Section: New Results
Algorithms for molecular docking
Prediction of Interface Water Molecules Using a Knowledge Base
Participants : Georgiy Derevyanko, Sergei Grudinin.
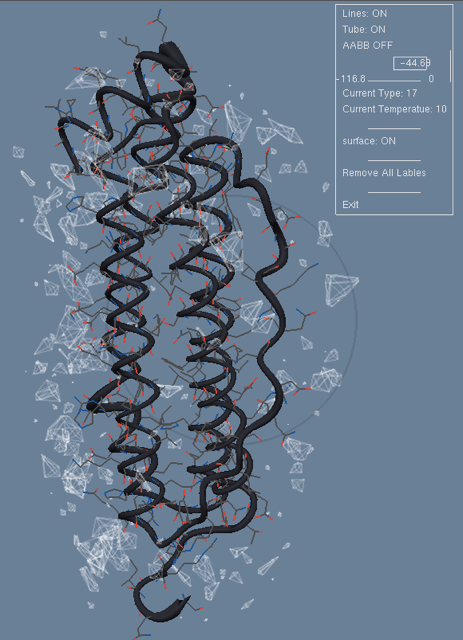
We developed a method to predict positions for interface water molecules as part of the predicted protein-protein complex. For this purpose we used a previously developed knowledge-base scoring methodology. First, we constructed a training set of non-homologous protein complexes with interfacial water. Then, we deduced the water-protein interaction energy using this training set. And finally, we positioned water molecules around a test protein complex on a regular grid and optimized their positions according to the knowledge-based water-protein interaction energy. This method was validated in a recent CAPRI competition. Figure 12 illustrates our method on a test protein.
|
Development of a Knowledge-Based Scoring Function
Participants : Georgiy Derevyanko, Sergei Grudinin.
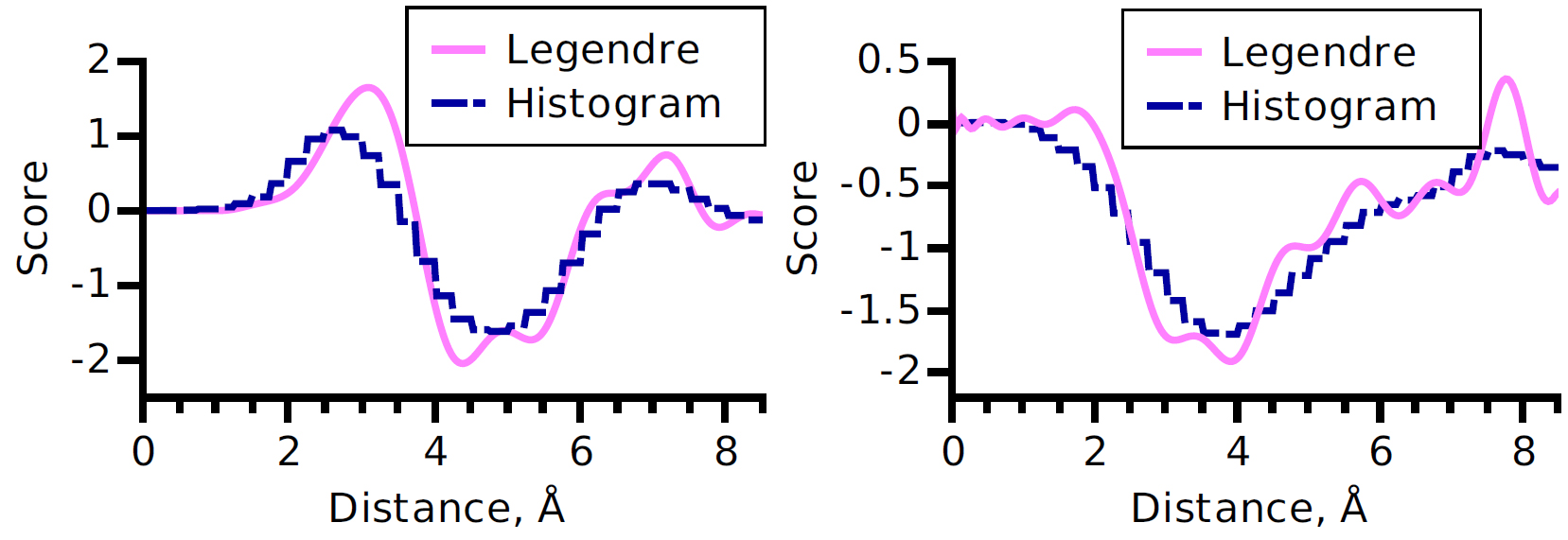
We developed a new method to obtain a knowledge-based potential function for protein-protein interactions. To derive such a potential, we formulated a convex quadratic programming problem with about 1,000,000 of linear constraints and developed a fast iterative solver to solve it. We validated this scoring function in the CAPRI competition Round 24, where our prediction of the Target 50 was ranked 4th.
