Section: New Results
Botrytis cinerea phenotype recognition and classification: toward the establishment of links between phenotypes and antifungal molecules
Participants : Sarah Laroui, Eric Debreuve, Xavier Descombes.
This work is made in collaboration with Aurelia Vernay and Florent Villiers (Bayer).
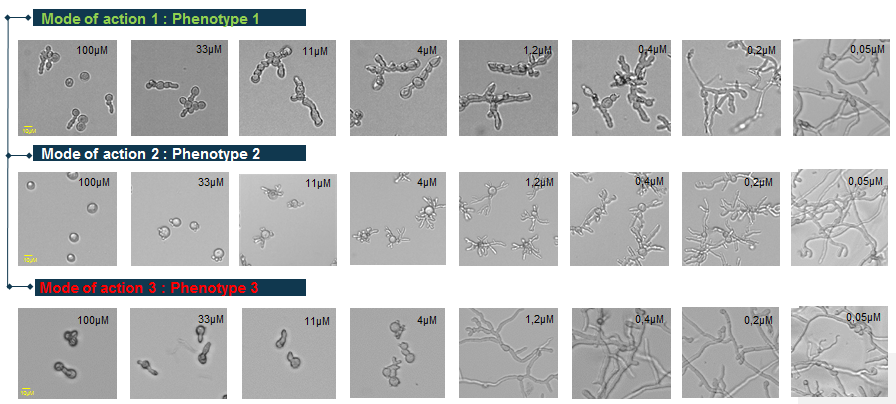
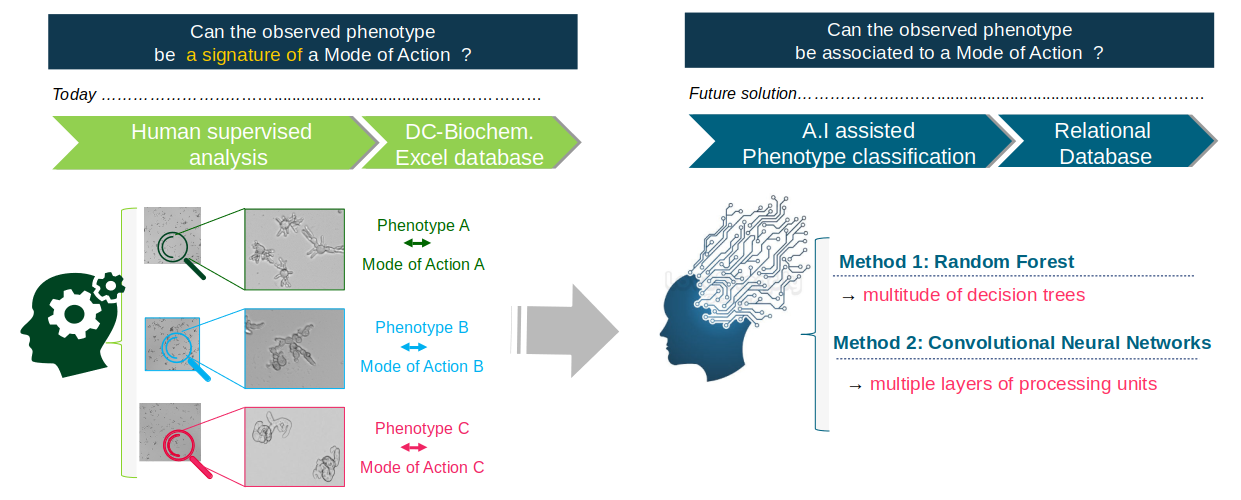
Botrytis cinerea is a reference model of filamentous phytopathogen fungi. Some chemical treatments can lead to characteristic morphological changes, or phenotypic signatures. These phenotypes could be associated with the treatment Mode of Action (Figure 12). In order to recognise and characterise different phenotypes and associate them with the different modes of action of the molecules (Figure 13), 24-hour images are taken by transmitted light microscopy. Because of the different dose-response effects, each given molecule is tested at ten concentrations.
We compared the results of classification of these images using two methods: random forests and convolutional neural networks (Deep Learning).
To learn the Random Forest classifier, we developed a robust image analysis and classification framework relying on morphometric and topological characteristics. A number of 16 features are extracted from three representations of the objects (binary mask, skeleton and graph). Some are calculated globally over all the objects of an image (ex: the skeleton length variance) while others are calculated on each object of an image (ex: the number of nodes of the graph). The second method uses a convolutional neural network. It has been implemented using Tensorflow, an open source library for Machine Learning, created by Google to develop applications in Deep Learning.
This method achieves better results than Random Forests, and it proved to be very robust to inter-experiment variations with an average classification accuracy of 88%. In addition, it does not require data pre-processing for feature extraction. However the explanatory aspect that exists with random forests is lost.