Keywords
Computer Science and Digital Science
- A3.1. Data
- A3.2. Knowledge
- A3.3.2. Data mining
- A3.3.3. Big data analysis
- A3.4. Machine learning and statistics
- A6.1. Methods in mathematical modeling
- A6.2. Scientific computing, Numerical Analysis & Optimization
- A8.2. Optimization
- A9.8. Reasoning
Other Research Topics and Application Domains
- B1.1.7. Bioinformatics
- B1.1.10. Systems and synthetic biology
- B3. Environment and planet
1 Team members, visitors, external collaborators
Research Scientists
- David Sherman [Team leader, Inria, Senior Researcher, HDR]
- Pascal Durrens [CNRS, Researcher, HDR]
- Alain Franc [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Senior Researcher, until Apr 2021, HDR]
- Clémence Frioux [Inria, Researcher]
- Simon Labarthe [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Researcher]
Post-Doctoral Fellows
- Guillaume Ravel [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, until Oct 2021]
- Pablo Ugalde Salas [Inria, from Nov 2021]
PhD Students
- Mohamed Anwar Abouabdallah [Inria]
- Maxime Lecomte [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement]
Technical Staff
- Ariane Badoual [Inria, Engineer]
- Philippe Chaumeil [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Engineer, until Apr 2021]
- Jean-Marc Frigerio [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Engineer]
- Franck Salin [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Engineer]
Interns and Apprentices
- Johan Baric-Monzat [Inria, Jan 2021]
- Mathieu Bolteau [Inria, from Feb 2021 until Aug 2021]
- Samuel Dutron [Inria, Jan 2021]
Administrative Assistants
- Catherine Cattaert Megrat [Inria, from Aug 2021]
- Roweida Mansour El Handawi [Inria, until Sep 2021]
External Collaborator
- Alain Franc [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, from May 2021, HDR]
2 Overall objectives
Diversity, evolution, and inheritance form the heart of modern biological thought. Modeling the complexity of biological systems has been a challenge of theoretical biology for over a century 57 and flourished with the evolution of data for describing biological diversity, most recently with the transformative development of high-throughput sequencing. However, most concepts and tools in ecology and population genetics for capitalizing on this wealth of data are still not adapted to high throughput data production. A better connection between high-throughput data production and tool evolution is highly needed: computational biodiversity.
Paradoxically, diversity emphasizes differences between biological objects, while modeling aims at unifying them under a common framework. This means that there is a limit beyond which some components of diversity cannot be mastered by modeling. We need efficient methods for recognizing patterns in diversity, and linking them to patterns in function. It is important to realize that diversity in function is not the same as coupling observed diversity with function. Diversity informs both the study of traits, and the study of biological functions (Figure 1). The double challenge is to measure these links quickly and precisely with pattern recognition, and to explore the relations between diversity in traits and diversity in function through modeling.
Figure contains three words, “diversity”, “function”, and “traits” in a triangle, with double-ended arrows between paris of words
Pleiade links recognition of patterns, classes, and interactions with applications in biodiversity studies and biotechnology. We develop distance methods for NGS datasets at different levels of organization: between genomes, between individual organisms, and between communities; and develop high-performance pattern recognition and statistical learning techniques for analyzing the resulting point clouds. We refine inferential methods for building hierarchical models of networks of cellular functions, exploiting the mathematical relations that are revealed by large-scale comparison of related genomes and their models. NGS analysis outputs are further used to inform multi-scale models of population dynamics, with a special focus on microbes communities. These models need in turn new methodological improvements (machine learning, inference, PDE simplification) to fully integrate multi-omics data. We combine these methods into integrated e-Science solutions to place these tools directly in the hands of biologists.
Our methodology (Figure 2) is designed pragmatically to advance the state of the art in applications from biodiversity and biotechnology: molecular based systematics and community ecology, annotation and modeling for biotechnology.
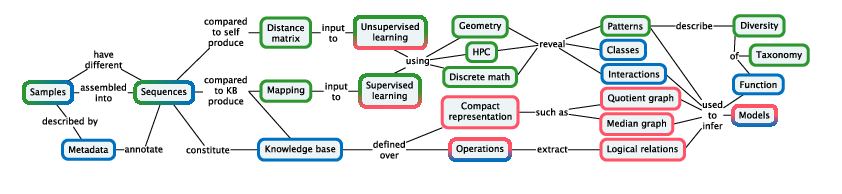
Figure is a bipartite concept map, linking concepts in rounded boxes by means of named relations.
3 Research program
3.1 A Geometric View of Diversity
Diversity may be studied as a set of dissimilarities between objects. The underlying mathematical construction is the notion of distance. Knowing a set of objects, it is possible, after computation of pairwise distances, or sometimes dissimilarities, to build a Euclidean image of it as a point cloud in a space of relevant dimension. Then, diversity can be associated with the shape of the point cloud. The human eye is often far better than an algorithm at recognizing a pattern or shape. One objective of our project is to narrow the gap between the story that a human eye can tell, and that an algorithm can tell. Several directions will be explored. First, this requires mastering classical tools in dimension reduction, mainly algebraic tools (PCA, NGS, Isomap, eigenmaps, etc ...). Second, neighborhoods in point clouds naturally lead to graphs describing the neighborhood networks. There is a natural link between modular structures in distance arrays and communities on graphs. Third, points (representing, say, DNA sequences) are samples of diversity. Dimension reduction may show that they live on a given manifold. This leads to geometry (differential or Riemannian geometry). It is expected that some properties of the manifold can tell something of the constraints on the space where measured individuals live. The connection between Riemannian geometry and graphs, where weighted graphs are seen as mesh embedded in a manifold, is currently an active field of research 56, 55. See as well 58 for a link between geometric structure, linear and nonlinear dimensionality reduction.
Figure shows three views of a point cloud with varying local densities with colored islands
Biodiversity and high-performance computing: Most methods and tools for characterizing diversity have been designed for datasets that can be analyzed on a laptop, but NGS datasets produced for metabarcoding are far too large. Data analysis algorithms and tools must be revisited and scaled up. We will mobilize both distributed algorithms like the Arnoldi method and new algorithms, like random projection or column selection methods, to build point clouds in Euclidean spaces from massive data sets, and thus to overcome the cubic complexity of computation of eigenvectors and eigenvalues of very large dense matrices. We will also link distance geometry 49 with convex optimization procedures through matrix completion 38, 40.
Intercalibration: There is a considerable difference between supervised and unsupervised clustering: in supervised clustering, the result for an item is independent from the result for an item , whereas in unsupervised clustering, the result for an item (e.g. the cluster it belongs to, and its composition) depends on nearby items . Which means that the result may change if some items are added to or subtracted from the sample. This raises the more global problem of how to merge two studies to yield a more comprehensive view of biodiversity?
See 44 for some of our recent work linking the distance geometry problem, nonlinear mapping, and weighted least-squares scaling.
