digital competencies, 6M active users), about optimizing human learning using reinforcement learning. Samuel Girard 's PhD is currently on this funding (105k from région Île-de-France, 20k from Pix).
Plan de relance with Dataiku
Soda had a 24 months post-doc funded by “plan de relance” jointly with Dataiku on using embeddings for database analytics. The post-doc started beginning of November 2022 and ended in October 2024. Gaël Varoquaux is in charge at Soda.
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Inria associate team not involved in an IIL or an international program
RED
-
Title:
Recommendations Encouraging Diversity
-
Duration:
2024 -> 2026
-
Coordinator:
Koh Takeuchi (takeuchi@i.kyoto-u.ac.jp)
-
Partners:
- Kyoto University (Japan)
-
Inria contact:
Jill Jenn Vie
-
Summary:
We want to create recommender systems that optimize for cultural diversity. Finding items that not only optimize click-through rate, or profit, but also encourage users to discover new things. The goal of this project is first, to borrow methods from causal inference to measure the treatment effect of recommendations (defined as the diversity after and before recommendation), and methods from reinforcement learning to optimize this treatment effect. One key element to achieve this project is that plenty of real data is available thanks to our current partnership with Pass Culture, an app used by the French government to provide a budget ranging from 20 to 300 euros for every 15 to 18 years old in order to purchase culture goods. These works will be done between Soda team and Kyoto University.
10.2 European initiatives
10.2.1 Horizon Europe
INTERCEPT-T2D
INTERCEPT-T2D project on cordis.europa.eu
-
Title:
Early Interception of Inflammatory-mediated Type 2 Diabetes
-
Duration:
From January 1, 2023 to December 31, 2027
-
Partners:
- INSTITUT NATIONAL DE RECHERCHE EN INFORMATIQUE ET AUTOMATIQUE (INRIA), France
- UNIVERSITA DEGLI STUDI DI VERONA (UNIVR), Italy
- INSTITUT NATIONAL DE LA SANTE ET DE LA RECHERCHE MEDICALE (INSERM), France
- UNIVERSITAT BASEL, Switzerland
- ASSISTANCE PUBLIQUE HOPITAUX DE PARIS, France
- DEUTSCHE DIABETES FORSCHUNGSGESELLSCHAFT EV (DDFG), Germany
- FEDERATION FRANCAISE DES DIABETIQUES, France
- INSERM TRANSFERT SA, France
- Olatec Therapeutics, BV (Olatec Therapeutics, BV), Netherlands
- CENTRE HOSPITALIER UNIVERSITAIRE DE LIEGE (CHUL), Belgium
- KAROLINSKA INSTITUTET (KI), Sweden
- UNIVERSITATSSPITAL BASEL (KANTONSSPITAL BASEL), Switzerland
- TECHNISCHE UNIVERSITAET DRESDEN (TUD), Germany
-
Inria contact:
Gael Varoquaux
-
Summary:
The overall concept of INTERCEPT-T2D is to establish whether an inflammatory-mediated profile contributes to the onset of Type 2 Diabetes (T2D) complications, thus enabling the identification of patients most at risk of complications and the design of personalized prevention measures.
T2D is a heterogeneous disease, which is an obstacle to the delivery of an optimal tailored treatment. Consequently, patients’ individual trajectories of progressive hyperglycemia and risk of chronic complications are so far difficult to predict. In this context, onset of diabetic complications represents the most important transitional phase of T2D development toward premature disability and mortality.
Chronic systemic inflammation has been suggested to be a major contributor to the onset and progression of T2D complications. INTERCEPT-T2D will bring a new and clinically relevant dimension in T2D care considering at diagnosis inflammatory parameters that are of importance for the transition to T2D-related complications. The combination of state-of-the-art genomics and cell-biology technologies with targeted clinical interventions should lead to potent patients’ stratification. It should allow the identification and prognosis of a novel class or subclass of patients characterized by an “Inflammatory-mediated T2D” endotype.
The project has access to the best-documented longitudinal human European cohorts of patients with T2D, with reliable clinical and biological data allowing to trace the transition and evolution towards organ complications. This, added to the exploitation of an extensive health data warehouse, will enable us to establish the inflammatory trajectory of citizens with T2D from diagnosis to the development of complications.
To explore the ability to prevent the transition phase of T2D towards organ complications, INTERCEPT-T2D will conduct a phase II clinical trial with an anti-inflammatory therapy targeting NLRP3 Inflammasome activity in patients with T2D.
