Section: New Results
Computational Cardiology & Image-Based Cardiac Interventions
Population-based priors for group-wise Personalisation
Participants : Roch Molléro [Correspondant] , Hervé Delingette, Xavier Pennec, Nicholas Ayache, Maxime Sermesant.
The authors acknowledge the partial funding by the MD-Paedigree EU Project.
Personalised cardiac model, Parameter observability, Statistical modeling, Dimensionality reduction, Heterogeneous clinical data, Imputation
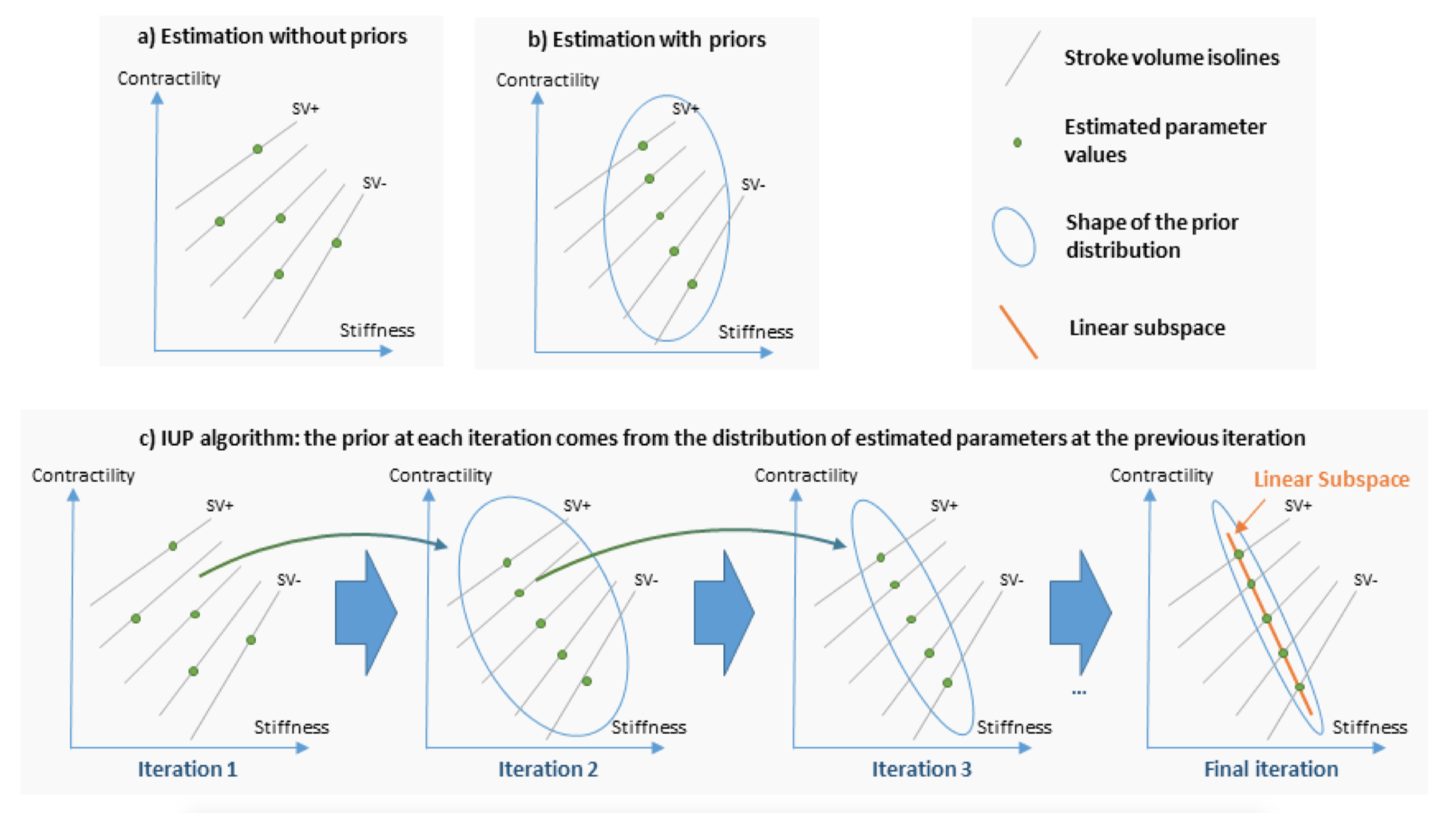
Personalised cardiac models have a large number of parameters while the available data for a given patient is typically limited to a small set of measurements, thus the parameters cannot be estimated uniquely. This is a practical obstacle for clinical applications, where accurate parameter values can be important. Here we explore an original approach based on an algorithm called Iteratively Updated Priors (IUP), in which we perform successive personalisations of a full database through Maximum A Posteriori (MAP) estimation, where the prior probability at an iteration is set from the distribution of personalised parameters in the database at the previous iteration (Figure 17). At the convergence of the algorithm, estimated parameters of the population lie on a linear subspace of reduced (and possibly sufficient) dimension in which for each case of the