Section: Research Program
Research Program
The scientific foundations of our team concern the development of new processing algorithms in the field of medical image computing : image fusion (registration and visualization), image segmentation and analysis, management of image related information. Since this is a very large domain, which can be applied on numerous types of application; for seek of efficiency, the purpose of our methodological work primarily focuses on clinical aspects and for the most part on head and neck related diseases. In addition, we emphasize our research efforts on the neuroimaging domain. Concerning the scientific foundations, we have pushed our research efforts:
-
In the field of image fusion and image registration (rigid and deformable transformations) with a special emphasis on new challenging registration issues, especially when statistical approaches based on joint histogram cannot be used or when the registration stage has to cope with loss or appearance of material (like in surgery or in tumor imaging for instance).
-
In the field of image analysis and statistical modeling with a new focus on image feature and group analysis problems. A special attention was also to develop advanced frameworks for the construction of atlases and for automatic and supervised labeling of brain structures.
-
In the field of image segmentation and structure recognition, with a special emphasis on the difficult problems of i) image restoration for new imaging sequences (new Magnetic Resonance Imaging protocols, 3D ultrasound sequences...), and ii) structure segmentation and labelling based on shape, multimodal and statistical information.
-
Following past national projects where we had leading roles (e.g., Neurobase, NeuroLog, . . . ), we wanted to enhance the development of distributed and heterogeneous medical image processing systems.
|
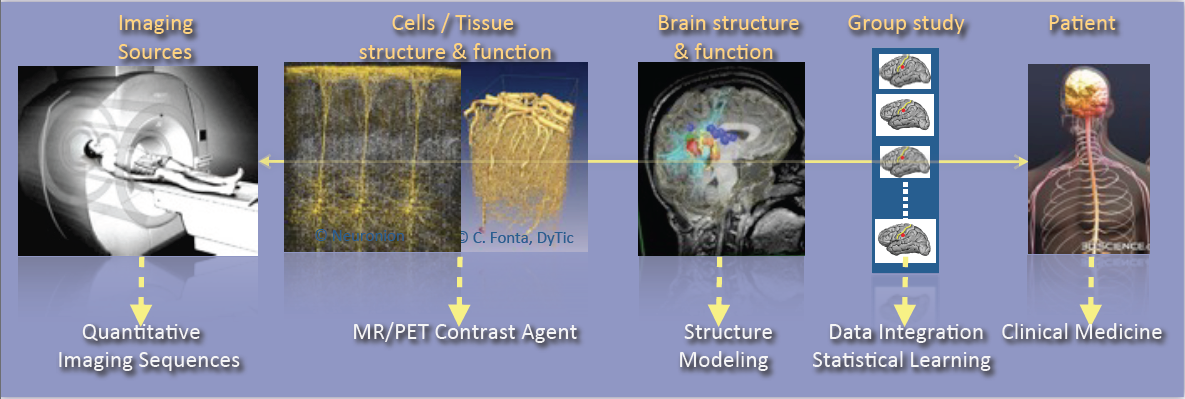
As shown in Fig. 1, research activities of the VisAGeS U1228 team are tightly coupling observations and models through integration of clinical and multi-scale data, phenotypes (cellular, molecular or structural patterns). We work on personalized models of central nervous system organs and pathologies, and intend to confront these models to clinical investigation studies for quantitative diagnosis, prevention of diseases, therapy planning and validation. These approaches are developed in a translational framework where the data integration process to build the models inherits from specific clinical studies, and where the models are assessed on prospective clinical trials for diagnosis and therapy planning. All of this research activity is conducted in tight links with the Neurinfo imaging platform environments and the engineering staff of the platform. In this context, some of our major challenges in this domain concern:
-
The elaboration of new descriptors to study the brain structure and function (e.g., variation of brain perfusion with and without contrast agent, evolution in shape and size of an anatomical structure in relation with normal, pathological or functional patterns, computation of asymmetries from shapes and volumes).
-
The integration of additional spatio-temporal imaging sequences covering a larger range of observation, from the molecular level to the organ through the cell (Arterial Spin Labeling, diffusion MRI, MR relaxometry, MR cell labeling imaging, PET molecular imaging, …). This includes the elaboration of new image descriptors coming from spatio-temporal quantitative or contrast-enhanced MRI.
-
The creation of computational models through data fusion of molecular, cellular, structural and functional image descriptors from group studies of normal and/or pathological subjects.
-
The evaluation of these models on acute pathologies especially for the study of degenerative, psychiatric or developmental brain diseases (e.g., Multiple Sclerosis, Epilepsy, Parkinson, Dementia, Strokes, Depression, Schizophrenia, …) in a translational framework.
In terms of methodological developments, we are particularly working on statistical methods for multidimensional image analysis, and feature selection and discovery, which include:
-
The development of specific shape and appearance models, construction of atlases better adapted to a patient or a group of patients in order to better characterize the pathology;
-
The development of advanced segmentation and modeling methods dealing with longitudinal and multidimensional data (vector or tensor fields), especially with the integration of new prior models to control the integration of multiscale data and aggregation of models;
-
The development of new models and probabilistic methods to create water diffusion maps from MRI;
-
The integration of machine learning procedures for classification and labeling of multidimensional features (from scalar to tensor fields and/or geometric features): pattern and rule inference and knowledge extraction are key techniques to help in the elaboration of knowledge in the complex domains we address;
-
The development of new dimensionality reduction techniques for problems with massive data, which includes dictionary learning for sparse model discovery. Efficient techniques have still to be developed to properly extract from a raw mass of images derived data that are easier to analyze.