Keywords
Computer Science and Digital Science
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.1.4. Multiscale modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.6. Optimization
- A6.2.7. High performance computing
- A6.2.8. Computational geometry and meshes
- A6.3. Computation-data interaction
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.3.4. Model reduction
- A6.3.5. Uncertainty Quantification
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1.1.2. Molecular and cellular biology
- B1.1.8. Mathematical biology
- B2.2.1. Cardiovascular and respiratory diseases
- B2.2.6. Neurodegenerative diseases
- B2.4.1. Pharmaco kinetics and dynamics
- B2.4.3. Surgery
- B2.6.1. Brain imaging
- B2.6.2. Cardiac imaging
1 Team members, visitors, external collaborators
Research Scientists
- Jacques Henry [Inria, Emeritus, HDR]
- Amel Karoui [Univ de Bordeaux, Researcher, until Aug 2021]
- Peter Langfield [Inria, Researcher, from Oct 2021]
- Michael Leguèbe [Inria, Researcher]
- Mark Potse [Univ de Bordeaux, Researcher]
- Bachar Tarraf [Univ de Bordeaux, Researcher, from Oct 2021]
- Nejib Zemzemi [Inria, Researcher]
Faculty Members
- Yves Coudiere [Team leader, Univ de Bordeaux, Professor, HDR]
- Mostafa Bendahmane [Univ de Bordeaux, Associate Professor, HDR]
- Jerome Fehrenbach [Univ Paul Sabatier, Associate Professor, until Aug 2021, HDR]
- Lisl Weynans [Univ de Bordeaux, Associate Professor, HDR]
Post-Doctoral Fellow
- Mohamadou Malal Diallo [Univ de Bordeaux]
PhD Students
- Andony Arrieula [Inria]
- Oumayma Bouhamama [Univ de Bordeaux]
- Syed Hassaan Ahmed Bukhari [Université de Saragosse - Espagne]
- Zeina Chehade [Univ de Bordeaux, from Oct 2021]
- Narimane Gassa [Univ de Bordeaux]
- Niami Nasr [Univ de Bordeaux]
- Valentin Pannetier [Univ de Bordeaux, from Oct 2021]
- Bachar Tarraf [Inria, until Sep 2021]
Technical Staff
- Mehdi Juhoor [Inria, Engineer, until Oct 2021]
- Pauline Migerditichan [Inria, Engineer]
Interns and Apprentices
- Felix Dusart [Inria, May 2021]
- Valentin Pannetier [Inria, from Feb 2021 until Jul 2021]
- Marion Panouilleres [Univ de Bordeaux, from Apr 2021 until Sep 2021]
Administrative Assistant
- Nathalie Robin [Inria]
Visiting Scientists
- Abir Amri [Ecole Nationale d’Ingénieur de Tunis - Tunisie, from Oct 2021]
- Khouloud Kordoghli [Ecole Nationale d’Ingénieur de Tunis - Tunisie, from Oct 2021]
External Collaborator
- Edward Vigmond [Univ de Bordeaux]
2 Overall objectives
The Carmen team develops and uses models and numerical methods to simulate the electrophysiology of the heart from the molecular to the whole-organ scale, and its relation to measurable signals inside the heart and on the body surface. It aims at:
- improving understanding of normal and pathological cardiac electrophysiology,
- improving the efficiency and accuracy of numerical models,
- exploiting all available electrical signals for diagnosis,
- improving understanding and guidance of ablative treatment of cardiac arrhythmia.
The numerical models developed, analyzed, and used by the team incorporate essentially the gating dynamics of the ion channels in the cardiac cell membranes and the heterogeneities of the cardiac tissue, coupling processes on the cellular scale into macroscopic reaction-diffusion models. The team also work on incorporating any new biological knowledge, at any scale, that helps to understand the mechanisms of arrythmias, their diagnosis or treatment. At the same time we use simpler or reduced models to solve the inverse problems related to non-invasive electrical imaging of the heart.
The fields involved in our research are: ordinary and partial differential equations (PDE), inverse problems, numerical analysis, high-performance computing, image segmentation, and mesh construction.
A main goal of the team is to contribute to the work packages defined in the project of IHU Liryc, an institute founded in 2011 that focuses on cardiac arrhythmia.
We cooperate with physiologists and cardiologists on several projects. The team is building new models and powerful simulation tools that will help to understand the mechanisms behind cardiac arrhythmias and to establish personalized and optimized treatments. A particular challenge consists in making the simulations reliable and accessible to the medical community.
3 Research program
3.1 Complex models for the propagation of cardiac action potentials
The contraction of the heart is coordinated by a complex electrical activation process which relies on about a million ion channels, pumps, and exchangers of various kinds in the membrane of each cardiac cell. Their interaction results in a periodic change in transmembrane potential called an action potential. Action potentials in the cardiac muscle propagate rapidly from cell to cell, synchronizing the contraction of the entire muscle to achieve an efficient pump function. The spatio-temporal pattern of this propagation is related both to the function of the cellular membrane and to the structural organization of the cells into tissues. Cardiac arrythmias originate from malfunctions in this process. The field of cardiac electrophysiology studies the multiscale organization of the cardiac activation process from the subcellular scale up to the scale of the body. It relates the molecular processes in the cell membranes to the propagation process through the multiscale structure of the tissue and organ, to measurable signals in the heart and to the electrocardiogram, an electrical signal on the torso surface.
Several improvements of current models of the propagation of action potentials are being developed in the Carmen team, based on previous work 36 and on the data available at IHU Liryc:
- Enrichment of the current monodomain and bidomain models 36, 49 by accounting for structural heterogeneities of the tissue at cellular and intermediate scales. Here we focus on multiscale analysis techniques applied to the various high-resolution structural data available at IHU Liryc.
- Coupling of the tissues from the different cardiac compartments and conduction systems. Here, we develop models that couple 1D, 2D and 3D phenomena described by reaction- diffusion PDEs.
These models are essential to improve our understanding of cardiac electrical dysfunction. To this aim, we use high-performance computing techniques in order to numerically explore the complexity of these models.
We use these model codes for applied studies in two important areas of cardiac electrophysiology: atrial fibrillation 42 and sudden-cardiac-death (SCD) syndromes 7, 645. This work is performed in collaboration with several physiologists and clinicians both at IHU Liryc and abroad.
3.2 Simplified models and inverse problems
The medical and clinical exploration of the cardiac electric signals is based on accurate reconstruction of the patterns of propagation of the action potential. The correct detection of these complex patterns by non-invasive electrical imaging techniques has to be developed. This involves solving inverse problems that cannot be addressed with the more complex models. We want both to develop simple and fast models of the propagation of cardiac action potentials and improve the solutions to the reconstruction questions of cardiac electrical imaging techniques.
These questions concern the reconstruction of diverse information, such as cardiac activation maps or, more generally, the whole cardiac electrical activity, from high-density body surface electrocardiograms. It is a possibly powerful diagnosis technique, which success would be considered as a breakthrough. Although widely studied during the last decade, the reconstructed activation maps, for instance, are highly inacurate and have a poor clinical interest. It remains a challenge for the scientific community to understand how body surface signals can better inform on the fine details of arrhythmic mechanisms.