database, there is a (possibly unique) parameter value for which the simulation fits the measurements. We first show how this property can help the modeler select a relevant parameter subspace for personalisation. In addition, since the resulting priors in this subspace represent the population statistics in this subspace, they can be used to perform consistent parameter estimation for cases where measurements are possibly different or missing in the database, which we illustrate with the personalisation of a heterogeneous database of 811 cases [18].
|
Fast Personalized Computer Simulation of Electrical Activation from CT Imaging in Post-infarction Ventricular Tachycardia
Participants : Nicolas Cedilnik [Correspondant] , Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
This work is funded by the IHU Liryc, in Bordeaux.
Cardiac modeling, Personalised simulation, ablation, intervention guidance.
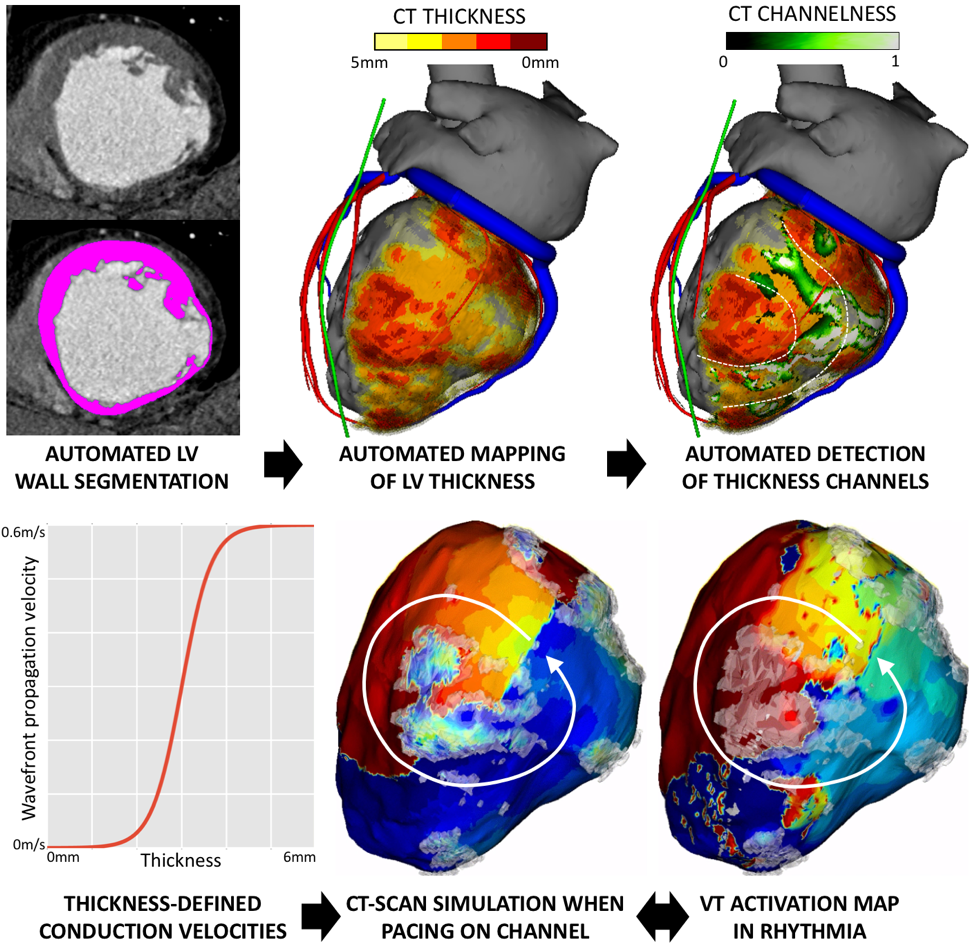
In the vast majority of post-MI VT ablation procedures, VT is either non inducible or non mappable. We introduce a fast and robust model of cardiac electrophysiology that can be directly parameterized from CT images to predict activation maps (Figure 18). The model is based on the Eikonal equation for wave propagation, where local conduction velocities are estimated from CT. A fully automated method is used to segment the LV