3.2 Knowledge Management for Biology
The heterogenous data generated in computational molecular biology and ecology are distinguished not only by their volume, but by the richness of the many levels of interpretation that biologists create. The same nucleic acid sequence can be seen as a molecule with a structure, a sequence of base pairs, a collection of genes, an allele, or a molecular fingerprint. To extract the maximum benefit from this treasure trove we must organize the knowledge in ways that facilitate extraction, analysis, and inference. Our focus has been on the efficient representation of relations between biological objects and operations on those representations, in particular heuristic analyses and logical inference.
Pleiade will develop applications in comparative genomics of related organisms, using new mathematical tools for representing compactly, at different scales of difference, comparisons between related genomes. New methods based on distance geometry will refine these comparisons. Compact representations can be stored, exchanged, and combined. They will form the basis of new simultaneous genome annotation methods, linked directly to abductive inference methods for building functional models of the organisms and their communities.
Since a goal of Pleiade is to integrate diversity throughout the analysis process, it is necessary to incorporate diversity as a form of knowledge that can be stored in a knowledge base. Diversity can be represented using various compact representations, such as trees and quotient graphs storing nested sets of relations. Extracting structured representations and logical relations from integrated knowledge bases (Figure 2) will require domain-specific query methods that can express forms of diversity.
3.3 Community-scale metabolic modeling
The emergent metabolism of microbial communities can be qualitatively modeled using a boolean approximation of metabolic dynamics7. In this approach the behavior of the system is described by logical rules that activate a given reaction as soon as its substrates become available; numerical parameters such as stochiometry or enzyme kinetics are ignored in favor of graph topology and paths. The advantage is that such qualitative models, unlike quantitative methods such as flux balance analysis, do not require the assumption that the system is stationary and can model systems where cells are constantly growing or constantly reproducing.
Network expansion, introduced in 43 as a recursive traversal of the structure of a metabolic graph, lends itself to concise definition using answer set programming48 and thus to efficient implementation using SAT solvers 47. In practice, using ASP for metabolic modeling makes it possibe to define both the activation of metabolic reactions in different conditions, and the constraints and optimizations needed to find solutions in a combinatorically large state space.
We focus in particular on the key question of determining minimal communities, subsets of the organisms present in an environment that are sufficient to produce a chosen behavior 45. The methodological goal here is to identify key species in a community through use of ASP to rapidly explore the state space and thus, through heuristic resolution of combinatorial problems, provide the guarantees an exhaustive search with a greatly reduced computational cost 4.
3.4 Multi-scale omic-based models of microbial comunities
Functional and taxonomic diversities, beyond intrinsic specificities encoded in the genetic material, are also strongly shaped by their environment. Spatial nutritional niches, microbial interactions and abiotic constraints lead to complex spatial structures in the microbial community that impact its overall dynamics. PDE-based models of the microbiota in its environment allow to include in the model these multiple mechanisms in order to decipher their influence on the community faith.
The main methodological developments are related to mathematical modeling (in particular the correct level of simplification in the multi-physic description of the microbial environment), model simplification (asymptotic approximation), inference from multi-omics data (including dimension reduction, statistical learning) and numerical developments (in particular fast approximation of metabolic models with machine learning methods). Strong interactions with community-scale metabolic models as developed in section 3.3 are sought, specially for multi-omics inference and knowledge-based machine learning constraints.
The goal is to achieve accurate models of microbial communities that could be used as digital twins of controlled experiments in microbial ecology. Culturomic facilities allow for the acquisition of multi-omics time-series in controlled conditions useful to build and fit population dynamics models, that can be used in turn to explore numerically biological assumptions, to help in experimental planing and data analysis.
3.5 Modeling by successive refinement
Describing the links between diversity in traits and diversity in function will require comprehensive models, assembled from and refining existing models. A first level of refinement is inferring a new model for a specific organism, on the basis of an annotated projection and knowledge of genome-to-genome relations (figure 4).
Beyond that, a recurring difficulty in building comprehensive models of biological systems is that accurate models for subsystems are built using different formalisms and simulation techniques, and hand-tuned models tend to be so focused in scope that it is difficult to repurpose them 36. Our belief is that a sustainable effort in building efficient behavioral models must proceed incrementally, rather than by modeling individual processes de novo. Hierarchical modeling33 is one way of combining specific models into networks. Effective use of hierarchical models requires both formal definition of the semantics of such composition, and efficient simulation tools for exploring the large space of complex behaviors. We have previously shown that this approach can be effective for certains kinds of systems in biotechnology 2, 37 and medicine 35. Our challenge is to adapt incremental, hierarchical refinement to modeling organisms and communities in metagenomic and comparative genomic applications.
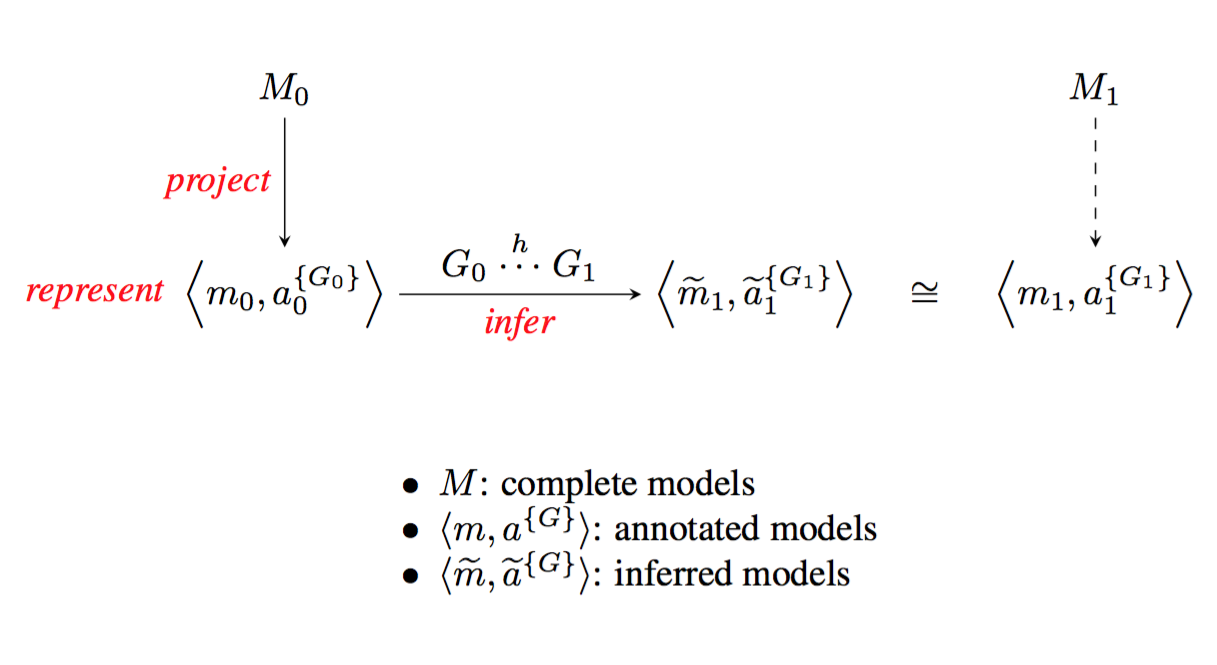
Figure shows M 0 and M 1 terms at the top left and top right, linked vertically by arrows labelled “project” to the bracketed expressions in the caption, linked left to right by a right-pointing arrow labelles “infer”.