The most usual method consists in solving a Laplace equation on the volume delimited by the body surface and the epicardial surface, forwhich we contribute by:
- studying in depth the dependance of the inverse problem on inhomogeneities in the torso, conductivity values, the geometry, electrode positions, etc.,
- improving the solution to the inverse problem by using new regularization strategies, factorization of boundary value problems, and the theory of optimal control.
In addition, we have started to explore many alternative approaches including:
- using complete propagation models in the inverse problem, like the bidomain or monodomain equations, for instance in order to localize electrical sources,
- constructing data-based models using e.g. statistical learning techniques, which would accurately represent some families of well-identified pathologies, or allow to combine physics and biology-informed models and clinical data, and
- constructing simpler models of the propagation of the activation front, based on eikonal or level-set equations.
3.3 Numerical techniques
We want our numerical simulations to be efficient, accurate, and reliable with respect to the needs of the medical community. Based on previous work on solving the monodomain and bidomain equations 38, 37, 48, 28, we will focus on:
- High-order numerical techniques with respect to the variables with physiological meaning, like velocity, AP duration and restitution properties,
- Efficient, dedicated preconditioning techniques coupled with parallel computing.
Existing simulation tools used in our team rely, among others, on mixtures of explicit and implicit integration methods for ODEs, hybrid MPI-OpenMP parallellization, algebraic multigrid preconditioning, and Krylov solvers. New developments include high-order explicit integration methods and task-based dynamic parallellism.
3.4 Cardiac electrophysiology at the microscopic scale
Traditional numerical models of whole-heart physiology are based on the approximation of a perfect muscle using homogenisation methods. However, due to aging and cardiomyopathies, the cellular structure of the tissue changes. These modifications can give rise to life-threatening arrhythmias. For our research on this subject and with cardiologists of the IHU Liryc, we aim to design and implement models that describe the strong heterogeneity of the tissue at the cellular level and to numerically explore the mechanisms of these diseases.
The literature on this type of model is still very limited 54. Existing models are two-dimensional 46 or limited to idealized geometries, and use a linear (purely resistive) behaviour of the gap-juction channels that connect the cells. We propose a three-dimensional approach using realistic cellular geometry (Fig. 1), nonlinear gap-junction behaviour, and a numerical approach that can scale to hundreds of cells while maintaining a sub-micrometer spatial resolution (10 to 100 times smaller than the size of a cardiomyocyte). Following preliminary work in this area by us 31, 30, 29 and by others 54 we proposed a European project with 10 partner institutes and a 5.8M€ budget to develop software that can simulate such models, with micrometer resolution, on the scale of millions of cells, using future exascale supercomputers (microcard.eu). This project runs from April 2021 to October 2024, and involves also the Inria teams CAMUS, STORM and CARDAMOM as well as the Inria-led MMG Consortium.
3.5 Models and tools for ablative therapies
Today, the most effective way to treat arrhythmias is to ablate selected regions of the cardiac tissue. As the lesions have no particular electric property, this creates conduction blocks that stop the disorganized propagation of action potentials. The ablation procedure consists in placing a catheter in contact with the targeted site and deliver energy into the tissue. The energy can be overheating by radio-frequency current, electroporating electric pulses or temperature drop (cryotherapy). In practice, the choice of the ablation site is done by the clinician based on previous signal measurements and imagery, and is also guided during the procedure with real-time measurement of the electric signal.
Our team works on several subjects related to ablation techniques that may improve the success rate of the treatments:
- accurate computation of electric fields generated by catheters : complex catheter shapes, contact models, tissue heterogeneities,
- models of creation of the lesions, either through temperature rise (radio-frequency) or electroporation,
- localization tools to help clinicians target the optimal ablation sites, based on both data of previously ablated patients and synthetic data.
4 Application domains
4.1 Scientific context: IHU Liryc
The University Hospital of Bordeaux (CHU de Bordeaux) is equipped with a specialized cardiology hospital, the Hôpital Cardiologique du Haut-Lévêque, where the group of Professor Michel Haïssaguerre has established itself as a global leader in the field of cardiac electrophysiology 44, 43, 33. Their discoveries in the area of atrial fibrillation and sudden cardiac death syndromes are widely acclaimed, and the group is a national and international referral center for treatment of cardiac arrhythmia. Thus the group also sees large numbers of patients with rare cardiac diseases.
In 2011 the group has won the competition for a 40 million euro Investissements d'Avenir grant for the establishment of IHU Liryc, an institute that combines clinical, experimental, and numerical research in the area of cardiac arrhythmia. The institute works in all areas of modern cardiac electrophysiology: atrial arrhythmias, sudden death due to ventricular fibrillation, heart failure related to ventricular dyssynchrony, and metabolic disorders. It is recognized worldwide as one of the most important centers in this area.
The Carmen team was founded as a part of IHU Liryc. We bring applied mathematics and scientific computing closer to experimental and clinical cardiac electrophysiology. In collaboration with experimental and clinical researchers at Liryc we work to enhance fundamental knowledge of the normal and abnormal cardiac electrical activity and of the patterns of the electrocardiogram, and we develop new simulation tools for training, biological, and clinical applications.
4.2 Basic experimental electrophysiology
Our modeling is carried out in coordination with the experimental teams from IHU Liryc. It helps to write new concepts concerning the multiscale organisation of the cardiac action potentials that will serve our understanding in many electrical pathologies. For example, we model the structural heterogeneities at the cellular scale 32 (the MICROCARD project), and at an intermediate scale between the cellular and tissue scales.
At the atrial level, we apply our models to understand the mechanisms of complex arrythmias and the relation with the heterogeneities at the insertion of the pulmonary veins. We will model the heterogeneities specific to the atria, like fibrosis or fatty infiltration 51, 42. These heterogeneities are thought to play a major role in the development of atrial fibrillation.
At the ventricular level, we focus on (1) modeling the complex coupling between the Purkinje network and the ventricles, which is supposed to play a major role in sudden cardiac death, and (2) modeling the heteogeneities related to the complex organization and disorganization of the myocytes and fibroblasts, which is important in the study of infarct scars for instance.
4.3 Clinical electrophysiology
Treatment of cardiac arrhythmia is possible by pharmacological means, by implantation of pacemakers and defibrillators, and by curative ablation of diseased tissue by local heating, freezing or electroporation. In particular the ablative therapies create challenges that can be addressed by numerical means. Cardiologists would like to know, preferably by noninvasive means, where an arrhythmia originates and by what mechanism it is sustained.
We address this issue in the first place using inverse models, which attempt to estimate the cardiac activity from a (high-density) electrocardiogram. A new project aims at performing this estimation on-site in the catheterization laboratory and presenting the results, together with the cardiac anatomy, on the screen that the cardiologist uses to monitor the catheter positions 47.