wall and assign local conduction velocity according to local LV thickness. Then, a “channelness” filter automatically detects potential VT isthmuses as channels of preserved thickness within severely thinned scar. The model can then be paced within each channel to produce simulated activation maps within seconds. A neural network for automated LV wall segmentation was trained on 450 CTs segmented by experts. Validation, performed on another 50 cases, showed excellent accuracy (Dice score vs. expert 0.95). In 11 patients undergoing post-MI VT ablation (age 58±13, 9 men), simulated activation maps were validated vs. 25 high density maps acquired in Rhythmia (16 paced, 9 VT). Quantitative differences between predicted and measured local activation times remained substantial, particularly in dense scar (> 50ms). Nonetheless, activation patterns were well predicted in most cases (22/25), the 3 poor correlations being observed in patients with fewer scar. Personalized simulation of activation maps from CT scan is feasible and reliably reproduces activation patterns in post-MI VT. The method is fast enough to be used clinically in an interactive fashion for procedural planning [4].
Cardiac Modeling, Medical Imaging and Machine Learning for Electrostructural Tomography
Participants : Tania Marina Bacoyannis [Correspondant] , Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
This work is funded within the ERC Project ECSTATIC with the IHU Liryc, in Bordeaux.
Machine Learning, Cardiac modeling, Personalised simulation, Inverse problem of ECG, Electrical simulation.
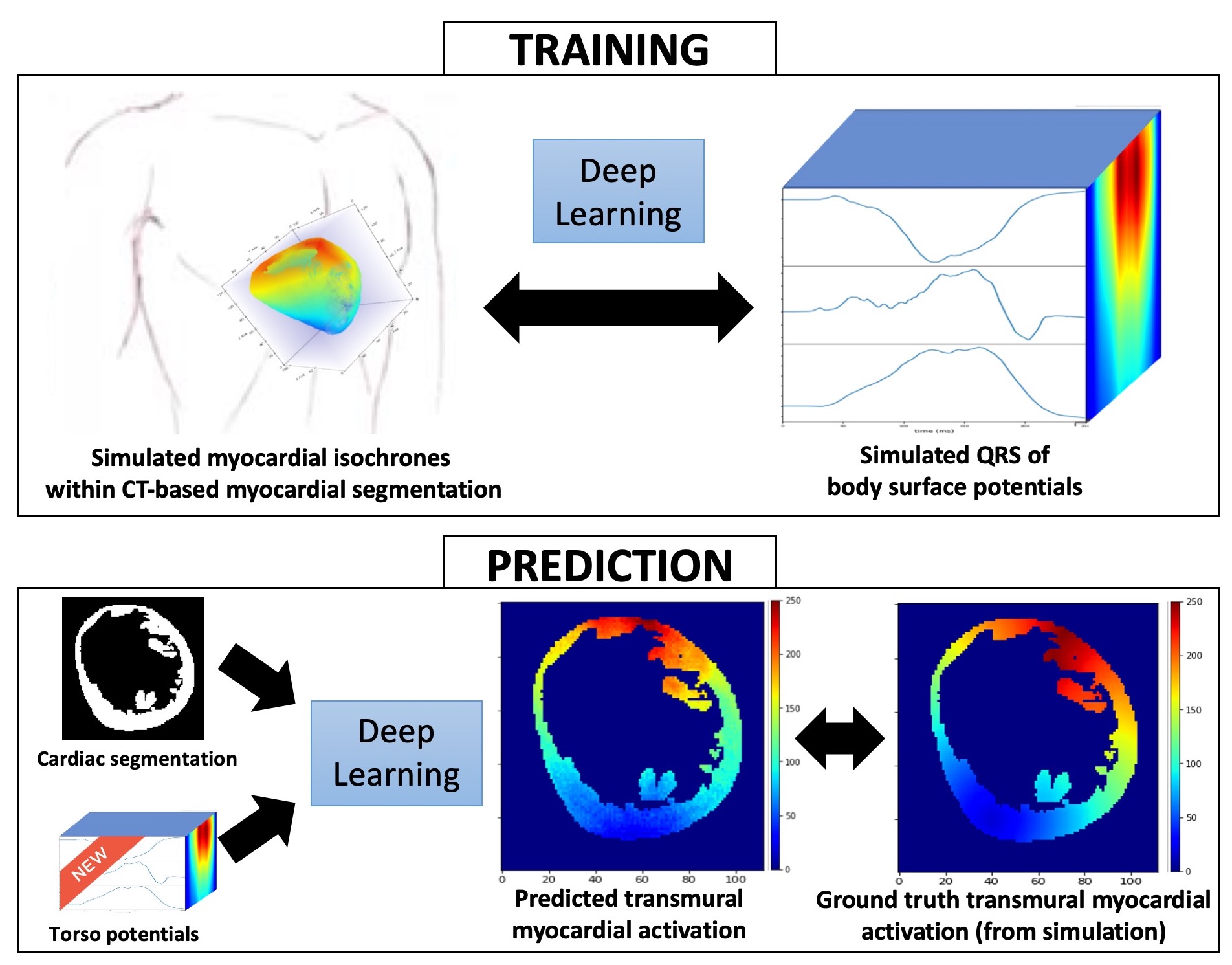
Electrocardiographic imaging (ECGI) aims at reconstructing cardiac activity from torso measurements.To achieve this one has to solve the ill-posed inverse problem of the torso propagation. We propose a novel application for Deep Learning Networks to learn spatio-temporal correlations on ECGI (Figure 19). We developped a conditional variational auto-encoder (CVAE).The input are activation maps and the model takes two conditions: on one hand the cardiac shape(cardiac segmentation) and the other one the ECG signals.
The model currently involves simulated data: 120 activations maps were simulated from one cardiac geometry along with simulated body surface potential maps.80% of the data was used for training and the remaining 20% for testing. As a result we were able to observe a good prediction of the activation pattern.
Next, we will test the model with real data provided by the IHU Liryc.
Discovering the link between cardiovascular pathologies and neurodegeneration through biophysical and statistical models of cardiac and brain images
Participants : Jaume Banus Cobo [Correspondant] , Maxime Sermesant, Marco Lorenzi.
