Section: New Results
Exploring Chemical Reaction Paths
Participant : Jaillet Leonard.
Context
In the past, we have developed a methodology to explore chemical reaction paths based on stochastic trees. One difficulty was the assessment of the quality of the paths found, and the comparison with existing state of the art methods.
To address these limitations, we have developed several new modules in SAMSON that propose state of the art methods and helpful tools to find and manipulate the paths and the important states of the considered systems. One can classify these modules into three categories: interpolation methods, minima and saddle point search methods and supporting tools. More details regarding these modules are provided below.
Interpolation methods
We have implemented the artificial force induced reaction (AFIR) method [59], that helps to find a transition path from a given initial state made of two compounds A and B, towards a goal compound X. In the futur, we would like to combine AFIR with our exploration methods.
The Linear Synchronous Transit (LST) and the Quadratic Synchronous Transit (QST) methods [42] have also been integrated in SAMSON. These methods generate paths such that each atom-pair distance in an intermediate structure is the interpolated value between those in the initial and target structures. The QST variant differs of the LST one from that the interpolated path also passes through a third intermediate point. Moreover, we have implemented the MINIMAX method as in [50] that alternates phases of minimization and QST interpolation to search for a transition path. These three methods appear to show a better behavior than a simple linear interpolation approach. In the future, we are also planning to combine them with our exploration methods.
Search for minima and saddle points
We have developed a SAMSON module to describe the energy basins associated to the various conformations of a given set. For this, conformations are first minimized and then clustered. This tool is also convenient when analyzing conformations along a given path to search for the states the closest to the saddle points.
In collaboration with the post doc Clément Betone, we have implemented two modules. First, the Dimer method as proposed in [45]. This method allows to find from a given point the closest saddle points in high dimensional potential surfaces, using only first derivatives. Some works remains to be done to solve the sensibility problem related to the initial orientation of the dimmer. Second, the freezing string method as proposed in [24]. This method searches efficiently for a saddle point between two given end states through a combination of interpolatoin and optimization.
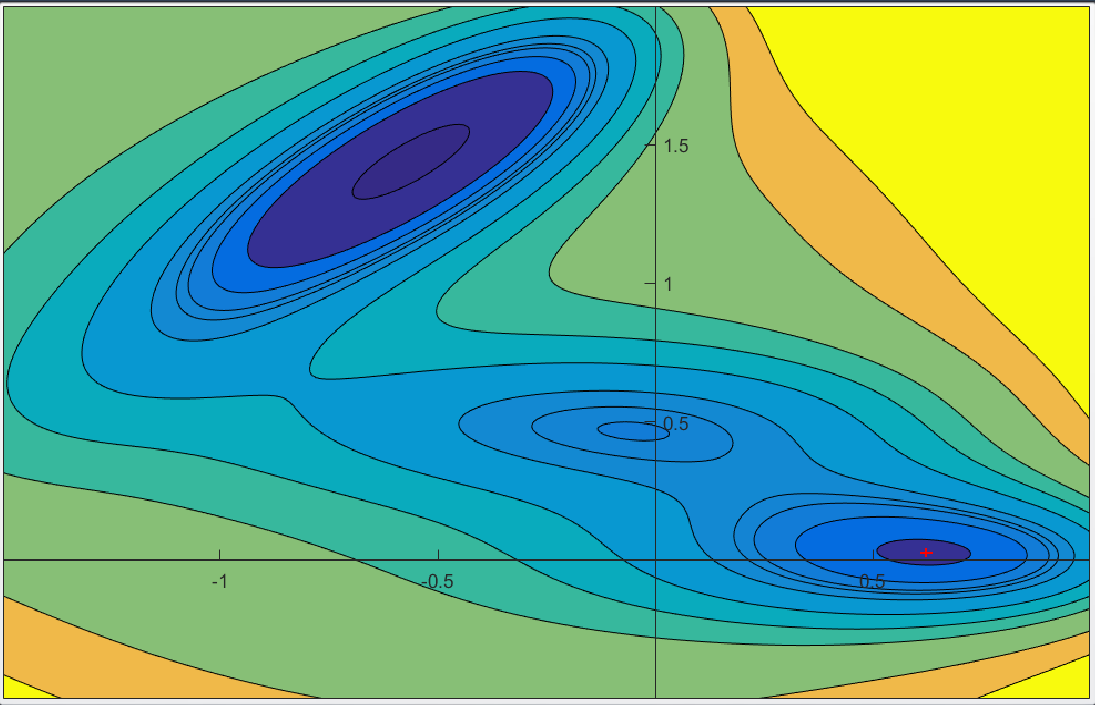
To assess the various methods and obtain a visual feedback of their behavior, we have developed several 2D force fields where the energetical landscape can easily be apprehended. Such an example of landscape is illustrated in Figure 8.
|
Supporting tools
We have proposed in SAMSON a set of tools to manipulate and perform some measures associated to set of conformations, that could potentially represent molecular paths.
A first tool was proposed to perform given measures for a given set of conformations, such as bond lengths, angle bends, or torsion angles. A second tool allows to align various conformations onto a reference on. Finally, another tool allows to compute the RMSD distance between pairs of conformations.