Section: New Results
Software development of SAMSON
Development of SAMSON Connect
Participants : Mohamed Yengui, Jocelyn Gate, Stephane Redon.
We have continued the development of SAMSON Connect, the web site that will contribute to the diffusion and promotion of SAMSON and SAMSON Elements (modules for SAMSON).
SAMSON Elements are adapted to different application domain and help users build new models, perform calculations, run interactive or offline simulations, visualize and interpret results, etc. The goal of SAMSON Connect is to bring together a set of users and developers of SAMSON Elements in all areas of nanoscience (physics, biology, chemistry, electronics, etc...). It offers a set of features available depending on the user role:
-
Developers (who have obtained the SAMSON-SDK) can develop SAMSON Elements and upload them to SAMSON Connect through the tools provided.
-
Users (who have obtained the SAMSON Core application) can add SAMSON Elements to their instance of SAMSON Core in one click. The download process is performed during startup of SAMSON and without outside intervention.
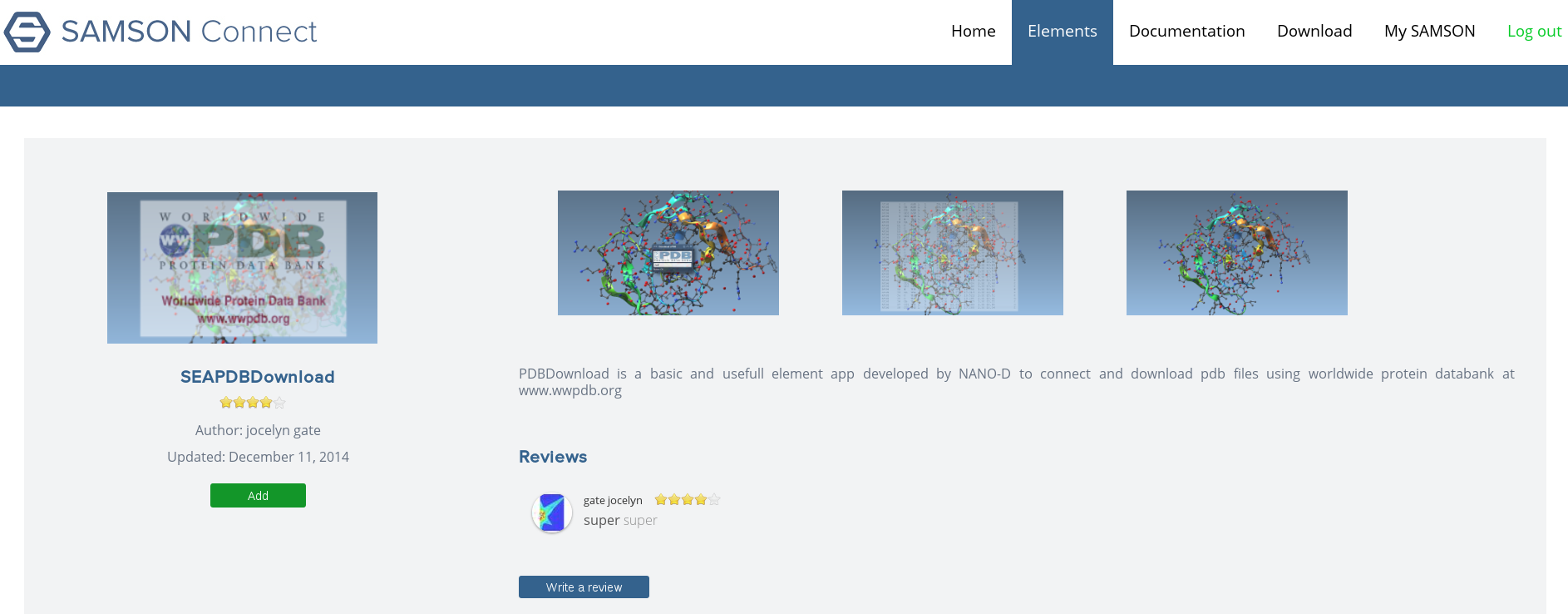
All users can give feedbacks, review and rate SAMSON Elements after adding them to their SAMSON Core (Figure 7 ).