Université Côte d'Azur (UCA)
Lumped models - Biophysical simulation - Statistical learning
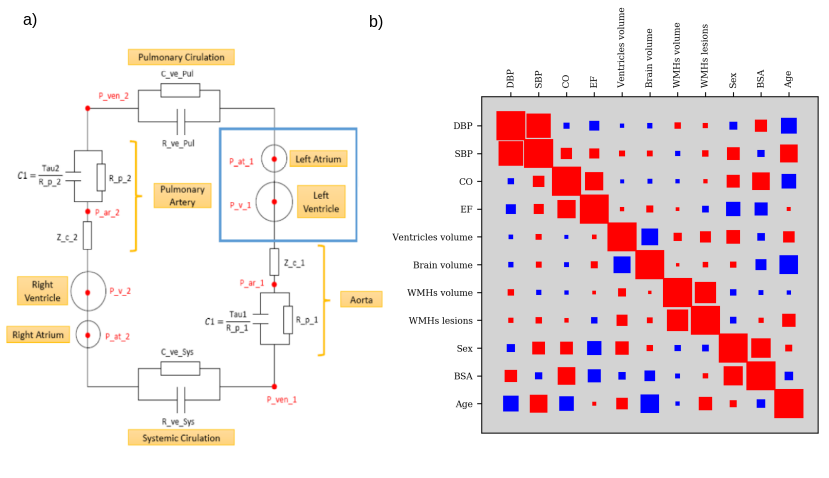
The project aims at developing a computational model of the relationship between cardiac function and brain damage from large‐scale clinical databases of multi‐modal and multi‐organ medical images. The model is based on advanced statistical learning tools for discovering relevant imaging features related to cardiac dysfunction and brain damage; these features are combined within a unified mechanistic framework to providing a novel understanding of the relationship between cardiac function, vascular pathology and brain damage (Fig. 20). We are also testing data-driven statistical learning models for the discovery of associations between cardiac function and brain damage. For example, by applying CCA we identified a first component that shows a positive correlation between the volume of white matter hyper intensities (WMHs), the number of WMHs lesions, the brain ventricles volume and high blood pressure values. On the other side we observed a second component, inversely associated to the first one, in which we observe a strong correlation between ejection fraction (EF) and total brain volume (white matter plus grey matter).
|
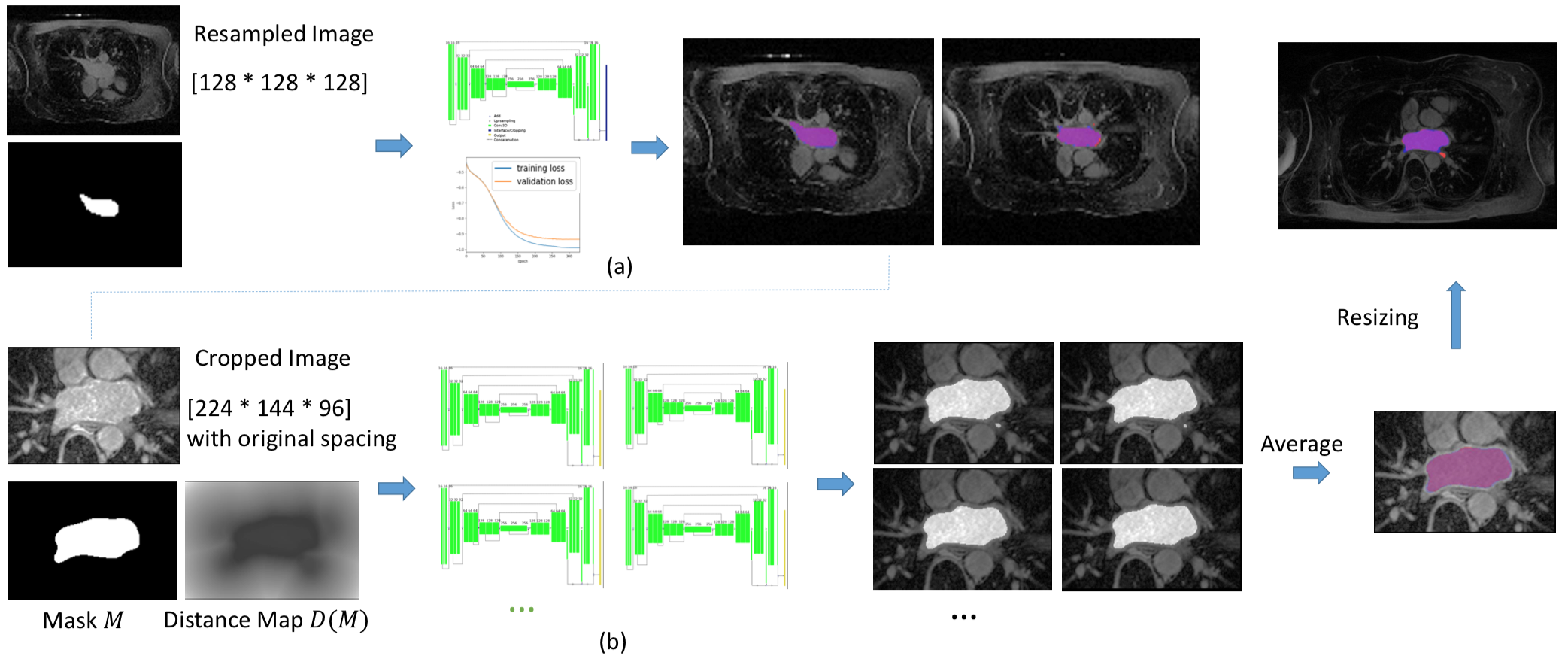
Automatic Image Segmentation of cardiac structures with Adapted U-Net
Participants : Shuman Jia [Correspondant] , Antoine Despinasse, Zihao Wang, Hervé Delingette, Xavier Pennec, Hubert Cochet, Maxime Sermesant.