4.4 Application in Deep Brain Stimulation
Since 2017, we have been working with neurosurgeons from the Bordeaux University Hospital (Pr Cuny and Dr. Engelhardt) on improving the planning technique for deep brain surgery (DBS) for Parkinson's and Essential tremor diseases. DBS is the last resort to treat the symptoms of Parkinson's disease after the drug Levodopa. The surgery consists in placing electrodes in very specific regions of the patient's brain. These regions are unfortunately not visible on the 1.5 Tesla MRI, the most widely available MRI machines in hospitals. The most effective solution to date is to introduce 5 micro-electrodes (MER) to record the activity of neurons in the patient's brain and to prospect by moving the electrodes in order to find the best location. However, this approach renders the surgery very cumbersome because the patient must be awake during the exploration phase. In addition, this phase takes at least 3 hours and mobilizes a neurologist with his staff. The total duration of the operation is between 7 and 8 hours. Many elderly patients do not tolerate this surgery. We have proposed an approach that avoids the prospecting phase and performs surgery under general anesthesia. The idea is to learn on pairs of clinical landmarks and the position of active electrodes in order to predict the optimal position of the DBS from a pre-operative image. This approach simplifies and standardizes surgery planning. We tested several approaches doi:10.3389/fneur.2021.620360. We continue to seek approaches to fully automate the targeting process. We carried out a proof of concept by learning on the clinical database of the Bordeaux University Hospital. The clinical validation of our approach is in progress through a clinical trial which includes patients from the University Hospitals of Bordeaux and Lyon. Pr Cuny has submitted a phase 3 national clinical research hospital project (PHRCN) including 11 CHUs in France which has been accepted by the General Directorate for Care Offers (DGOS). The aim is to compare our new approach to the ones used in the other centers. Inria Bordeaux is a partner in this project and we maintain the OptimDBS software and solve any technical problem related to the compatibility of the MRIs exported by our software and the surgical robots in the different centers.
5 Highlights of the year
- Two european H2020 projects started in 2021 : SimCardioTest and MICROCARD (see section 8.3) for details about each project. The MICROCARD project is coordinated by Mark Potse.
- Start of a new associate team called SPICY, following the previous EPICARD associate team (section 8.1.1). Start of a partnership within the ECOS Chile program.
- Nejib Zemzemi co-founded the Rebrain Startup with collaborators from the CHU de Bordeaux. The startup employs 2 engineers and will recruit Louise-Amelie Schmidt after the end of the maturation contract signed between Inria and AST.
- Lisl Weynans initiated the mentoring program which started this year at the IMB, Mathematics Institute of Bordeaux. The program consists in forming pairs of scientists in which a newcomer to the institute, either junior or senior, can get advice from a more experienced colleague about their everyday work or carreer.
- Narimane Gassa won the machine-learning challenge organized by IHU Liryc in April 2021. This challenge consisted in proposing a machine-learning-based model that is able to tell from implantable cardiac devices (ICD) recordings, if the recorded signals are relevant for interpretation or are too noisy due to extra-cardiac muscular activity or interference from electrical devices. This noise can modify an ECG and thus reduce the quality of ICD monitoring. The aim of this project was to improve the distant monitoring of ICDs, using automated learning, allowing an automated analysis of cardiac arrhythmia signals prior to human assessment. The management of patients could be potentially optimized by providing only relevant signals to the medical staff. Results were presented at the Computing In Cardiology 2021 conference 25.
- The team participated in the IHU Liryc workshop on Dec. 1st-2nd, with presentations and posters of results and projects, targetted towards the International Scientific Advisory Board of Liryc in view of its annual evaluation, and the whole institute.
6 New software and platforms
The two last parts of this section are generated automatically. The first section lists the new features of the software listed afterwards.
6.1 2021 Facts and Features
6.1.1 CEPS
- first public release,
- automation of numerical convergence tests, for any (time evolving) PDE problem,
- new implementation for more generic boundary conditions and more realistic stimulations,
- utilization in the H2020 project SimCardioTest in the framework of a collaboration with industrial partners (Microport CRM and InSilicoTrials).
6.1.2 CEMPACK and Propag
- New approaches to parallelism are tested in a simplified version of Propag, as part of the ANR Exacard project.
6.1.3 MUSIC - Carmen Plugins
New features:
- Create, visualize and export annotations for meshes and map.
- Perform point data interpolation on meshes and EP maps.
- Project any mesh or map onto another mesh or map.
- Compute electroanatomical mapping point distribution (mean ans standard deviation) in user defined regions.
- Finite elements method for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve.
- Export masks of segmented structures to DICOM-RTSTRUCT format (used in radiotherapy planning).
6.1.4 OptimDBS
Inria gave a licence to the SATT AST on OptimDBS for maturation and valorization. A licencing contract was signed by AST for the transfer of the software to the Rebrain startup company. Therefore, the website is not public anymore.
6.2 New software
6.2.1 CEPS
-
Name:
Cardiac ElectroPhysiology Simulation
-
Keywords:
Simulation, Health, Mesh, Cardiac, 3D, Cardiac Electrophysiology
-
Scientific Description:
As compared to other existing softwares, CEPS aims at providing a more general framework of integration for new methods or models and a better efficiency in parallel. CEPS is designed to run on massively parallel architectures, and to make use of state-of-the-art and well known computing libraries to achieve realistic and complex heart simulations. CEPS also includes software engineering and validation tools.
-
Functional Description:
CEPS is a modular high-performance computing software for performing numerical simulations in cardiac electrophysiology. It is based on modules : - management of geometries represented by meshes in 3D, 2D or 1D (volumes, surfaces, trees), - model simulation of cellular electrophysiology, - calculating the tissue propagation of the action potentials in the cardiac geometries, - calculation of extracardiac potentials, - time approximation methods in order 2, 3 and 4 specific to electrocardiography.
- URL:
-
Contact:
Michael Leguebe
-
Participants:
Mehdi Juhoor, Nejib Zemzemi, Antoine Gerard, Charlie Douanla Lontsi, Pierre-Elliott Bécue, Marc Fuentes, Yves Coudière, Michael Leguebe, Andjela Davidovic, Pauline Migerditichan, Florian Caro
-
Partners:
Université de Bordeaux, Fondation Bordeaux Université, CHU de Bordeaux, Inria
6.2.2 OptimDBS
-
Name:
Optimizing the Deep Brain Stimulation
-
Keywords:
Image analysis, Deep brain stimulation, Statistical learning
-
Functional Description:
Targeting software for deep brain stimulation
- URL:
-
Contact:
Nejib Zemzemi
-
Participants:
Nejib Zemzemi, Louise-Amelie Schmitt, Emmanuel Cuny, Julien Engelhardt
-
Partner:
CHU de Bordeaux
6.2.3 MUSIC - Carmen plugins
-
Name:
Carmen plugins for multi-modality imaging in Cardiology
-
Keywords:
Image segmentation, Mesh generation, Image filter, Numerical simulations, Cardiac Electrophysiology, Inverse problem, Finite element modelling, Visualization, 3D interaction, Registration
-
Scientific Description:
Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
-
Functional Description:
Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
- Publications:
-
Contact:
Nejib Zemzemi
-
Participants:
Hubert Cochet, Florent Collot, Mathilde Merle, Maxime Sermesant, Julien Castelneau, Mehdi Juhoor, Pauline Migerditichan, Nejib Zemzemi, Yves Coudière
-
Partners:
Université de Bordeaux, IHU - LIRYC
6.3 New platforms
CEMPACK
CEMPACK is a new collection of software that was previously archived in different places. It includes the high-performance simulation code Propag and a suite of software for the creation of geometric models, preparing inputs for Propag, and analysing its outputs. In 2017 the code was collected in an archive on Inria's GitLab platform. The main components of CEMPACK are the following.
