Section: New Results
Software Engineering
SAMSON User interface
Participants : Jocelyn Gate, Maria Werewka, Stephane Redon.
We have continued the development of SAMSON, our open-architecture platform for modeling and simulation of nanosystems (SAMSON: Software for Adaptive Modeling and Simulation Of Nanosystems):
-
We have moved to Qt5 to handle the Graphical User Interface.
-
We now compile SAMSON is 64 bits only. This removes limitations of 32 bits applications, in particular concerning memory limits.
-
We now have a complete installer mechanism for both users and developers.
-
We extended the set of development tools (action generators, UUID generators, etc.)
-
We have changed the windowing system to allow windows to move outside SAMSON.
-
We have designed a coherent style for icons, windows, menus, etc.
There are now more than 40 modules in SAMSON (parsers, editors, models, apps, etc.).
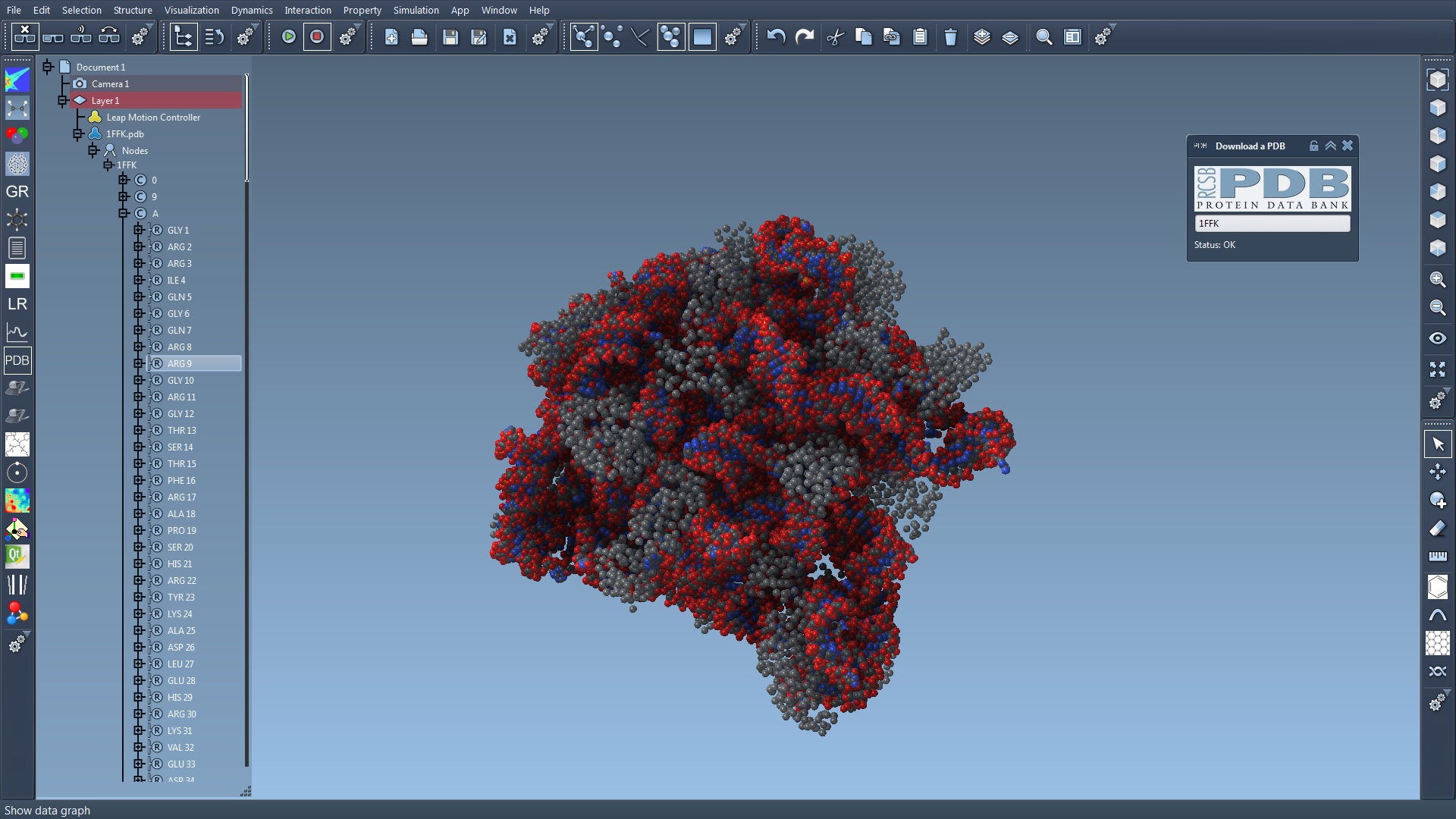
The current user interface of SAMSON is visible in Figure 11 .
|
SAMSON Elements
Participants : Svetlana Artemova, Stephane Redon.
We have added new SAMSON Elements (modules).
We have been working on input and output for SAMSON. Precisely, we now have the possibility to download molecules to SAMSON and save them to external files in three possible formats:
-
pdb (Protein Data Bank format, containing experimentally determined 3d structures and widely used for applications in biology);
-
mol2 (Sybyl chemical modeler input file, containing chemical compounds and small ligands);
Basic properties of atoms, residues, and molecules have been determined and structures storing these properties were implemented.
Finally, since energy minimization is crucial for providing physically-correct structures while interactively editing molecules in SAMSON, we have implemented several fast and stable algorithms to perform such energy minimization in SAMSON.
SAMSON Website
Participants : Mohamed Yengui, Jocelyn Gate, Stephane Redon.
We are developing a web application aiming at distributing and valorizing SAMSON and SAMSON Elements (modules). The goal of the website is to develop a community of users and developers in all areas of nanoscience (physics, biology, chemistry, electronics, etc.). The website will:
-
allow users and developers to create and manage accounts on the website.
-
allow visualizing, searching and downloading SAMSON and SAMSON Elements.
-
allow the creation, validation and dissemination of SAMSON Elements.
-
provide tracking requests for the arrival of new SAMSON Elements or the modification of an existing SAMSON Element.
To achieve this, we have designed the architecture in a way that speeds development effort for a faster product release, while keeping in mind scalability, security and high reliability.
We have also implemented and tested locally the account validation process. A user can now sign up, confirm the registration from the received email and authenticate with the registered account to download SAMSON and SAMSON elements from the website. We will make the site public when we release SAMSON.