4 Application domains
4.1 Genome and transcriptome annotation, to model function
Sequencing genomes and transcriptomes provides a picture of how a biological system can function, or does function under a given physiological condition. Simultaneous sequencing of a group of related organisms is now a routine procedure in biological laboratories for studying a behavior of interest, and provides a marvelous opportunity for building a comprehensive knowledge base of the relations between genomes1, 11. Key elements in mining these relations are: classifying the genes in related organisms and the reactions in their metabolic networks, recognizing the patterns that describe shared features, and highlighting specific differences.
Pleiade develops applications in comparative genomics of related organisms, using new mathematical tools for representing compactly, at different scales of difference, comparisons between related genomes. New methods based on computational geometry refine these comparisons. Compact representations can be stored, exchanged, and combined. They will form the basis of new simultaneous genome annotation methods, linked directly to abductive inference methods for building functional models of the organisms and their communities.
Our ambition in biotechnology is to permit the design of synthetic or genetically selected organisms at an abstract level, and guide the modification or assembly of a new genome. Our effort is focused on two main applications: genetic engineering and synthetic biology of oil-producing organisms (biofuels in CAER, palm oils), and improving and selecting starter microorganisms used in winemaking (collaboration with the ISVV and the BioLaffort company).
4.1.1 Oil Palm lipid synthesis
The largest source of vegetable oil 1 is the fruit mesocarp of the oil palm Elaeis guineensis, a remarkable tissue that can accumulate up to 90% oil, the highest level observed in the plant kingdom. The market share of oil palm is expected to increase in order to meet increased demand for vegetable oil, predicted to double by 2030 41, be it as food or as a source of biofuels in Africa. A significant proportion of palm oil is produced on small estates that do not have access to efficient milling facilities, and run a great risk of spoilage through oil acidification. Improving palm oil quality through genetics and selection will result in economic gains 51 by addressing several targets such as improvement of oil yield, tuning of oil quality through the rate of unsaturated fatty acids or impairment of degradation processes. Furthermore, as genome biodiversity resides mostly in Africa, oil from African oil palms can vary greatly in fatty acid composition according to cultivar genetic differences and to weather conditions, and the precise mechanisms regulating this variability are not yet understood.
A growing body of molecular resources for studying oil palm fruit are making it possible to study and improve the quality and quantity of oil produced by oil palms. In particular, these oils can vary greatly in fatty acid composition, and while the precise mechanisms regulating this variability are not completely understood, establishing a link between oil palm genotype and phenotype appears increasingly feasible. Pleiade will work with the CNRS/UB UMR 5200 (LBM), a laboratory with an established reputation in studying fatty acid metabolism in E. guineensis, to improve understanding of the links between genetic diversity and oil production, and participate in developing applications.
4.2 Molecular based systematics and taxonomy
Defining and recognizing the myriads of species occuring in the biosphere has been the focus of phenomenal energy over the past centuries and remains a major goal of Natural History. It is an iconic paradigm in pattern recognition (clustering has coevolved with numerical taxonomy many decades ago). Developments in evolution and molecular biology, as well as in data analysis, have over the past decades enabled a profound revolution, where species can be delimited and recognized by data analysis of sequences. We aim at proposing new tools, in the framework of E-science, which make possible () better exploration of the diversity in a given clade, and () assignment of a place in these patterns for new, unknown organisms, using information provided by sets of sequences. This will require investment in data analysis, machine learning, and pattern recognition to deal with the volumes of data and their complexity.
One example of this project is about the diversity of trees in Amazonian forest, in collaboration with botanists in French Guiana. Protists (unicellular Eukaryots) are by far more diverse than plants, and far less known. Molecular exploration of Eukaryotes diversity is nowadays a standard in biodiversity studies. Data are available, through metagenomics, as an avalanche and make molecular diversity enter the domain of Big Data. Hence, an effort will be invested, in collaboration with other INRIA teams (GenScale, HiePACS) for porting to HPC algorithms of pattern recognition and machine learning, or distance geometry, for these tools to be available as well in metagenomics. This will be developed first on diatoms (unicellular algae) in collaboration with INRAE team at Thonon and University of Uppsala, on pathogens of tomato and grapewine, within an existing network, and on bacterial communities, in collaboration with University of Pau. For the latter, the studies will extend to correlations between molecular diversity and sets of traits and functions in the ecosystem.
4.3 Community ecology and population genetics
Community assembly models how species can assemble or diassemble to build stable or metastable communities. It has grown out of inventories of countable organisms. Using metagenomics one can produce molecular based inventories at rates never reached before. Most communities can be understood as pathways of carbon exchange, mostly in the form of sugar, between species. Even a plant cannot exist without carbon exchange with its rhizosphere. Two main routes for carbon exchange have been recognized: predation and parasitism. In predation, interactions–even if sometimes dramatic–may be loose and infrequent, whereas parasitism requires what Claude Combes has called intimate and sustainable interactions 42. About one decade ago, some works 53 have proposed a comprehensive framework to link the studies of biodiversity with community assembly. This is still incipient research, connecting community ecology and biogeography.
We aim at developping graph-based models of co-occurence between species from NGS inventories in metagenomics, i.e. recognition of patterns in community assembly, and as a further layer to study links, if any, between diversity at different scales and community assemblies, starting from current, but oversimplified theories, where species assemble from a regional pool either randomly, as in neutral models, or by environmental filtering, as in niche modeling. We propose to study community assembly as a multiscale process between nested pools, both in tree communities in Amazonia, and diatom communities in freshwaters. This will be a step towards community genomics, which adds an ecological flavour to metagenomics.
Convergence between the processes that shape genetic diversity and community diversity–drift, selection, mutation/speciation and migration–has been noted for decades and is now a paradigm, establishing a continuous scale between levels of diversity patterns, beyond classical approaches based on iconic levels like species and populations. We will aim at deciphering diversity pattern along these gradients, connecting population and community genetics. Therefore, some key points must be adressed on reliability of tools.
Next-generation sequencing technologies are now an essential tool in population and community genomics, either for making evolutionary inferences or for developing SNPs for population genotyping analyses. Two problems are highlighted in the literature related to the use of those technologies for population genomics: variable sequence coverage and higher sequencing error in comparison to the Sanger sequencing technology. Methods are developed to develop unbiased estimates of key parameters, especially integrating sequencing errors 50. An additional problem can be created when sequences are mapped on a reference sequence, either the sequenced species or an heterologous one, since paralogous genes are then considered to be the same physical position, creating a false signal of diversity 46. Several approaches were proposed to correct for paralogy, either by working directly on the sequences issued from mapped reads 46 or by filtering detected SNPs. Finally, an increasingly popular method (RADseq) is used to develop SNP markers, but it was shown that using RADseq data to estimate diversity directly biases estimates 34. Workflows to implement statistical methods that correct for diversity biases estimates now need an implementation for biologists.