-
Propag-5.1
Applied modeling studies performed by the Carmen team in collaboration with IHU Liryc and foreign partners 752, 42, 41, 34 rely on high-performance computations on the national supercomputers Irene, Occigen, and Turing. The Propag-5 code is optimized for these systems. It is the result of a decades-long development first at the Université de Montréal in Canada, then at Maastricht University in the Netherlands, and finally at the Institute of Computational Science of the Università della Svizzera italiana in Lugano, Switzerland. Since 2016 most of the development on Propag has been done by M. Potse at the Carmen team 53. The code scales excellently to large core counts and, as it is controlled completely with command-line flags and configuration files, it can be used by non-programmers. It also features:
- a plugin system for membrane models,
- a completely parallel workflow, including the initial anatomy input and mesh partitioning, which allows it to work with meshes of more than nodes,
- a flexible output scheme allowing hundreds of different state variables and transient variables to be output to file, when desired, using any spatial and temporal subsampling,
- a configurable, LUSTRE-aware parallel output system in which groups of processes write HDF5/netCDF files, and
- CWEB documentation of the entire code base.
The code has been stable and reliable for several years. It can be considered the workhorse for our HPC work until CEPS takes over.
-
Gepetto
The Gepetto suite, named after a famous model maker, transforms a surface mesh of the heart into a set of (semi-)structured meshes for use by the Propag software or others. It creates the different fiber orientations in the model, including the transmurally rotating ventricular fibers and the various bundle structures in the atria (figure 2), and creates layers with possibly different electrophysiological properties across the wall. A practically important function is that it automatically builds the matching heart and torso meshes that Propag uses to simulate potentials in the torso (at a resolution of 1 mm) after projecting simulation results from the heart model (at 0.1 to 0.2 mm) on the coarser torso mesh 50. Like Propag, the Gepetto software results from a long-term development that started in Montreal, Canada, around 2002. The code for atrial fiber structure was developed by our team.
-
Blender plugins
Blender is a free software package for the production of 3-D models, renderings, and animations, comparable to commercial software such as Cinema4D. CEMPACK includes a set of plugins for Blender that facilitate the production of anatomical models and the visualization of measured and simulated data. It uses the MMG remeshing library, which is developed by the CARDAMOM team at Inria Bordeaux.
7 New results
7.1 Partial differential equations analysis
- Three Scale Unfolding Homogenization Method Applied to Cardiac Bidomain Model 12, 11. In this work, we are dealing with a rigorous homogenization result at two different levels for the bidomain model of cardiac electrophysiology. The first level associated with the mesoscopic structure such that the cardiac tissue consists of extracellular and intracellular domains separated by an interface (the sarcolemma). The second one related to the microscopic structure in such a way that the intracellular medium can only be viewed as a periodical layout of unit cells (mitochondria). At the interface between intra- and extracellular media, the fluxes are given by nonlinear functions of ionic and applied currents. A rigorous homogenization process based on unfolding operators is applied to derive the macroscopic (homogenized) model from our meso-microscopic bidomain model. We apply a three-scale unfolding method in the intracellular problem to obtain its homogenized equation at two levels. The first level upscaling of the intracellular structure yields the mesoscopic equation. The second step of the homogenization leads to obtain the intracellular homogenized equation. To prove the convergence of the nonlinear terms, especially those defined on the microscopic interface, we use the boundary unfolding method and a Kolmogorov-Riesz compactness’s result. Next, we use the standard unfolding method to homogenize the extracellular problem. Finally, we obtain, at the limit, a reaction-diffusion system on a single domain (the superposition of the intracellular and extracellular media) which contains the homogenized equations depending on three scales. Such a model is widely used for describing the macroscopic behavior of the cardiac tissue, which is recognized to be an important messengers between the cytoplasm (intracellular) and the other extracellular inside the biological cells.
- Optimal control for nonlocal reaction-diffusion system describing calcium dynamics in cardiac cell 13. In this work, we introduce an optimal control for a nonlocal calcium dynamic model in a cardiac cell acting on ryanodine receptors. The optimal control problem is considered as a coupled nonlocal reaction-diffusion system with a transmission boundary condition covering the sarcoplasmic reticulum and cytosolic domain. We establish the well-posedness result of the adjoint problem using Faedo-Galerkin approximation, a priori estimates, and compactness arguments. The numerical discretization of direct and adjoint problems is realized by using the implicit Euler method in time and the finite element for spatial discretization. Moreover, we obtain the stability result in the L2-norm for the direct and the adjoint discrete problems. Finally, in order to illustrate the control of our calcium dynamic model, we present some numerical experiments devoted to constant and nonlocal diffusions using the proposed numerical scheme.
7.2 Numerical analysis and development of numerical methods
- Development of an Eulerian method to numerically solve incompressible bifluid problems with high density ratios with the specificity of a sharp second-order numerical scheme for the spatial resolution of the discontinuous elliptic problem for the pressure 15.
- A positive cell vertex Godunov scheme for a Beeler-Reuter based model of cardiac electrical activity. 14. The monodomain model is a widely used model in electrocardiology to simulate the propagation of electrical potential in the myocardium. In this work, we investigate a positive nonlinear control volume finite element scheme, based on Godunov's flux approximation of the diffusion term, for the monodomain model coupled to a physiological ionic model (the Beeler–Reuter model) and using an anisotropic diffusion tensor. In this scheme, degrees of freedom are assigned to vertices of a primal triangular mesh, as in conforming finite element methods. The diffusion term which involves an anisotropic tensor is discretized on a dual mesh using the diffusion fluxes provided by the conforming finite element reconstruction on the primal mesh and the other terms are discretized by means of an upwind finite volume method on the dual mesh. The scheme ensures the validity of the discrete maximum principle without any restriction on the transmissibility coefficients. By using a compactness argument, we obtain the convergence of the discrete solution and as a consequence, we get the existence of a weak solution of the original model. Finally, we illustrate by numerical simulations that the proposed scheme successfully removes nonphysical oscillations in the propagation of the wavefront and maintains conduction velocity close to physiological values.
7.3 Modeling
-
We proposed a model of cardiac mitochondria using a simplification approach in order to calibrate the model parameters to experimental data 21. The model describes the major features of mitochondrial activity willingly omitting detailed descriptions of these features in order to reduce its complexity (Fig. 3). In the end, our model consists of 6 state variables and 32 parameters, which is much less than the current models proposed by biologists. Preliminary sensitivity analysis was done on our model using Sobol indices, so as to run the calibration with identifiable parameters only. Our calibrated model was able to reproduce robustly respiration data.