RECeSS
RECeSS project on cordis.europa.eu
-
Title:
Robust Explainable Controllable Standard for drug Screening
-
Duration:
From May 1, 2023 to January 31, 2025
-
Partners:
- INSTITUT NATIONAL DE RECHERCHE EN INFORMATIQUE ET AUTOMATIQUE (INRIA), France
- UNIVERSITAET ROSTOCK (UROS), Germany
-
Inria contact:
Jill-Jênn Vie
-
Summary:
In 2021, drug development pipelines last 10 years in average, and cost around $2 billion, while facing high failure rates, as only around 10% of Phase 0 drug candidates reach the commercialization stage. These issues can be mitigated through drug repurposing, where existent compounds are systematically screened for new therapeutic indications. Collaborative filtering is a semi-supervised learning framework that leverages known drug-disease matchings to make novel recommendations. However, prior works cannot be leveraged because of their lack of focus on human oversight and robustness to biological data.
This project aims at bridging the gap between drug research and collaborative filtering by implementing a RECeSS classifier, that is
(1) Robust: deals with class imbalance in drug-disease matchings, and missing drug/disease features, by semi-supervised learning;
(2) Explainable: connects predicted matchings to perturbed biological pathways through enrichment analyses, based on the learnt importance of features in the model;
(3) Controllable: guarantees a bound on the false positive rate using an adaptive learning scheme;
(4) Standard: algorithms are trained and tested by a standardized open-source pipeline.
Predicted matchings will be independently validated by structure-based methods. This innovative interdisciplinary project relies on a solid basis of newly curated data (up to 1,386 drugs, 1,599 diseases, 12 feature types).
In the short term, this would yield the first method that fully integrates biological interpretation and risk assessment to collaborative filtering-based repurposing. Long-term outcomes might help define sustainable and transparent drug development for rare diseases.
10.3 National initiatives
PEPR Santé Numérique
Soda is part of the “PEPR Santé Numérique” in the SMATCH subgroup that focuses on evidence of clinical efficacy. Soda will address two questions. The first question, addressed in collaboration with the PreMedical team, is that of external validity of randomized trials: how much is the treatment effect measured in a randomized clinical trial affected by the sampling bias of the trial, the difference between the study population and the intended target population. The second question, addressed in collaboration with the Heka team, is that of defining guidelines to evaluate software as a medical device. One particular challenge that we will tackle is to give procedures and recommendations to evaluate an update to a software used in clinical decision making using historical data rather than a trial. The project started end of 2023. Gaël Varoquaux is in charge at Soda, and Judith Abecassis is also supervising.
Project Partages
“Partages” is a large project funded by BPI France to develop digital commons for medical text analysis. In particular, the project will create material suitable for fine-tuning or aligning language models to perform best on French medical texts. Beyond the medical terms, there are specific challenges of clinical texts: these often result from scanning notes that have been taken fast, full of context-specific abbreviations and typos. The role of Soda is to design data-augmentation routine that help making language models robust to these challenges. The project started end of 2024. Gaël Varoquaux is in charge at Soda, and Judith Abecassis is also supervising.
10.4 Public policy support
French National IA commission
Gaël Varoquaux was an expert at the French National commission that lasted from September 2023 to March 2024. The commission auditioned hundreds of experts and did bibliographic research to advise on all aspects of AI and society, having been tasked by the government to advise on public policy around AI. In March, the commission handed in a general report, with 150 pages of high-level analysis, as well as technical recommendations for the French administration.
International Scientific Report on the Safety of Advanced AI
Gaël Varoquaux is an expert for the UK AI Safety Institute and for the International Scientific Report on the Safety of Advanced AI 36. A panel of international experts from all countries are working together with a team of experts to consolidate analysis around AI safety. The report touches upon all aspects of risks of AI (privacy, fairness, cybersecurity, bio weapons, loss of control...) as well as the current scientific evidence on their evolutions and possible mitigations. It is meant to ground public policy across the world on scientific evidence. An interim report is available 36, but the final report is still in progress.