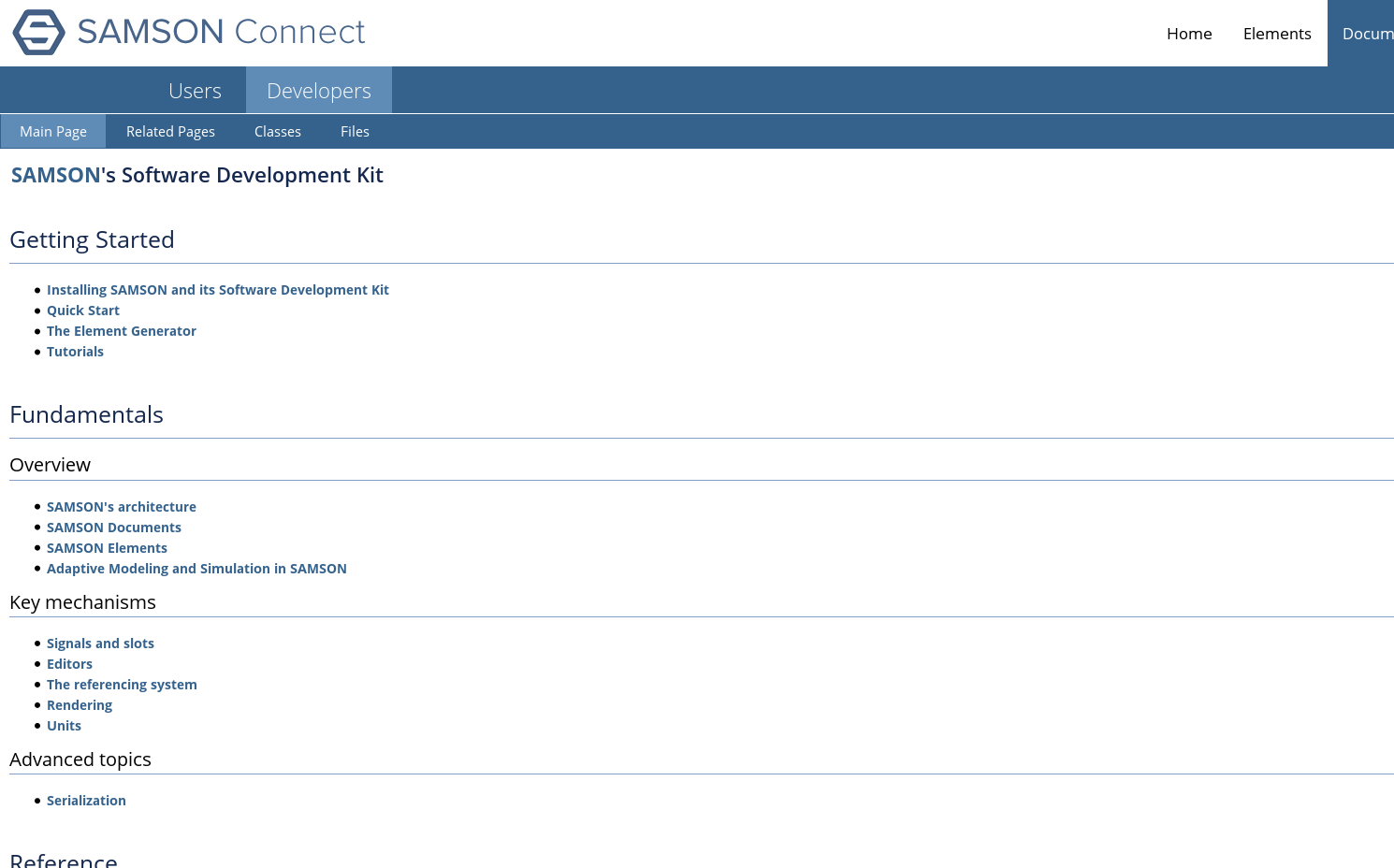
SAMSON Connect also features some documentation to develop new elements for SAMSON (Figure 8 ).
SAMSON Connect will be available at http://samson-connect.net .
Deployment of SAMSON and the SAMSON SDK
Participants : Jocelyn Gate, Mohamed Yengui, Stephane Redon.
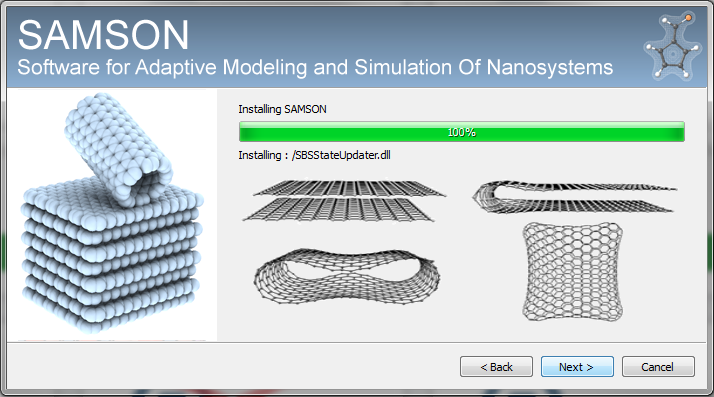
The SAMSON installer has been split in two parts: SAMSON-setup (installation of the SAMSON application, Figure 9 ) and SAMSON-Developer-setup (installation of the SAMSON SDK). internet. It is very useful to increase security.