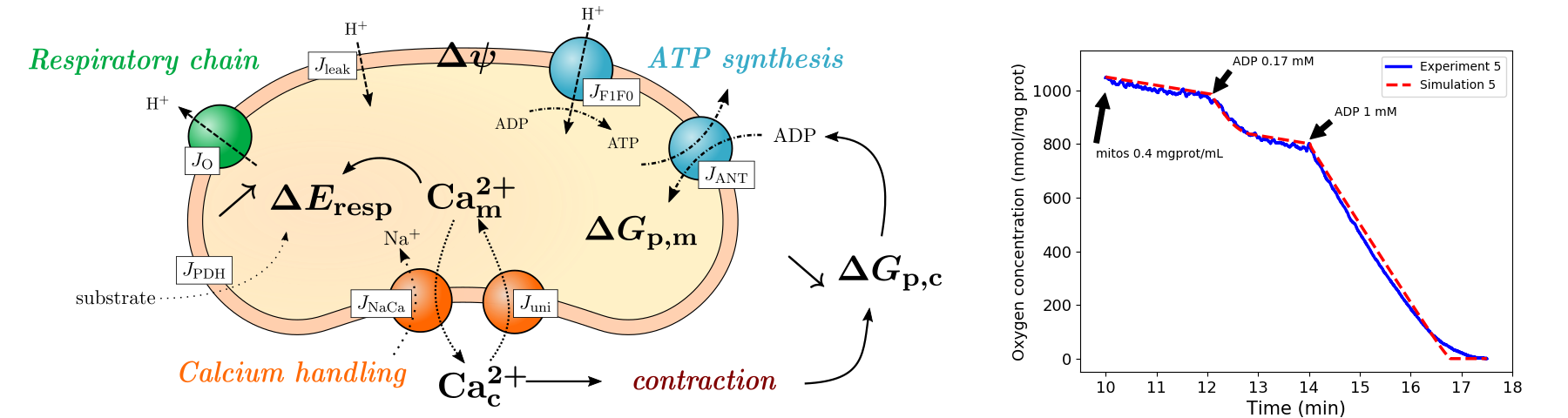
A cardiac mitochondron and the flux considered in the model; graph of the respiration rates (model vs data)
Figure 3: Left : Schematic of the phenomena taken into account in our mitochondrial model. Right : reproduction of respiration data using our calibrated model. This model is now being extended to include the effects of Reactive Oxygen Species (ROS) and the mitochondrial permeability transition pore (mPTP). Further data analysis and likelihood function tuning are required to calibrate this extended model to fluorescence data of individual mitochondria, using bayesian techniques. Ph.D. thesis of Bachar Tarraf. Interdisciplinary work between Liryc teams (modeling and metabolism) and other laboratories (ISM Bordeaux and LASS Toulouse), in the framework of the ANR Mitocard project.
- We modeled microstructural heterogeneities in the cardiac tissue by mean of a homogenized model. It enables to include, into macroscopic equivalent conductivity coefficients, quantitative information about the microstructure available from high resolution imaging modalities, such as fibrosis or diverse types of inclusions (follow up of Andjela Davidovic Ph.D thesis). The complete procedure to derive the macroscopic coefficients from MR images, first published online in 2020, was finally published in a special issue of the International Journal on Numerical Methods in Biomedical Engineering 40.
- Spiral Waves Generation Using an Eikonal-Reaction Cardiac Electrophysiology Model 27 In this work, we used the conduction velocity adapted eikonal model 35 in order to simulate reentry spiral waves. The new model that we present here combines the CV-adapted eikonal model with a specific parametrized stimulation current obtained from the diffusion term of the monodomain model. The monodomain model is computed once off-line and the generated diffusion current has been approximated, parametrized and used to stimulate the MS ode system. The preliminary results showed that with this model we have been able to simulate spiral waves with the same pattern as the monodomain model. To the best of our knowledge, this is the first work able to generate spiral waves using the eikonal model. Differently from the monodomain model, [8], the EK-CV-DIFF formulation captures the correct value of the conduction velocity even using a coarse resolution. This eikonal model is easy to parametrize and thus could be a good candidate to conduct personalized simulations in order to simulate the electrical wave of a given patient. Our future work will be to parametrize this model in order to provide a new electrocardiography imaging tool especially in atrial fibrillation conditions
7.4 Modeling and inverse problems
- Cardiac Activation Maps Reconstruction: A Comparative Study Between Data-Driven and Physics-Based Methods 20. Proof of concept of AI based methods for the development of new ECGi algorithms: second part of Amel Karoui Ph.D. thesis. The PhD was defended in 2021.
- Validation on experimental data of the patchwork method, a ECGI method selecting among several numerical method the one minimizing the residuals on the torso 23. (Ph.D thesis of Oumayma Bouhamama, co-supervised with Laura Bear from Signal Processing team).
- Resolution of the inverse problem of Electrical Impedance Tomography with an immersed boundary method on a cartesian grid in 2 dimensions (Ph.D thesis of Niami Nasr, co-supervised by J. Dardé, IMT Toulouse).
- Development and validation on academic test-cases of an eikonal model to solve the inverse problem of the propagation of the front on the heart surface from body-surface measurements, in collaboration with J. Fehrenbach (IMT, Toulouse).
- We developed a new machine-learning approach for the localization of tissue that is provoking premature ventricular contractions (PVC) 22. The approach is original in several ways. Firstly, as it is meant to be used during an intervention, it has to be very fast – eventually close to real-time. Secondly, it was trained on simulated data only, but then showed good performance for real patients. This was possible due to the use of highly realistic patient-based models to produce the training data. Finally, it uses only the 8 ECG electrodes that are needed to record a standard electrocardiogram, in contrast to traditional inverse models which use hundreds of electrodes. This limited lead set could be used because the problem of PVC localisation is intrinsically very constrained. The use of a limited lead set is very important for clinical applicability.
- A volume source method for solving ECGI inverse problem. 26 In this paper, we present a volumic model to solve the electrocardiography inverse problem that takes into account structural informations obtained by imaging techniques. The direct problem maps a volumic current in the cardiac muscle (ventricles) to the body surface electrical measures. The model is based on coupling bidomain heart model with torso conduction. The corresponding inverse problem is solved with the Tikhonov regularization. Simulated public database EDGAR is used for the evaluation of this method and we compared them to standard method of fundamental solutions (MFS). The sensitivity to noise is also assessed. The correlation coefficients and the relative error of activation times were computed. Results show that the volume method performs better than MFS for any noise level, and reconstruct in addition endocardial information.
7.5 High performance computing
- For the MICROCARD project, we developed a prototype software that creates artificial geometric models of cardiac muscle tissue at the cellular scale. We need such models for our simulations, because current imaging data at this scale is limited to a handful of cells, while we require thousands to, ultimately, billions of cells (each represented by roughly a thousand tetrahedral elements). Our software defines a level-set function on a tailored mesh. The MMG software is then used to discretize the level set and improve the mesh quality. With the current prototype it is feasible to produce networks of a few hundred cells, and using replication techniques we can make these meshes as large as can be stored: our largest test so far required 16TB storage, while a full human heart would require in the order of 2PB. These results will be presented at the ECCOMAS meeting in June 2022.

A view of a set of cardiac cell surface meshes.
Figure 4: Synthetic model of cardiac tissue created with our software. Colors identify individual cells. - With R. Turpault and K. Khadra, collaborators from IMB (Institut de Mathématiques de Bordeaux), we continue the development of a very high order finite volume method for the monodomain equation, following the method published in 39. During 2021, we developped ands started to test the MPI parallel version that will be used ot extend our numerical experiments to large three-dimensional models. The numerical scheme also serves as a sandbox for the method to be developped in the MICROCARD project. This foreseen method will have to account for the Laplace equation in the whole computational domain, like the previous one, and in addition with jumps on all the cell membranes, which are spread among the whole computational domain.
8 Partnerships and cooperations
8.1 International initiatives
8.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
SPICY
-
Title:
Stochastic forward and inverse Problems In Cardiac electrophysiologY
-
Duration:
2021 ->
-
Coordinator:
Mourad Bellasoued (mourad.bellassoued@enit.utm.tn)
-
Partners:
- Université de Tunis El Manar
-
Inria contact:
Mostafa Bendahmane
-
Summary:
Compared to classical method in electrophysiology, in this project we use the stochastic bidomain model on the heart to add constraints when solving the inverse problem. The problem is then reduced to an optimal control question. We intend to construct a general stochastic monodomain or stochastic bidomain models type, to identify the parameters that correspond to the working hypothesis and see that it is possible to reproduce the observed ECG signal in patients with arrythmias. From these stochastic models we will questions and problems of control and stabilization in order to find strategies to prevent or reverse the arrhythmic activity.
