Section: New Results
As-rigid-as-possible shape interpolation for molecular modeling
Participants : Minh Khoa Nguyen, Leonard Jaillet, Stephane Redon.
Computer-aided methods play an important role in the study of molecular structures and interactions. Inspired by the as-rigid-as-possible approaches in the field of computer graphics, we created a tool for studying large deformation of molecular structures. This tool generates interpolated structures between two known conformations of a molecule while satisfying physical constraints. The users may use it for exploring, preprocessing, or combining their model with other biological algorithms. The developed method is flexible and can be extended to include physical properties of molecular structures.
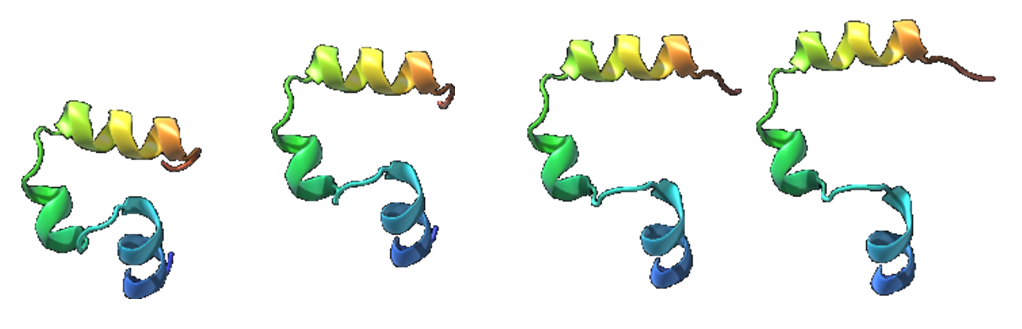
We tested our method on a graphene sheet folding into a nanotube (Figure 8 ) and a few biological molecules, one of which is shown in Figure 9 . The results show realistic transition motions compared to those from the linear interpolation approach.
The ARAP interpolation method has two main advantages: simplicity and preservation of local rigidity. The method is totally geometrical, yet can be extended to include physical or biological properties such as bond strength. It will be proposed as a SAMSON Element for the SAMSON software platform for computational nanoscience.