5 Social and environmental responsibility
5.1 Footprint of research activities
Pleiade uses high-performance computing resources as an integral part of its research program.
6 Highlights of the year
Alain Franc authored a book on Tensor Ranks 26, released as an Inria Research Report and on ArXiv.
Pleiade members contributed to a large meta-analysis of more than 5,000 metagenomic samples associated to the human faecal microbiome in 13. Temporal, geographical and family associations of persistently occurring species suggest explanations of their behavior with respect to human hosts.
Pleiade members curated and optimized metabolic models of bacterial communities contributing to organoleptic qualities of cheese. A model of the microbial community dynamics was developed and compared to metabolomics data, revealing the qualitative and quantitative impact of community association compared to individual culture 23.
7 New software and platforms
7.1 New software
7.1.1 Metage2Metabo
-
Keywords:
Metabolic networks, Microbiota, Metagenomics, Workflow
-
Scientific Description:
Flexible pipeline for the metabolic screening of large scale microbial communities described by reference genomes or metagenome-assembled genomes. The pipeline comprises several main steps. (1) Automatic and parallel reconstruction of metabolic networks. (2) Computation of individual metabolic potentials (3) Computation of collective metabolic potential (4) Calculation of the cooperation potential described as the set of metabolites producible by species only in a cooperative context (5) Computation of minimal-sized communities sastifying a metabolic objective (6) Extraction of key species (essential and alternative symbionts) associated to a metabolic function
-
Functional Description:
Metabolic networks are graphs which nodes are compounds and edges are biochemical reactions. To study the metabolic capabilities of microbiota, Metage2Metabo uses multiprocessing to reconstruct metabolic networks at large-scale. The individual and collective metabolic capabilities (number of compounds producible) are computed and compared. From these comparisons, a set of compounds only producible by the community is created. These newly producible compounds are used to find minimal communities that can produce them. From these communities, the keytstone species in the production of these compounds are identified.
-
News of the Year:
(1) Improvements of the pipeline and its continuous integration (2) Release of version 1.5.0 (3) Development of m2m-analysis subpipeline
- URL:
- Publication:
-
Contact:
Clemence Frioux
-
Participants:
Clemence Frioux, Arnaud Belcour, Anne Siegel
7.1.2 MiSCoTo
-
Name:
Microbiota Screening and COmmunity Selection with TOpology
-
Keywords:
Metabolic networks, ASP - Answer Set Programming, Logic programming
-
Scientific Description:
MiSCoTo solves combinatorial problems using Answer Set Programming. It aims at minimizing either the number of selected species or both the number of selected species and the cost of the interaction between them, characterized by the number of metabolic exchanges. In the first case, the level of modeling is called lumped or mixed-bag, in the latter, it is compartmentalized.
-
Functional Description:
Metabolic networks are composed of biochemical reactions and gather the expected metabolic capabilities of species. For organisms that live in interaction altogether (microbiotas), complementarity between these networks can be exploited to predict cooperation events. This software takes as inputs metabolic networks for various species (host, symbionts of the microbiota), components of the growth medium and a metabolic objective (metabolites to be produced), and aims at selecting a minimal set of symbionts to ensure the metabolic objective can be achieved. The software can use two types of modelings: a simplified one and another that takes into account the cost of metabolic exchanges and aims at minimizing it.
-
Release Contributions:
Memory usage optimization. Fix issues with input file formats.
-
News of the Year:
(1) Release of version 3.1.1 (2) New functionality: miscoto-focus determines the metabolic potential of symbionts of interest in the community
- URL:
- Publication:
-
Contact:
Clemence Frioux
-
Participants:
Clemence Frioux, Anne Siegel, Enora Fremy, Camille Trottier, Arnaud Belcour
7.1.3 MeneTools
-
Name:
Metabolic networks Topological tools
-
Keywords:
Metabolic networks, Graph, Topology, Bioinformatics, Systems Biology, ASP - Answer Set Programming
-
Scientific Description:
MeneTools are a set of tools for the exploration of the producibility potential in a metabolic network using the network expansion algorithm. The MeneTools can: - assess whether targets are producible starting from nutrients (Menecheck) - get all compounds that are producible starting from nutrients (Menescope) - get all reactions that are activable from nutrients (Meneacti) - get production paths of specific compounds (Menepath) - obtain compounds that if added to the nutrients, would ensure the producibility of targets (Menecof) - identify metabolic deadends, i.e. metabolites that act as reactants of reactions but never as products, or metabolites that act as products of reactions but never as reactants. This is a purely structural analysis. All MeneTools using modelling follow the producibility in metabolic networks as defined by the network expansion algorithm.
-
Functional Description:
MeneTools consists in four topological tool to analyze metabolic models in a graph-based perspective. Menecheck verifies the producibility of target compounds from available substrates (growth medium) of the metabolic network. Menescope gives the whole range of accessible compounds in the metabolic network starting from substrates. Menepath give the production paths of given compounds in the model. Menecof proposes compounds that need to be produced or added as substrate for ensuring the producibility of targets.
-
News of the Year:
(1) Release of version 3.2.0 (2) New functionality: mene-seed identifies external compounds from the topology of the network
- URL:
- Publications:
-
Contact:
Clemence Frioux
-
Participants:
Clemence Frioux, Anne Siegel, Arnaud Belcour
7.1.4 Fluto
-
Keywords:
ASP - Answer Set Programming, Answer Set Programming, Metabolic networks, Flux Balance Analysis, Linear programming
-
Scientific Description:
Fluto performs metabolic network completion with respect to topological and linear reaction rate constraints based on the stoichiometry of metabolic reactions.
-
Functional Description:
Fluto relies on Answer Set Programming (ASP) and a hybrid modelling that associates to ASP a Linear Programming (LP) constraint propagator. Models satisfying the qualitative constraints of network expansion are tested for satisfiability of flux constraints with the LP propagator. Resulting answer sets permit the completion of a metabolic network that ensures the metabolic reaction of interest is activated according to both formalisms.
-
News of the Year:
Reorganisation of the code. Implementation of continuous integration. Addition of the Sagot & Acuna formalism in the software.
- URL:
- Publications:
-
Contact:
Clemence Frioux
-
Participant:
Sven Thiele
-
Partners:
Max Planck Institute Magdeburg, University of Potsdam
7.1.5 Emapper2GBK
-
Keywords:
Bioinformatics, Metabolic networks, Functional annotation
-
Functional Description:
Starting from FASTA and Eggnog-mapper annotation files, Emapper2GBK builds a GBK file that is suitable for metabolic network reconstruction with Pathway Tools, and adds the GO terms and EC numbers annotations in the GenBank file.