8.1.2 Participation in other International Programs
ECOS Nord
-
Title:
Quantitative and numerical analysis of inverse problems in cardiology
-
Partner Institution(s):
- University of Puebla, Mexico, with Prof. A. Fraguella Colar
-
Date/Duration:
2020, reconducted in 2021
ECOS Sud
-
Title:
Virtual Element Methods for Bidomain Model For Cardiac Electrophysiology
-
Partner Institution(s):
- University of the Bío-Bío, Concepción, Chile
-
Summary:
This project is framed within the area of mathematical modeling and numerical simulations in physiology, particularly we explore the cardiac electrical potentials and we develop several applications using VEM discretizations to bidomain model. For instance, primal and mixed formulations will be considered. The main motivation to study this model is that it constitutes a stepping stone towards the more complex models with important applications in cardiac dynamics where we could have complex domains. Thus, we can explote the capability of VEM to use general meshes. We also plan to derive a posteriori error estimators for the VEM and use it to drive space/time adaptive schemes.
Note that there was no activity in the ECOS nord project, due to the Covid crisis, and the postdoctoral researchers involved in the project not being present anymore. There has been a very reduced activity in the ECOS sud project, mainly through a small number of visioconferences. In both projects, no visit could be carried out.
8.2 International research visitors
8.2.1 Visits of international scientists
Khouloud Kordoghli
-
Status
PhD
-
Institution of origin:
Université de Tunis El Manar
-
Country:
Tunisia
-
Dates:
October to December 2021
-
Context of the visit:
the visit was part of the SPICY associated team.
-
Mobility program/type of mobility:
research stay
Abir Amri
-
Status
PhD
-
Institution of origin:
Université de Tunis El Manar
-
Country:
Tunisia
-
Dates:
October to December 2021
-
Context of the visit:
the visit was part of the SPICY associated team.
-
Mobility program/type of mobility:
research stay
8.2.2 Visits to international teams
Narimane Gassa
-
Visited institution:
EP solutions SA (industrial partner)
-
Country:
Swiss
-
Dates:
September to October 2021 (2 months)
-
Context of the visit:
The visit is part of Narimane Gassa's secondment in the PersonalizedAF project, see below.
-
Mobility program/type of mobility:
research stay
8.3 European initiatives
EuroHPC MICROCARD.
The MICROCARD project, co-funded by EuroHPC, ANR, and five other national organizations, is a medium-size European project (10 partners, 5.8M€ budget) aiming to build software that can simulate cardiac tissue cell by cell, on future exascale supercomputers. It is led by the CARMEN team (Mark Potse, scientific coordinator; Yves Coudière, technical and scientific manager). The project was granted in June 2020 and started in April 2021 when all national co-funding procedures had been completed. Although the project is coordinated by Université de Bordeaux, Inria is strongly involved with four of its project teams (CARMEN, CAMUS, CARDAMOM, STORM) and the Inria-led MMG Consortium. The project further involves Inria's strategic partner SIMULA (Norway), five more universities, and three industrial partners including one manufacturer of large-scale computers. The software built in this project will be used for research into life-threatening cardiac arrhythmia mechanisms, in particular those that involve subtle structural tissue damage.
The scale of simulations envisaged in the MICROCARD project is enormous. The human heart has two billion muscle cells. A cell-by-cell model, in which each of these cells would be discretized with in the order of a thousand tetrahedral elements, would thus lead to a problem with unknowns. Initial tests have shown that the geometrical model, if it could be produced, would require 2PB storage. To represent the complex electrophysiology on each model element, the computation would involve in the order of variables, which will have to be integrated over thousands to millions of time steps. The project is therefore challenging from a numerical as well as an informatics point of view. The MICROCARD consortium involves applied mathematicians, specialists in numerical science, software engineers, and biomedical engineers representing a large spectre of expertise to meet these challenges.
H2020 RIA SimCardioTest.
The SimCardioTest project is a collaborative project between 10 organizations from 6 European countries and United States funded by the European Commission (EU H2020). This project will provide new insight into designing new predictive tools in cardiac pathologies and will accelerate the uptake of computer simulations for testing medicines and medical devices. It is lead by Inria, and the Carmen team participates in this project as affiliated to the University of Bordeaux (UBx). Yves Coudière is the coordinator for the UBx partner, and in particular work package leader for WP2.
WP2 aims at designing and implementing numerical models of cardiac pacemakers or implantable defibrillators, in view of designing in-silico trials useful for assessing the security or efficacy of these devices. This work is based on clinical images, on our software CEPS and the platform SOFA. It is carried out in collaboration with the Inria teams EPIONE (Sophia-Antipolis) and DEFROST (Lille), and with the industrial partners Microport CRM and InSilicoTrials.
Within the Carmen team, we focus on the simulation of the electrical behavior of industrial pacemakers, in order to output numerical quantities related to their pacing and sensing functions in a heart.
H2020 MSCA-ITN PersonalizeAF.
The PersonalizeAF project is a Marie-Curie translational training project. It addresses the challenge of understanding and treating atrial arrythmias, by delivering an innovative multinational, multi-sectorial, and multidisciplinary research and training programme in new technologies and novel strategies for individualized characterization of AF substrate to and increase treatments’ efficiency.
From the research point of view, PersonalizeAF integrates data and knowledge from in-vitro, in silico, ex vivo and in vivo animal and human models to: 1) generate an individual description of the state of the atrial muscle identifying the disease mechanisms and characteristics; 2) understanding the potential effect that different therapies have on different atrial substrates; and 3) combining this information to generate a specific profile of the patient and the best therapy for each patient.
In the Carmen team, we supervise 1 PhD student (ESR11, Narimane Gassa) from PersonalizeAF. She is working on simplified models of atrial fibrillation based on eikonal equations coupled to the Mitchell-Schaeffer ionic model. She aim at personalizing such a simple model using aclinical anatomical data and body surface potential recordings. Her first secondment, carried out in 2021, was to vissit the company EP Solutions SA that produces a technology able to record such electrical maps.
8.4 National initiatives
ANR Exacard.
Granted by ANR in July 2018, EXACARD is a collaborative project with computer scientists from Labri that targets hexascale computing in cardiac electrophysiology, it serves as a sandbox to prepare the project EuroHPC MICROCARD. PI for Bordeaux University/Liryc: Yves Coudière.
ANR MITOCARD.