Several helper tools related to SAMSON Elements management were developed to facilitate Element deployment. For example, the element packager is a tool useful for developers who want to distribute a new SAMSON Element on the SAMSON Connect platform. With this packager we can control many things: check whether the file is valid, if the SAMSON Element is readable with SAMSON, add a description file that contains useful information (name, author ID, checksum, element version, SDK version, operating system, etc.).
We added a service requester to SAMSON to communicate with SAMSON Connect and
Universal Force Field
Participants : Svetlana Artemova, Leonard Jaillet, Stephane Redon.
We have implemented a version of the Universal Force Field (UFF) [19] in SAMSON, as a SAMSON Element embedding an interaction model. UFF is a classical force field, which can take as input almost every atom of the periodic table. Such flexibility allows to potentially use UFF on a large spectrum of systems and since its introduction, it has been applied to simulate problems involving main group compounds, organic molecules, metal complexes and has even been recently extended to MOF (Metal Organic Framework) [11] . The general energy expression for UFF as described in [19] is:
where stands for bond stretching, describes angle bending, is dihedral angle torsion term, represents inversion, stands for van der Waals interactions and represents electrostatics (this last term is rarely considered for UFF, we do not study it neither). Forces involved in the atoms interactions can then be derived from the previous expression. Each energetic term in UFF can be computed based on simple rules deduced from a set of parameters. This set is based on the atoms' elements, their hybridization, and the overall connectivity of the molecular system.
In our implementation, we took into account several corrections and refinements that have been lately proposed in the literature for Universal Force Field. Our contribution also concerns the development of algorithms to automatically perceive the system's topology (covalent bonds and bond orders assignments). Moreover, we have introduced a method to automatically find the correct typization of the atoms. Precisely, atoms' hybridizations and oxidation states are computed, and resonance groups within or out of cycles are detected and treated. The implementation provided is computationally efficient enough to allow interactive simulation in SAMSON. The validity of the force field was tested on several groups of molecules proposed as benchmarks in the literature.
Integration of existing tools
Participants : Nadhir Abdellatif, Svetlana Artemova, Stephane Redon.
We have obtained funding from the Nanosciences Foundation in Grenoble to integrate in SAMSON some tools developed and used by the Grenoble community, in the form of SAMSON Elements, i.e. modules that integrate into SAMSON and may interact with SAMSON's main data graph. In particular, we have been meeting with some biologists and physicists to determine which tools and methods used (or developed) in Grenoble would be most appropriate for integration.
We integrated our first Element which is Babel, a chemical toolbox designed to “speak the many languages of chemical data”, i.e. read, write and convert data files (over 110 chemical file formats) from molecular modeling, chemistry, solid-state materials, biochemistry, or related areas (see http://openbabel.org ). The corresponding SAMSON element is an app that delegates all calculations to the Babel external executable. The app also makes it possible to import the data files to SAMSON to visualize the molecular data and proceed with other SAMSON elements.
We have also integrated Clustal, a tool for multiple sequence alignment. Thanks to Clustal's license, all source code is wrapped into the SAMSON Element (whose source code will be made available as well), and SAMSON users do not need to install Clustal separately.
Various
Participants : Stephane Redon, Svetlana Artemova, Marc Aubert.
-
Units management was added to SAMSON. The mechanism relies on C++ template meta-programming techniques to perform dimensional analysis and automatic conversions at compile time, and has no runtime overhead. This was a significant undertaking, but one that will be very helpful to integrate in SAMSON different domains of nanoscience that have come to use different units for identical dimensions (e.g. kilocalories per mole in biology, electron volts in chemistry, etc.).
-
SAMSON's reflection mechanism was improved to perform type registration and casting, and facilitate scripting and pipelining of SAMSON Elements.
-
SAMSON has its own file format, which allows it to save the data graph information.
-
More data graph nodes are now visible in SAMSON's data graph view.
-
The split between classical and quantum interaction models was abandoned, for simplicity.
-
Selection methods have been improved, and selection is now undoable. Selections may be saved, retrieved, have boolean operations performed onto them, etc.
-
Controllers, a new type of data graph nodes, were added to SAMSON. Controllers are used to act on other data graph nodes (e.g. translate and rotate models).
-
Existing parsers for input and output of molecular information in SAMSON have been improved and accelerated, and property windows for these parsers have been added.
-
The Lennard-Jones potential has been added as an interaction model to SAMSON.
-
A new editor for adding atoms corresponding to a chemical formula (in disorder) has been created.
-
The work on a new editor containing functional groups and frequently-used molecular patterns has been started.
-
Periodic Boundary Conditions (an important concept in molecular simulations) were implemented in SAMSON.
-
We decided to use the Qt5 framework for shaders management, for some maintenance reasons especially. This structure implied some other type changes to adapt to Qt5, such as the vertex buffers.
-
We changed the way viewports display text. It is now possible to run SAMSON on every platform (Windows, Linux and Mac) and display text, and it provides Elements programmers a simple way to add text where they want in the 3D view.