Section: New Results
Modelling axon growth from in vivo data
Participants : Agustina Razetti, Xavier Descombes, Caroline Medioni, Florence Besse.
Axons develop embedded in mechanically constrained environments. Thus, to fully understand this dynamical process, one must take into account collective mechanisms and mechanical interactions within the axonal populations. However, techniques to directly measure this from living brains are today lacking or heavy to implement. This interdisciplinary work intends to close the gap between classic in vitro experimental assumptions and real in vivo situations, where the final neuronal morphology is acquired through a dynamical and environmental-dependent process. We use as biological model Drosophila axon remodeling and analyze, for the first time to our knowledge, the mechanical situation of a whole population of neurons (650 individuals) growing together in a constraint space (i.e. medial lobe of the Mushroom Body).
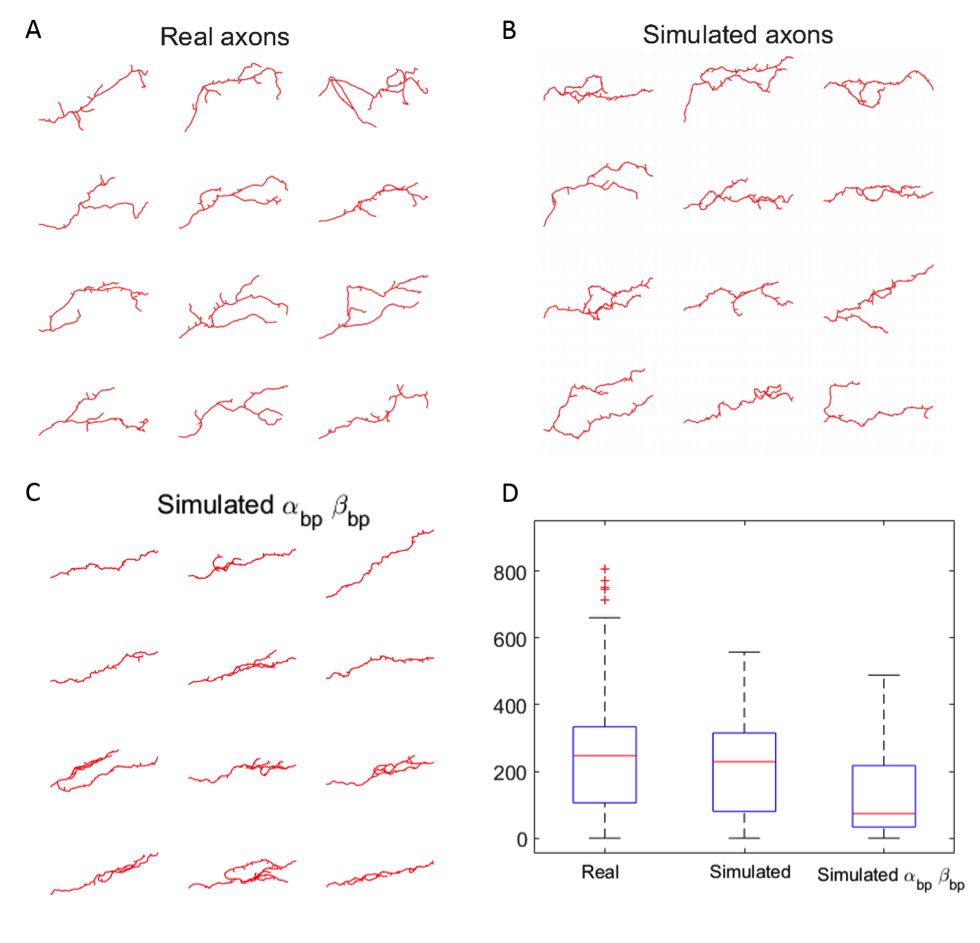
We have designed a mathematical model of single axon growth based on Gaussian Markov Chains with two parameters, accounting for axon rigidity and attraction to the target field. We used this model to simulate the growing axons embedded in space constraint populations to test our hypothesis. We explored new branch formation mechanisms to mimic the growth of wild type axons population , as well as predict different mutant phenotypes. This approach allowed also to analyze dynamical aspects of the neuron collective growth process such as speed and density in function of space and time, which help to explain several characteristics of the neuron morphology and behavior during development. Among the obtained results, the proposed model is able to reproduce the intra-population morphological variability. Interestingly, applying the ESA distance between trees previously developed in the team [22] showed that real axons present shapes that showcase a compromise between collective elongation and morphological variability, essential for axonal connectivity (Figure 15). Finally, we explored other branch occurrence strategies –from uniformly random to occurrence upon mechanical interactions- to contrast and validate with previously developed hypothesis on the importance of branching for axonal elongation in vivo.
|