The MITOCARD project (Electrophysiology of Cardiac Mitochondria), coordinated by S. Arbault (Université de Bordeaux, ISM), was granted by the ANR in July 2017. The objective of MITOCARD is to improve understanding of cardiac physiology by integrating the mitochondrial properties of cell signaling in the comprehensive view of cardiac energetics and rhythm pathologies. It was recently demonstrated that in the heart, in striking contrast with skeletal muscle, a parallel activation by calcium of mitochondria and myofibrils occurs during contraction, which indicates that mitochondria actively participate in Ca2+ signaling in the cardiomyocyte. We hypothesize that the mitochondrial permeability transition pore (mPTP), by rhythmically depolarizing inner mitochondrial membrane, plays a crucial role in mitochondrial Ca2+ regulation and, as a result, of cardiomyocyte Ca2+ homeostasis. Moreover, mitochondrial reactive oxygen species (ROS) may play a key role in the regulation of the mPTP by sensing mitochondrial energetics balance. Consequently, a deeper understanding of mitochondrial electrophysiology is mandatory to decipher their exact role in the heart's excitation-contraction coupling processes. However, this is currently prevented by the absence of adequate methodological tools (lack of sensitivity or selectivity, time resolution, averaged responses of numerous biological entities). The MITOCARD project will solve that issue by developing analytical tools and biophysical approaches to monitor kinetically and quantitatively the Ca2+ handling by isolated mitochondria in the cardiomyocyte.
MITOCARD is a multi-disciplinary project involving 4 partners of different scientific fields: the CARMEN team as well as:
-
ISM,
the largest chemistry laboratory of the Université de Bordeaux, where the necessary measurement methods will be developed,
-
Liryc,
where mitochondria are studied at all levels of integration from the isolated mitochondrion to the intact heart,
-
LAAS,
the MiCrosystèmes d'Analyse (MICA) group at the Laboratory of Analysis and Architecture of Systems, which develops the biological microsensors for this project.
The project will:
- develop chips integrating 4 different electrochemical microsensors to monitor in real-time key mitochondrial signaling parameters: Ca2+, membrane potential, quinone reduction status, O2 consumption, and ROS production,
- develop microwell arrays integrating ring nanoelectrodes to trap single mitochondria within micrometric chambers and measure locally by combined fluorescence microscopy and electrochemical techniques intra- (by fluorescence) and extra-mitochondrial (electrochemistry) metabolites,
- develop a mathematical model of mitochondrial Ca2+ and ROS handling built on existing knowledge, new hypotheses, and the measured data.
The model may serve both to assess biological assumptions on the role of mitochondria in Ca2+ signaling and to integrate pathological data and provide clues for their global understanding.
ANR MAESTRO.
The ANR project MAESTRO (Magnetic Signal detection of ventricular arrhythmOgenic substrates), coordinated by Prof. Michel Haïssaguerre (IHU Liryc), has a computational component for which we hope to recruit a postdoc in early 2022, to be directed by Mark Potse.
GENCI.
GENCI project A0110307379, “Interaction between tissue structure and ion-channel function in cardiac arrhythmia,” coordinated by Mark Potse, comprises 4 million core-hours on the supercomputer Jean-Zay at IDRIS. Compared to previous years it is a modest allocation. This is because most of our computational needs in 2022 are either smaller or larger than the national scale.
8.5 Regional initiatives
Labex CIMI (Toulouse).
"Electrical Impedance Tomography for the Inverse Problem of Electrocardiographic Imaging" PI Jérémi Dardé (IMT, Toulouse) The project gathers complementary areas of applied mathematics and signal processing to address all relevant aspects of the problem, including modelling, numerical experiments and statistical analysis. Furthermore, if these new methods prove to increase the accuracy of ECGi, the project would lead to in-vivo experiments in collaboration with the Electrophysiology and Heart Modelling Institute (IHU-Liryc)
9 Dissemination
9.1 Promoting scientific activities
9.1.1 Scientific events: organisation
General chair, scientific chair
- Peter Langfield : session chair for the World Congress on Biomechanics 2022
9.1.2 Scientific events: selection
Member of the conference program committees
- Yves Coudière : program committee member, FIMH 2021, the international conference of Functional Imaging and Modeling of the Heart
- Nejib Zemzemi : program committee member, FIMH 2021, the international conference of Functional Imaging and Modeling of the Heart
9.1.3 Journal
Member of the editorial boards
- Mark Potse: associate editor of Frontiers in Cardiac Electrophysiology and Journal of Electrocardiology; guest associate editor for Frontiers in Cardiovascular Medicine.
Reviewer - reviewing activities
- Mark Potse: Frontiers in Computational Electrophysiology, Journal of Electrocardiology, IEEE Access, Computing in Cardiology (congress),
- Peter Langfield: Journal of Computational Dynamics,
- Lisl Weynans: ESAIM: Mathematical Modelling and Numerical Analysis, Computers in Biology and Medicine,
- Yves Coudière: Mathematical Biosciences and Engineering, Computers in Biology and Medicine, Annals of Biomedical Engineering, FIMH 2021 (conference).
9.1.4 Research administration
- Yves Coudière: executive director of IMB (Institut de Mathématiques de Bordeaux, UMR 5251 CNRS, UB, Bdx-INP)
- Yves Coudière: member of the Scientific Committee of IHU Liryc
- Mark Potse: council member of the International Society of Electrocardiology
- Mark Potse: member of the user board (Comité d'utilisateurs) of IDRIS
- Mark Potse: management of the website of the International Society of Electrocardiology.
9.2 Teaching - Supervision - Juries
9.2.1 Teaching
The 2 assistant professors and 1 professor of the team teach at several levels of the Bordeaux University programs in Mathematics, Neurosciences, and Medicine (respectively, 192, 192 and 96 h/year on average). The researchers also have a regular teaching activity, contributing to several courses in the Applied Mathematics at the Bachelor and Master levels (between 16 and 72 h/year).
The PhD students who ask for it are used to teach between 32 and 64 h/year, usually courses of general mathematics in L1 or mathematics for biologists in L1 or L2.
Teaching responsibilities at the University of Bordeaux:
- Yves Coudière: Master MAS (Mathématiques Appliquées, Statistiques), parcours MNCHP (Modélisation Numérique Calcul Haute Performance),
- Yves Coudière then Lisl Weynans: Licence Mathématique parcours ingénierie mathématique,
- Lisl Weynans: Mineure Mathématiques du parcours International de la Licence,
- Mostafa Bendahmane: Responsable de la mobilité internationale des étudiants de Licence MIASHS.
Courses (L for Bachelor level, M for Master level):
- Numerical analysis (L2)
- Programming for scientific computing with C++ (L3)
- Programming projects with Python (M1)
- Numerical approximation of PDEs: Finite Differences, Finite Elements, Finite Volumes (M1, M2)
- Supervision of programming projects (L3, M1)
- Mathematical modelling in L2 parcours medicine and physics
- Linear Algebra, Optimization under constraints (L2 and Essca school)
- Analysis, L2
- Computational Neurosciences, M2
- Neuropsychology and Psychophysiology, L3
- Graduate program EUR Digital Public Health, Bordeaux University, March 2019. Philosophy of science and the role of numerical models: introductory course in the module Modeling in life science
9.2.2 Supervision
In addition to the PhD students of the team (7 on Jan 1st, 8 on Dec 31st), team members were the co-supervizors of the following students:
- Fakhrielddine Bader (Ecole centrale de Nantes) : "Homogéneisation et convergence double échelle du modèle bidomaine en électrocardiologie". Defended in 2021.
9.2.3 Juries
- Lisl Weynans: reviewer of PhD thesis of Georis BILLO (M2P2, Marseille)
- Yves Coudière: reviewer and jury member of Tania Bacoyannis (Université Côte d'Azur); jury member of Fakhrielddine Bader (Ecole centrale de Nantes).