The authors acknowledge the partial funding by the Agence Nationale de la Recherche (ANR)/ERA CoSysMedSysAFib and ANR MIGAT projects.
We proposed automated, two-stage, three-dimensional U-Nets with a contour loss, to segment the left atrium, as explained in [28], which obtained state-of-the-art results in the STACOM international challenge (Figure 21). Using similar method, we participated in Data Challenge organised at Journées Francophones de Radiologie and obtained a second place for renal cortex segmentation.
|
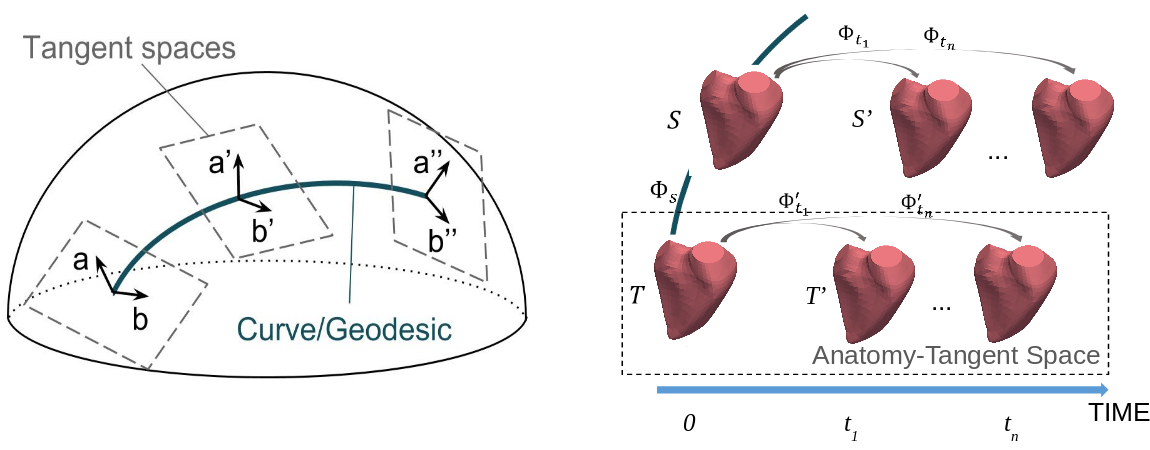
Parallel Transport of Surface Deformation
Participants : Shuman Jia [Correspondant] , Nicolas Duchateau, Pamela Moceri, Xavier Pennec, Maxime Sermesant.
The authors acknowledge the partial funding by the Agence Nationale de la Recherche (ANR)/ERA CoSysMedSysAFib and ANR MIGAT projects.
We looked into normalization of temporal deformation and proposed a more symmetric mapping scheme for pole ladder, which relies on geodesic symmetries around mid-points, as illustrated in [29] (Figure 22)). This modified parallel transport method method was shown to be of order 4 on general manifolds and exact in symmetric spaces [58].