11 Dissemination
11.1 Promoting scientific activities
11.1.1 Scientific events: organisation
General chair, scientific chair
-
Judith Abécassis
Workshop MECOSA: Methodological and Computational Advances in Survival Analysis on November 26th. Joint organization with teams HeKA (Agathe Guilloux and Linus Bleistein) and Premedical (Julie Josse)
-
Jill-Jênn Vie
Workshop WASL 2024: Optimizing Human Learning, on March 19 in Kyoto, Japan. Co-located with the Learning Analytics & Knowledge conference. Joint organization with Yizhu Gao (U. Georgia), Samuel Girard (Inria), Hisashi Kashima (Kyoto U.), Fabrice Popineau (CentraleSupélec & LISN), Yong Zheng (Illinois Institute of Technology).
11.1.2 Scientific events: selection
Member of the conference program committees
-
Gaël Varoquaux
Area chair: ICML, NeurIPS, ICLR
Reviewer
-
Gaël Varoquaux
AIstats, IJCAI, NeurIPS workshop selection
-
Judith Abécassis
NeurIPS (Top Reviewer), ICLR
-
Marine Le Morvan
AIstats
-
Jill-Jênn Vie
AAAI workshop AI4ED, ICLR, EDM (senior PC), LAK
11.1.3 Journal
Member of the editorial boards
-
Gaël Varoquaux
Machine Learning Journal
Reviewer - reviewing activities
-
Gaël Varoquaux
Machine Learning Journal
-
Judith Abécassis
Plos One, BMC Bioinformatics
-
Jill-Jênn Vie
International Journal of Artificial Intelligence in Education
11.1.4 Invited talks
-
Gaël Varoquaux
Keynotes: TRL workshop NeurIPS, ODSC Europe, Epiclin, Dataiku Technical Kick Off, Morocco AI, MIDL, ICCR, P16 Kick Off
Other invited talks dotAI, Health Data Hub scientific days, SESSTIM seminar, ENS Lyon physics seminar, Mila biomedical group
-
Judith Abécassis
Young Statisticians and Probabilists (YSP) Days, Sacl-AI for Science Workshop, workshop "Causal inference in Genetics", les Treilles.
-
Marine Le Morvan
MIA Paris-Saclay seminar (INRAE), Statistics and Computer Science Day (IHES)
-
Jill-Jênn Vie
Conseil scientifique de l'Éducation nationale, AI in education workshop (Inria Bordeaux)
11.1.5 Scientific expertise
-
Gaël Varoquaux
Comité Scientifique ANR Techniques Spécifiques de l'IA, Comité Scientifique DATAE
-
Judith Abécassis
Scientific expert for the call "AGIR EN SANTÉ PUBLIQUE (AGIR-SP)" INCa
-
Jill-Jênn Vie
Organisation internationale de la francophonie, conseil scientifique MonProjetSup
11.1.6 Research administration
-
Gaël Varoquaux
Scientific president of the ClusterAI submission for Université Paris Saclay
11.2 Teaching - Supervision - Juries
Courses
-
Gaël Varoquaux
- AI on tabular data, Ellis Doctoral Symposium, 1.5h
- Preparing tabular data for machine learning, tutorial, AutoML conference, 1.5h
- Learning on messy tabular data, Hi-Paris Summer School, 1.5h
- Machine learning, Inria academy, executives from the French ministry of defense 1h
-
Marine Le Morvan
- Deep Learning, Ecole Polytechnique, 27h
- Refresher Course in Artificial Intelligence, Ecole Polytechnique, 15h
- Statistics and machine learning with missing values, Université Paris Dauphine, 6h
-
Jill-Jênn Vie
- INF471S ICPC–SWERC training (advanced algorithms), École polytechnique, 60h
- Préparation au SWERC, ENS Paris-Saclay, 21h eq. TD
-
Judith Abécassis
- Causal Inference DS-UA 9201, NYU Paris, Spring 2024, 56h eq. TD
- Causal Inference DS-UA 9201 (with Houssam Zenati, MIND team), NYU Paris, Fall 2024, 48h eq. TD
E-learning
-
Machine learning with Scikit-learn MOOC
40 hours of learning starting as an introduction to machine learning and covering more advanced topics such as data preparation and model selection. Accessible on inria.github.io/scikit-learn-mooc, and designed by Loïc Esteve , Arturo Amor , Guillaume Lemaître , Olivier Grisel , Gaël Varoquaux .