9.3 Popularization
9.3.1 Education
- Yves coudière : Chiche! December 2021
10 Scientific production
10.1 Major publications
- 1 articleConvergence of discrete duality finite volume schemes for the cardiac bidomain model.Networks and Heterogeneous Media622011, 195-240
- 2 articleA mathematical model of Purkinje-Muscle Junctions.Mathematical Biosciences and Engineering842011, 915-930
- 3 articleExistence And Uniqueness Of The Solution For The Bidomain Model Used In Cardiac Electrophysiology.Nonlinear Anal. Real World Appl.1012009, 458-482URL: http://hal.archives-ouvertes.fr/hal-00101458/fr
- 4 articleA 2D/3D Discrete Duality Finite Volume Scheme. Application to ECG simulation.International Journal on Finite Volumes612009, URL: http://hal.archives-ouvertes.fr/hal-00328251/fr
- 5 articleStability And Convergence Of A Finite Volume Method For Two Systems Of Reaction-Diffusion Equations In Electro-Cardiology.Nonlinear Anal. Real World Appl.742006, 916--935URL: http://hal.archives-ouvertes.fr/hal-00016816/fr
- 6 articleThe Early Repolarization Pattern; A Consensus Paper.Journal of the American College of Cardiology66resulting from the Symposium on J Wave Patterns and a J Wave Syndrome, Glasgow, August 2013. Defines terminology for the J point: Jo, Jp, Jt. Provisionally accepted 19 May 2015.2015, 470-477URL: http://dx.doi.org/10.1016/j.jacc.2015.05.033
- 7 articleReduced Sodium Current in the Lateral Ventricular Wall Induces Inferolateral J-Waves.Front Physiol7365August 2016
- 8 articlePreconditioning the bidomain model with almost linear complexity.Journal of Computational Physics2311January 2012, 82--97URL: http://www.sciencedirect.com/science/article/pii/S0021999111005122
10.2 Publications of the year
International journals
International peer-reviewed conferences
Scientific book chapters
10.3 Cited publications
- 28 articleConvergence of discrete duality finite volume schemes for the cardiac bidomain model.Networks and Heterogeneous Media622011, 195-240
- 29 inproceedingsModélisation et simulation de l'électrophysiologie cardiaque à l'échelle microscopique.43e Congrès National d'Analyse Numérique (CANUM)oral presentationSMAIObernai, Alsace, FranceMay 2016, URL: http://smai.emath.fr/canum2016/resumesPDF/peb/Abstract.pdf
- 30 miscTheoretical and Numerical Study of Cardiac Electrophysiology Problems at the Microscopic Scale..PosterJuly 2016
- 31 inproceedingsA Three-Dimensional Computational Model of Action Potential Propagation Through a Network of Individual Cells.Computing in Cardiology 2017Rennes, FranceSeptember 2017, 1-4
- 32 inproceedingsMicroscopic Simulation of the Cardiac Electrophysiology: A Study of the Influence of Different Gap Junctions Models.Computing in CardiologyMaastricht, NetherlandsSeptember 2018
- 33 articleImpact of Septal Radiofrequency Ventricular Tachycardia Ablation; Insights From Magnetic Resonance Imaging.Circulation1302014, 716-718
- 34 inproceedingsDo we need to enforce the homogeneous Neuman condition on the Torso for solving the inverse electrocardiographic problem by using the method of fundamental solution ?Computing in Cardiology 201643Computing in Cardiology 2016Vancouver, CanadaSeptember 2016, 425-428
- 35 articleA conduction velocity adapted eikonal model for electrophysiology problems with re-excitability evaluation.Medical Image Analysis43January 2018, 186-197
- 36 articleOptimal monodomain approximations of the bidomain equations used in cardiac electrophysiology.Mathematical Models and Methods in Applied Sciences246February 2014, 1115-1140
- 37 articleA 2D/3D Discrete Duality Finite Volume Scheme. Application to ECG simulation.International Journal on Finite Volumes612009, URL: http://hal.archives-ouvertes.fr/hal-00328251/fr
- 38 articleStability And Convergence Of A Finite Volume Method For Two Systems Of Reaction-Diffusion Equations In Electro-Cardiology.Nonlinear Anal. Real World Appl.742006, 916--935URL: http://hal.archives-ouvertes.fr/hal-00016816/fr
- 39 articleA domain decomposition strategy for a very high-order finite volumes scheme applied to cardiac electrophysiology.Journal of Computational Science372019, 101025URL: http://www.sciencedirect.com/science/article/pii/S1877750319303862
- 40 articleModelling the action potential propagation in a heart with structural heterogeneities: From high-resolution MRI to numerical simulations.International Journal for Numerical Methods in Biomedical EngineeringFebruary 2020, e3322
- 41 articleSpatially Coherent Activation Maps for Electrocardiographic Imaging.IEEE Transactions on Biomedical Engineering64May 2017, 1149-1156
- 42 inproceedingsEpicardial Fibrosis Explains Increased Transmural Conduction in a Computer Model of Atrial Fibrillation .Computing in CardiologyVancouver, CanadaSeptember 2016
- 43 articleSudden cardiac arrest associated with early repolarization.N. Engl. J. Med.3582008, 2016--2023
- 44 articleSpontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins.N. Engl. J. Med.3391998, 659-666
- 45 articleMechanism of Right Precordial ST-Segment Elevation in Structural Heart Disease: Excitation Failure by Current-to-Load Mismatch.Heart Rhythm72010, 238-248URL: http://dx.doi.org/10.1016/j.hrthm.2009.10.007
- 46 articleA microstructural model of reentry arising from focal breakthrough at sites of source-load mismatch in a central region of slow conduction.Am. J. Physiol. Heart Circ. Physiol.3062014, H1341-1352
- 47 inproceedingsA new ECG-based method to guide catheter ablation of ventricular tachycardia.iMAging and eLectrical TechnologiesUppsala, SwedenApril 2018
- 48 articlePreconditioning the bidomain model with almost linear complexity.Journal of Computational Physics2311January 2012, 82--97URL: http://www.sciencedirect.com/science/article/pii/S0021999111005122
-
49
articleA Comparison of monodomain and bidomain
-diffusion models for action potential propagation in the human heart.IEEE Transactions on Biomedical Engineering53122006, 2425-2435URL: http://dx.doi.org/10.1109/TBME.2006.880875 - 50 articleCardiac Anisotropy in Boundary-Element Models for the Electrocardiogram.Medical and Biological Engineering and Computing472009, 719--729URL: http://dx.doi.org/10.1007/s11517-009-0472-x
- 51 inproceedingsAnatomically-induced Fibrillation in a 3D model of the Human Atria.Computing in CardiologyMaastricht, NetherlandsSeptember 2018
- 52 miscRegional conduction slowing can explain inferolateral J waves and their attenuation by sodium channel blockers.PosterSeptember 2016
- 53 articleScalable and Accurate ECG Simulation for Reaction-Diffusion Models of the Human Heart.Frontiers in Physiology9April 2018, 370
- 54 articleA Cell-Based Framework for Numerical Modeling of Electrical Conduction in Cardiac Tissue.Front. Phys.5This is very very similar to what we were doing with PEB... Looks like we're scooped at least for the approach, but we do have a few abstracts ([becue:cinc17], [becue16a], MMCE meeting in Ottawa November 2017). Note it's in Frontiers in (Biomedical) Physics, not Physiology.2017, 48