- URL:
- Publication:
-
Contact:
Clemence Frioux
-
Participants:
Clemence Frioux, Arnaud Belcour, Anne Siegel
7.1.6 Biodiversiton
-
Name:
Biodiversiton
-
Keywords:
Biodiversity, Comparative metagenomics, Clustering, Dimensionality reduction, Masses of data
-
Functional Description:
Biodiversiton is a suite of tools for biodiversity composed by Rsyst, pairwise_dis, diagno_syst, and yapotu. The global project provides tutorials, datasets, and a readme for the whole suite.
- URL:
-
Authors:
Alain Franc, Jean-Marc Frigerio, Franck Salin
-
Contact:
Alain Franc
7.1.7 Rsyst
-
Name:
Rsyst
-
Keywords:
Biodiversity, Metagenomics, Clustering, Dimensionality reduction, Masses of data
-
Functional Description:
Contains the R-Syst databases, in sqlite format, as well as python programs for querying them through a python interface for the most common queries.
- URL:
-
Authors:
Jean-Marc Frigerio, Franck Salin, Alain Franc
-
Contact:
Alain Franc
-
Partner:
INRAE
7.1.8 pydiodon
-
Name:
Pydiodon
-
Keywords:
Dimensionality reduction, Data analysis
-
Functional Description:
Most dimension reduction methods inherited from Multivariate Data Analysis, and currently implemented as elements in statistical learning for handling very large datasets (meaning the dimension of spaces is the number of features), rely on a chain of pretreatments, a core with a SVD for low rank approximation of a given matrix, and a post-treatment for interpreting results. The costly part in computations is the SVD, which is in cubic complexity. Diodon is a list of functions and drivers which implement (i) pre-treatments, SVD and post-treatments on a large diversity of methods, (ii) random projection methods for running the SVD which permits to bypass the time limit in computing the SVD, and (iii) an implementation in C++ of the SVD with random projection at prescribed rank or precision, connected to MDS.
Pydiodon is a deliverable of the ADT Diodon (see https://gitlab.inria.fr/diodon) which will provide an API in python (pydiodon) and C++ (cppdiodon), the former developed by Pleiade with the SED, the latter developped by the SED with Hiepacs (connections with FMR).
-
News of the Year:
In 2020, ADT Diodon has started with a fresh version of diodon as a starting point: new project in inria gitlab, renamed
- URL:
-
Contact:
Alain Franc
-
Participants:
Alain Franc, Jean-Marc Frigerio, Franck Salin, Florent Pruvost
-
Partner:
INRAE
7.1.9 Yapotu
-
Name:
Yet Another Pipeline for OTU building
-
Keywords:
Metagenomics, Biodiversity, Dimensionality reduction, Masses of data
-
Functional Description:
The main functionalities are as follows: 1) building OTUs from a fasta file (swarm, vsearch, ..) or a distannce file (yapotu) for an environmenal sample 2) building a fasta file and a distance file per OTU 3) checking the consistency of the OTUs by displaying them as a graph (see OTU as a graph below) 4) displaying the shape of an OTU or of a set of OTUs by Multidimensional Scaling 5) implementing Hierachical Aggregative Clustering of an OTU or a set of OTUs with various aggregation methods
-
News of the Year:
Ugraded from an older version, fusion with declic now deprecated, new functionalities for working with massive data sets
- URL:
-
Authors:
Alain Franc, Jean-Marc Frigerio, Franck Salin
-
Contact:
Alain Franc
-
Partner:
INRAE
7.2 New platforms
Participants: David Sherman, Ariane Badoual.
As a founding principle, Pleiade supports reproducible scientific analyses and promotes a declarative approach using reusable software modules, rigorous documentation of data provenance, and systematic recording of workflows. The latter is a challenge when interactive interfaces are used, but can be addressed, to cite two examples, in Galaxy by extracting workflows, and in other systems by using Jupyter notebooks. Part of Pleiade's mission is to automate the deployment of environments that support these goals, for non-technical end users.
Pleiade maintains specific computing resources to support our work and that of our collaborators. There are four main use cases:
- Fast deployment of containerized user environments, combining biological data and databases, software modules specified by version, a CWL executor, and interactive tools including web front ends, notebooks, or Galaxy. A user environment will provide at least one specific HTTPS endpoint, created dynamically. A single researcher may deploy several different environments in the course of one day.
- Support for development and testing of workflows, as above but configured for team members who are developing software modules or interfaces, and who must often deploy several different environments simultaneously.
- Dynamically allocated containerized compute tasks, including both individual analysis steps in workflows and GitLab runner containers used for continuous integration. These tasks arrive in bursts that often cannot be planned in advance.
- Long-running stream preprocessing, a low-priority background task that watches external databases for changes, chooses pertinent data, precomputes representations and ingests them into local data bases.
We support community best practices for reproducible computing in bioinformatics, using biocontainers generated by bioconda, in CWL or Galaxy workflows. For internal use we provide TES endpoints and host JupyterHub environments.
Pleiade's environment is built on OKD 4, the community distribution of Kubernetes developed alongside of RedHat Openshift. OKD4 in particular uses the CRI-O runtime, not Docker, and containers run unprivileged. Software-defined storage and S3 endpoints are provided by Ceph.
8 New results
8.1 Large-scale analyses of microbiome diversity
Participants: Clémence Frioux.
Shotgun metagenomics is becoming a routine analysis for the characterisation of human, animal and environmental microbiotas. Having access to the complete DNA sequences of microorganisms inhabiting an environment constitutes an invaluable resource for deciphering its diversity and organisation. As a result, the growing resources of available metagenomes for a variety of ecosystems make it possible to study the distribution of bacteria and fungi environmental or host-associated microbiomes. In 8, we discussed the different possibilities for assessing the functions of an ecosystem starting from sequences. We evaluated the applicability and pitfalls of metabolic modelling in the context of metagenomes.
We performed a first meta-analysis of metagenomic data in 3 in which more than 13,000 metagenomes from 25 ecosystems were compiled. We demonstrated the differences in bacteria-to-fungi relative abundance ratio between environmental and host-associated microbiotas. We were able to distinguish habitats based on their composition in bacteria and fungi, highlighting differences between environmental habitats, external host and human influenced habitats, and anaerobic habitats like the gut.
In 2021, we performed a large meta-analysis of more than 5,000 metagenomic samples associated to the human faecal microbiome in 13. In this work, we identified groups of species that are particularly persistent in metagenomes. We studied them in the light of temporal, geographical and family associations. Altogether, we proposed strategies that could explain the different behaviours exhibited by species with respect to their human host association.