11.2.1 Supervision
-
Gaël Varoquaux
PhD advisor for Celestin Eve (50%), Sébastien Melo (60%), Meilame Tayebjee (25%) Julie Alberge (30%), Jovan Stojanovic (50%), Félix Lefebvre (100%), Alexandre Perez (50%), Léo Grinsztajn (50%)
-
Judith Abécassis
PhD advisor for Julie Alberge (70%)
-
Marine Le Morvan
PhD advisor for Alexandre Perez (50%), Sébastien Melo (40%)
-
Jill-Jênn Vie
PhD advisor for Jean Vassoyan (33%), Marie Generali (33%), Samuel Girard (33%)
11.2.2 Juries
-
Gaël Varoquaux
PhD committee for Hugo Thimonier (éxaminateur)
-
Marine Le Morvan
- PhD comittee for Gabriel Damay (éxaminateur)
- jury pour concours CRCN/ISFP Saclay
- comité de sélection poste professeur assistant en apprentissage statistique à Polytechnique
11.3 Popularization
11.3.1 Productions (articles, videos, podcasts, serious games, ...)
-
Gaël Varoquaux
Scientific chronicles in Les Échos
Podcasts IA, pas que de la data; Data driven 101; Dialogue Machine
-
Judith Abécassis
article dans le journal TELECOM "Quels usages pour l’intelligence artificielle en oncologie ? de la recherche au patient"
11.3.2 Participation in Live events
-
Gaël Varoquaux
- Panel on AI at BPI's BIG forum, one of the largest forum on investments in Tech in France
- Panel on AI at the Paris “Chambre de Commerce et de l'Industrie”
- Panel on AI and creative and cultural industries, French embassy Berlin
- Panel on sustainability and AI, AI pulse
-
Judith Abécassis
- Round table "IA et parcours de soins", espace PariSanté Campus, salon SantExpo
- conference "IA et Santé : Des modèles prédictifs aux modèles prescriptifs, interprétabilité et explicabilité", Startup accelerator for health prevention, BPI France, PariSante Campus (via Inria Academy)
- webinar "IA et Santé : Des modèles prédictifs aux modèles prescriptifs" Ramsay Santé, Inria Academy
-
Jill-Jênn Vie
- Panel at UNESCO: “AI4T: AI for and by teachers & Pix: assessing digital skills in a changing world”, Digital competencies for teachers and school students seminar
- Atelier “Intelligence artificielle et éducation”, Colloque In Fine (plan de formation à destination des personnels de l'éducation nationale), Futuroscope
- Coach of École polytechnique at International Competitive Programming Contest (ICPC): 2 Silver medals (3rd & 4th places) at Southwestern Regionals 2024 in January 2024, admitted to ICPC World Finals in Astana, Kazakhstan in September 2024; won Southwestern Regionals 2025 in December 2024 (1st, 3rd, and 21st places), Gold and Silver medal.
- Panel on AI and human learning, Numérique en communs, November 2024
11.3.3 Others science outreach relevant activities
-
Jill-Jênn Vie
Girls Can Code! 2-day girls-only autumn school for discovering programming in November 2024 in École polytechnique. 50 participants from middle school and high school. Organized with non-profit organization Prologin.
12 Scientific production
12.1 Major publications
- 1 inproceedingsReconfidencing LLM Uncertainty from the Grouping Loss Perspective.EMNLP 2024 - Conference on Empirical Methods in Natural Language ProcessingMiami, United StatesarXiv2024HALDOIback to textback to text
- 2 articleConcomitant medication, comorbidity and survival in patients with breast cancer.Nature Communications151April 2024, 2966HALDOIback to text
- 3 inproceedingsBetter by Default: Strong Pre-Tuned MLPs and Boosted Trees on Tabular Data.Neural Information Processing SystemsVancouver (BC), Canada2024HALDOIback to text
- 4 inproceedingsCARTE: Pretraining and Transfer for Tabular Learning.Proceedings of Machine Learning ResearchForty-first International Conference on Machine Learning, ICML 2024235Vienna, AustriaJuly 2024HALback to text
- 5 miscImputation for prediction: beware of diminishing returns.July 2024HALback to text
- 6 inproceedingsA Pre-Trained Graph-Based Model for Adaptive Sequencing of Educational Documents.NeurIPS 2024 Workshop FM-EduAssess - The First Workshop p, Large Foundation Models for Educational AssessmentVancouver, CanadaDecember 2024HALback to text
- 7 inproceedingsTowards Scalable Adaptive Learning with Graph Neural Networks and Reinforcement Learning.EDM 2023 - 16th International Conference on Educational Data MiningBangalore, IndiaMay 2023HAL
12.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Scientific book chapters
Reports & preprints