Figure 13 shows the use of Legendre polynomials to fit statistics obtained on the knowledge base.
Development of a Local Knowledge-Based Potential for Structure Optimization and Prediction of Point Mutations in a Protein
Participants : Petr Popov, Sergei Grudinin.
We developed and validated a method that reconstructs the shape of the binding potential function between two proteins by locations of its global minima. After, we used the obtained potential function for optimization of positions of two docking partners. We demonstrated that using our method we can significantly improve the quality of predictions of such widely-used docking algorithms as HEX and ZDOCK. We validated this method in the CAPRI competition Round 26, where our re-scoring prediction of the Target 53 was ranked 3rd.
We have also developed a method to predict the influence of point mutations on the binding affinity constant of a protein complex. First, we made point mutations and reconstructed the sidechain of the mutated residue. Then, we repacked the sidechains that are within a certain cutoff distance from the mutated residue. After, we optimized the structure of two proteins using a smooth pair-additive knowledge-based potential function. We iteratively repeated the two previous steps until convergence of the binding energy. Finally, we converted the obtained binding energy into the binding affinity constant of two proteins. We validated this method in the CAPRI competition Round 26 with the Targets 55 and 56.
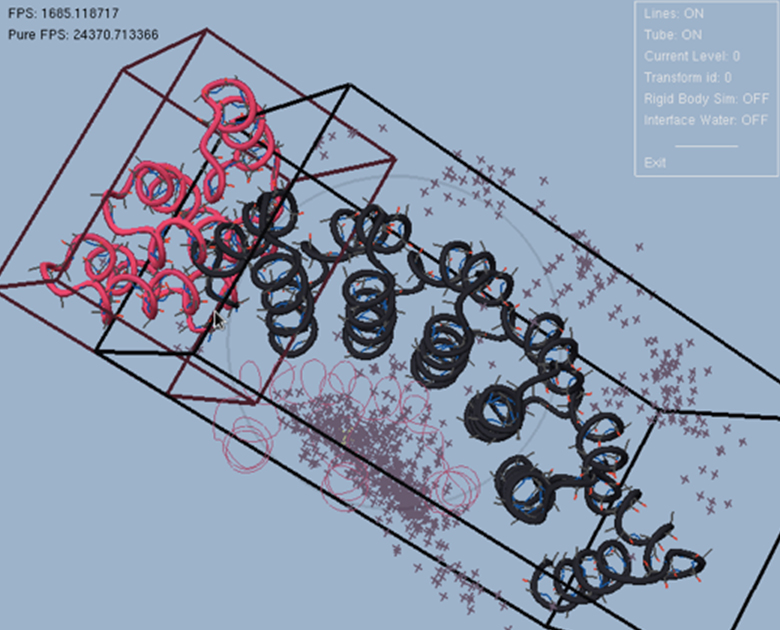
Figure 14 shows an interactive docking session using a knowledge-based potential for CAPRI Round 26 Target 53.