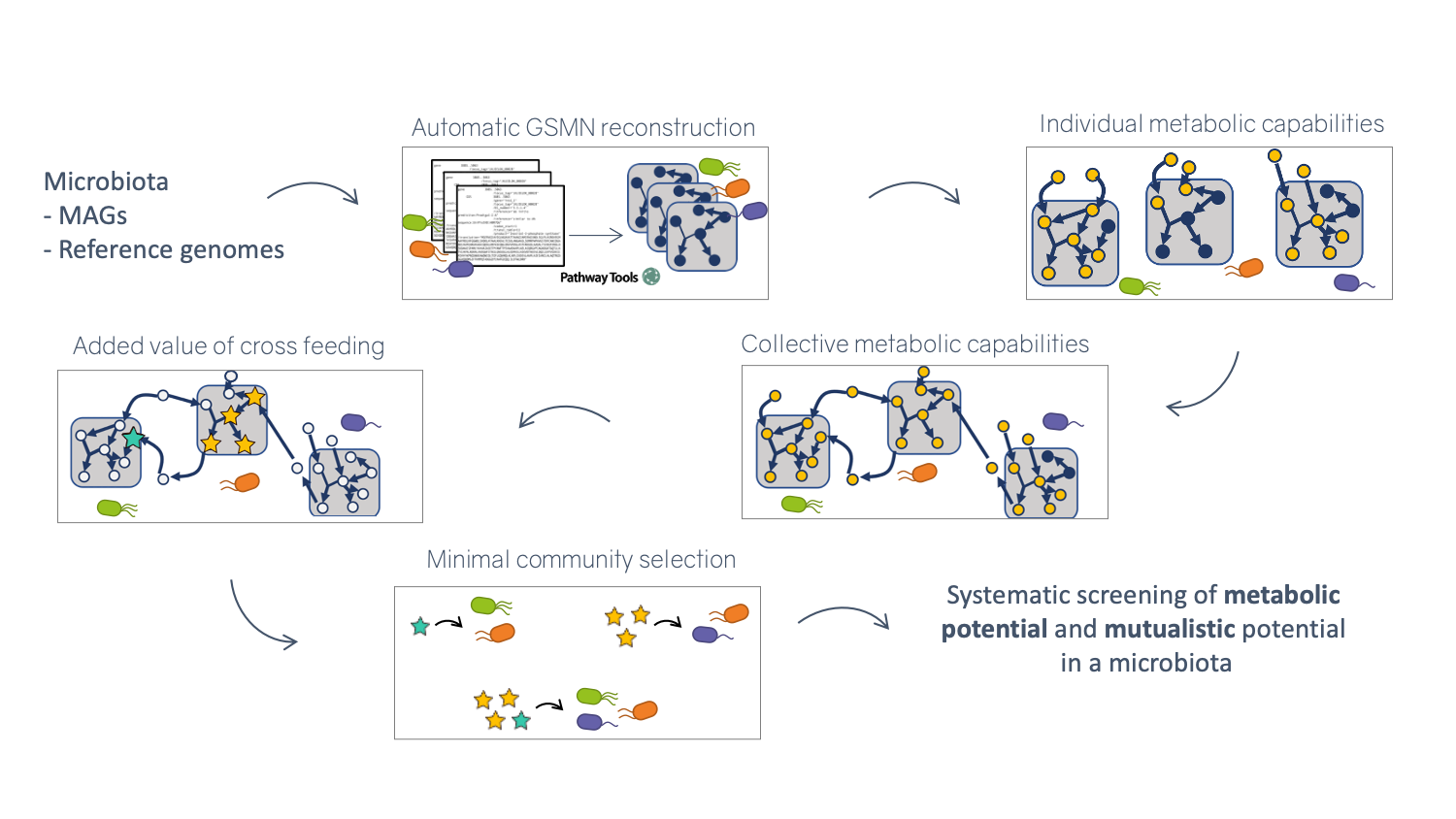
Illustration of the metage2metabo pipeline steps permitting metabolic screening of a microbial community
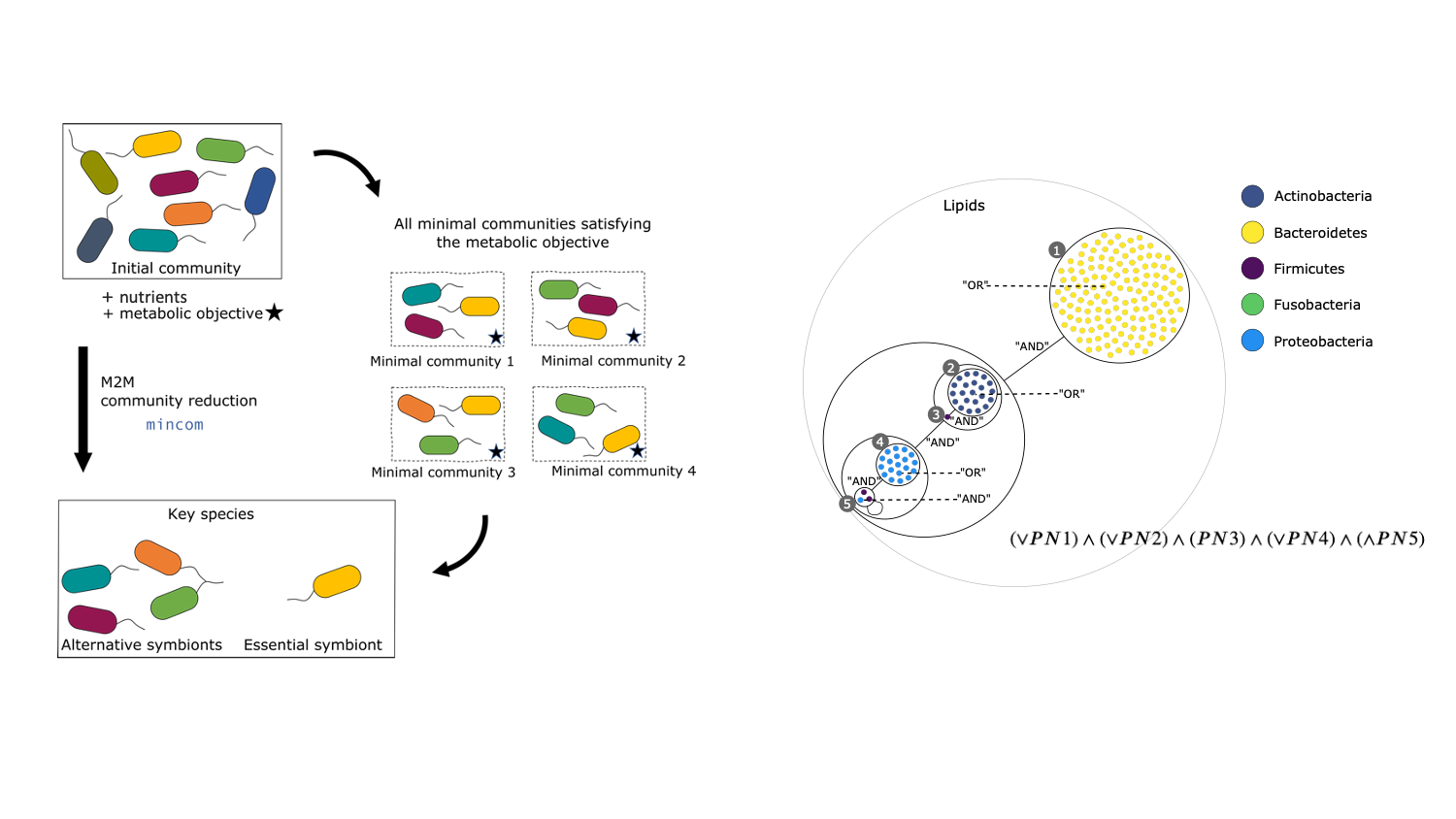
The figure has two parts. Part (a) on the left is a cycle between three boxes, labeled clockwise from the top “initial community”, “all minimal communities satisfying the metabolic objective”, and “key species”. Each box contains individual bacteria. A thick vertical arrow from “initial community” to “key species” is labeled “M2M”. Curved thin unlabeled arrows lead from “initial community” to “minimal communities” and thence to “key species”. Part (b) on the right is a circle containing nested circles in differing numbers, representing inclusions of sets of species. Lines between circles at the same level are labeled with “AND” or “OR”. At the bottom is written a boolean formula.
