Section: New Results
Incremental methods for long range interactions
Participants : Semeho Edorh, Stephane Redon.
Adaptively Restrained Molecular Dynamics (ARMD) were recently proposed with the purpose of speeding up molecular simulations. The main idea is to modify the Hamiltonian such that the kinetic energy is set to zero for low velocities, which allows to save computational time since particles do not move and forces need not be updated.
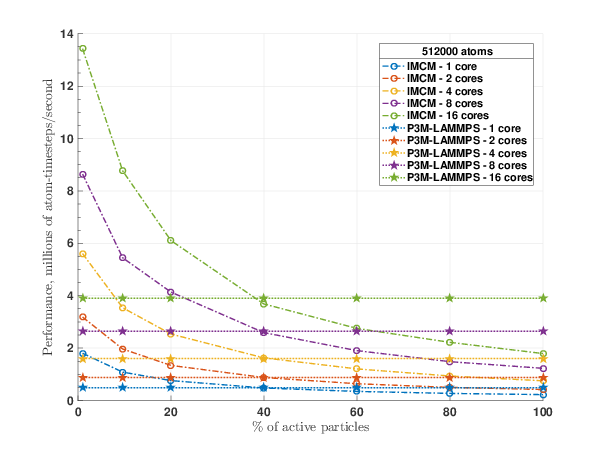
We continued our work on developing an extension of ARMD to electrostatic simulations. Therefore, we developed a fast method dedicated to the computation of the electrostatic potential in adaptively restrained systems. The proposed algorithm is derived from a multigrid-based alternative to the popular particle mesh methods. Our algorithm ,labeled as Incremental Mesh Continuum Method (IMCM),was implemented inside LAMMPS, a popular molecular dynamics simulation package. During ARMD simulations, IMCM scales with the number of active particles.
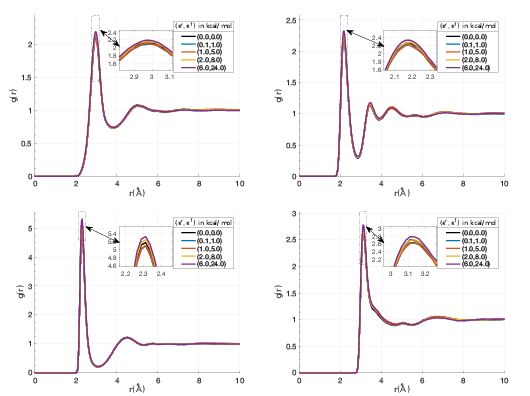
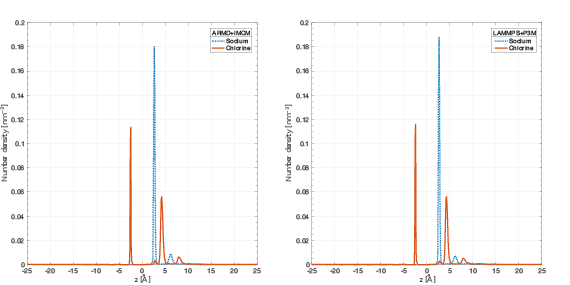
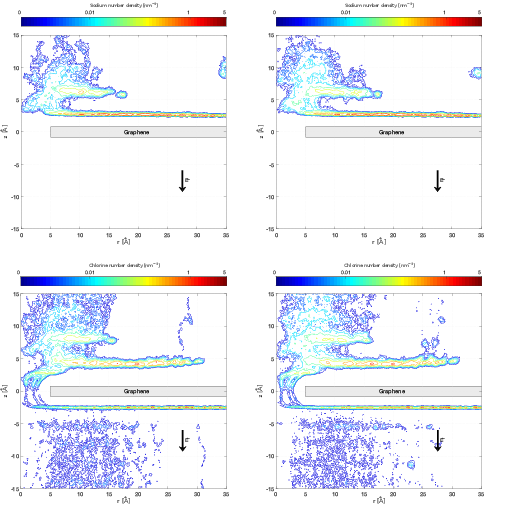
The performance of the new algorithm was accessed on various molecular systems. 1 showed that IMCM is able to outperform the well-established Particle Particle Particle Mesh (P3M) for adaptively restrained simulations. For an aqueous solution of sodium chloride, water molecules can be adaptively restrained. On this system, ARMD was able to reproduce static properties of sodium chloride (2). When a functionalized nanopore is placed at the center of the system, ARMD and IMCM were able to reproduce the ion selectivity property (3). For this benchmark, this positively charged nanopore acts like a sieve that blocks the flux of Sodium atoms, while promoting the crossing of Chlorine particles(4).
|
|