By assembling and binning the sequencing reads produced in metagenomics, it is possible to obtain metagenome-assembled genomes (MAGs) that can be assigned to taxonomic clades. These MAGs can be used to build predictions of the metabolism of the associated species, and in this way used as a proxy to understand the physiology of the underlying community. Metage2Metabo (M2M), a software system designed in Pleiade, aims at analysing the metabolic complementarity within a microbiota 5 or a large collection of reference genomes, and to identify key species among them. Key species are members of the ecosystem that appear in some (alternative symbionts) or in all (essential symbionts) minimal communities associated to a function (see Figure 6). M2M is a flexible pipeline that automatically performs metabolic network reconstruction from annotated genomes or MAGs, and analyses the resulting networks to capture the metabolic potential of associated species, both individually and as a community (see Figure 5). We demonstrated the applicability of M2M using large-scale collections of genomes and MAGs (see Figure 6 b.), promoting the use of such a screening workflow to screen the metabolism of metagenomes 4. We presented the software and its associated work as a highlight paper in the MPA 19, JOBIM 18 and CMSB 20 conferences in 2021, and as a poster at the iHMC conference 28. In addition, Clémence Frioux presented her work on discrete modelling of metabolic networks to the French community of metabolic modellers in Bordeaux during a dedicated workshop 21.
The developments in ADT MetagenoPic project with Ariane Badoual continued in 2021. This project aims at building a platform suitable for the analysis of raw metagenomic data, and bridging the resulting treated data to our existing methods for metabolic screening of communities (M2M).
Finally, a collaboration with the company Ysopia Bioscience as an Inria Tech contract was set up in 2021, permitting the analysis of metagenomic data and the connection to the metabolic screening provided by Metage2Metabo.
8.2 Modelling the metabolic of the gut microbiome in time and space
Participants: Pablo Ugalde-Salas, Simon Labarthe, Clémence Frioux.
In 2021, an Inria Exploratory Action - SLIMMEST – carried out by Simon Labarthe and Clémence Frioux – was initiated. This project aims at combining discrete reasoning models of metabolism to numerical metamodels and PDEs for the simulation of microbial communities in time and space. The selected methodology is the coupling between PDE-based microbial population dynamics model with metamodels of complex optimizations predicting their metabolism. The main difficulty here is to ensure the scalability of the simulation and the selection of relevant metabolic functions and species to be tracked over time.
The starting point of the project was the participation to the 2021 edition of the CEMRACS event. During five weeks, two PhD students - Julien Martinelli and Thibault Malou - worked on the modeling of the Salmonella infection in the healthy human gut microbiome. Simon Labarthe presented his work on biofilm simulation at the conference 22, and Clémence Frioux presented hers on metabolic modeling.
This project led to the recruitment of Pablo Ugalde-Salas in November, that will develop the coupling between PDE and statistical learning methods. Coralie Muller will join the team in 2022 to work on metabolic exploration.
8.3 Characterization of Molecular Biodiversity
Participants: Mohammed Anwar Abouabdallah, Romain Peressonni, Alain Franc.
In 2021, Pleiade continued the development and refinement of new methods for chacterizing molecular biodiversity. Two approaches are being pursued, each with a PhD student in their third year.
- The central focus of Mohammed Anwar Abouabdallah's PhD is building OTUs from a pairwise distance matrix using Stochastic Block Models (SBM). Building OTUs is traditionally seen as a form of unsupervised clustering. This work is done in collaboration with the MIAT INRAE research unit in Toulouse and HiePACS. It represents a connection between metabarcoding and statistical modeling, a topic which deserves investigation and is expanding (Figure 7 from 52).
- A major goal of Pleiade is to develop a geometric view of biodiversity. The tool selected up to now is to associate a point cloud to a dataset (pairwise distances between sequences) and to study its shape. This approach has expanded and been developed in 2020 as a collaboration with HiePACS through the cosupervision of Romain Peressonni's PhD, which aims to provide new approaches and algorithms for computing distances between two point clouds.
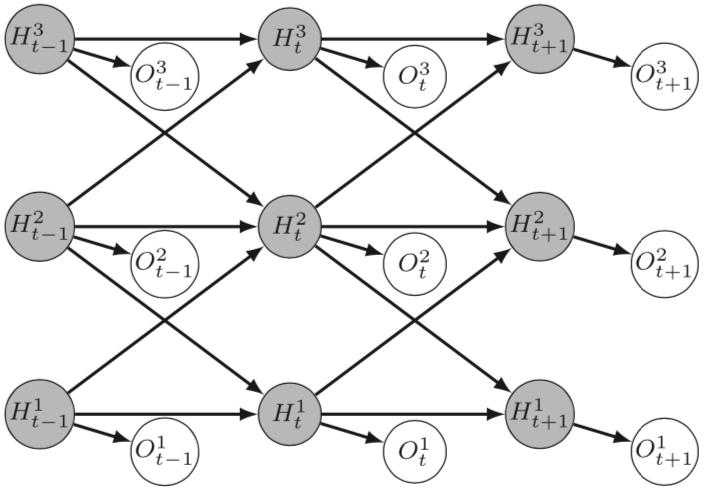
The figure is a 3 by 3 matrix of filled circles labeled with a capital H, with a subscript t-1, t, t+1 in columns left to right, and a superscript 3, 2, 1 in rows from top to bottom. Each circle is connected by a short arrow to an unfilled circle, down and to the right, labeled with a capital O and matching subscripts and superscripts. Between the filled circles are horizontal arrows in the same row, and diagonal arrows to the filled circles in the next column to the right and the neighboring row above or below.
8.4 Dimension Reduction
Participants: Alain Franc, Jean-Marc Frigério.
Metabarcoding is a series of technical procedures to build molecular based inventories from large datasets of amplicons. The underlying information needs to be compacted without losing its information content before it can be further processed with domain-specific tools. This links metabarcoding tools to dimension reduction techniques, which is an important topic in Pleiade. This has been implemented through a participation in following research projects:
- Contribution to and finalization of the ADT Gordon project in Inria BSO. The objective of this project (partners: Tadaam (coordinator), Storm, HiePACS, Pleiade) is to integrate SVD as a tool available in Chameleon, starPU and new Madeleine. The contribution of Pleiade is to bring in metabarcoding as a use case, and random projection as a method for scaling Multidimensional Scaling (which requires an SVD) in collaboration with HiePACS with a template implemented in Diodon. A MDS on a 106 x 106 matrix has been succesfully run at the end of ADT Gordon, on Occigen, in 900 seconds including I/O. The final report has been issued by Tadaam in December 2020.
- A consequence of this involvement is the submission in 2020 of a new ADT, called Diodon, for extending to a diversity of linear dimension reduction techniques what has been aquired in ADT Gordon for MDS, namely a significant progress in speed and memory management brought by random SVD, which can be integrated into a diversity of methods : PCA, CoA, etc.
- Pleiade is involved in the EU project EOSC-Pillar, in a task for better connecting data to calculation, currently data in Inrae Dataverse system connected to tools running on a INRAE local server or on PlaFRIM, on a testbed on biodiversity assessment with metabarcoding. This task in done in collaboration with INRAE DipSO (Direction Science Ouverte) and the Inria HiePACS project-team.
8.5 Multi-omic analysis of a cheese-derived bacterial community
Participants: Maxime Lecomte, David Sherman, Simon Labarthe, Clémence Frioux.
Understanding and controlling the interactions within bacterial communities has applications in multiple industrial domains, among which the food processing industry. The TANGO project, conducted by the INRAE department STLO (Rennes) aimed at following a controlled bacterial community during the process of cheese production. The project also involved studying the impact of changes in production processes on organoleptic properties of the cheese. Multi-omic data was generated all along the experiment, enabling the monitoring of gene expression in bacteria, but also the metabolite production in the cheese.
Pleiade is involved in the processing and integration of the multi-omic data into dynamic models of the communtity metabolism. This project is carried by Maxime Lecomte, PhD student INARE-Inria, in collaboration with Hélène Falentin (INRAE STLO Rennes), Clémence Frioux, Simon Labarthe and David Sherman.
In 2021, metabolic models were curated and optimised with respect to monoculture experimentations. A model of the microbial community dynamics was built, optimised, and predictions were compared to metabolomics data. The analysis of these models highlighted metabolic pathways used by the bacteria in a milk-based environment, and the qualitative and quantitative impacts of the community over the individual metabolisms. Preliminary results were presented by Maxime Lecomte at the MetaboDay, a conference dedicated to the scientists interested in metabolism in Bordeaux 23 and in the poster sessions of the CMSB 32, JOBIM 29 and MPA 31 conferences.
8.6 Metabolic analyses of marine algae
Brown algae, especially the species Ectocarpus siliculosus, are important models for deciphering the complex interactions within marine holobionts, with the goal of studying their metabolism together with the metabolism of the bacteria that inhabit their direct environment. Because performing wet lab experiments on such systems is technocally challenging, there is need for bioinformatic predictive methods for assessing the putative roles and interdependences between species. In parallel, addressing the difficulties brought by the study of these organisms is also a means to enhance and calibrate the tools devlopped in the team. Hence a fruitful collaboration for the past years has been developed with scientists from the Roscoff Biological station.
The recent publication of the genome of Ectocarpu subulatus6, constitutes a valuable resource for future studies that involve this stress-tolerant alga. A direct application of our methods for minimal community selection was performed in 5. In this work, we illustrated how predictions performed with MiSCoTo, a tool developped in the team, can meaningfully suggest metabolic dependencies between an alga and associated bacteria, and can help build controlled communities for laboratory cultures. A review papier summarising how the use of combinatorial optimisation problems such as the one solved in MiSCoTo can be applied to elucidate the physiology of brown algae is available 7. In 14, the impact of the annotation pipeline on the subsequent metabolic modelling and community selection was studied using the bacterial communities associated to Ectocarpus subulatus as an application.
9 Bilateral contracts and grants with industry
9.1 Bilateral contracts with industry
Participants: Clémence Frioux, Éloïse Guillem.
An Inria tech contract was conducted with the biotech Ysopia Bioscience between June and November 2021. This collaboration was carried out by Éloïse Guillem, engineer of the SED department, and Clémence Frioux.10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Inria associate team not involved in an IIL or an international program
Participants: Clémence Frioux, Maxime Lecomte.
Pleiade is a member of the SymBioDiversity associated team with the Mathomics department of Universidad de Chile in Santiago de Chile. This Inria associayed team is led by the Inria Dyliss project team.10.2 National initiatives
Participants: Alain Franc, Jean-Marc Frigério.
10.2.1 Agence Française pour la Biodiversité
The AFB is a public law agency of the French Ministry of Ecology that supports public policy in the domains of knowledge, preservation, management, and restoration of biodiversity in terrestrial, aquatic, and marine environments. Pleiade is a partner in two AFB projects developed with the former ONEMA: one funded by ONEMA, the second by labex COTE, where BioGeCo/Pleiade is responsible for data analysis, with implementaton of the tools recently developed for scaling MDS. Calculations have been made on CURTA at MCIA and PlaFRIM at INRIA.
11 Dissemination
Participants: Clémence Frioux, David Sherman.
Member of the organizing committees
- Clémence Frioux - CMSB 2021. Computational Methods in Systems Biology. September 22-24th, Bordeaux, France.
Member of the conference program committees
- Clémence Frioux - CMSB 2021. Computational Methods in Systems Biology. September 22-24th, Bordeaux, France.
Reviewer
- Clémence Frioux - CMSB 2021. Computational Methods in Systems Biology. September 22-24th, Bordeaux, France.
Reviewer - reviewing activities
- Clémence Frioux - Microbiome
- Clémence Frioux - Computers in Biology and Medicine
11.0.2 Invited talks
- Clémence Frioux - French workshop on metabolic modelling
11.0.3 Leadership within the scientific community
David Sherman is on the steering committee of Biosena, a regional research network of the New Aquitaine region dedicated to Biodiversity and Ecosystemic Services. Biosena associates actors from the academic and socio-economic sectors, with the goal of contributing to the understanding and preservation of biodiversity and to the improvement of ecosystemic services. Biosena contributes to this goal through research, knowledge dissemination, outreach, and skill transfer in the form of Research Action, in keeping with the recommendations of Ecobiose.
David Sherman is member of the board (membre du Conseil d'administration) and secretary of the Mobsya Association, Lausanne. Mobsya develops and commercializes the Thymio educational robot, geared towards K-12.
David Sherman is member of the board (membre du Conseil d'Administration) and lead advisor for software of the Poppy Station Association. Poppy Station develops open-hardware open-source humanoid robots for research and education.
11.1 Teaching - Supervision - Juries
11.1.1 Teaching
Clémence Frioux
- Master – ENSTBB Bordeaux INP - Bioinformatics
- Master – Master Bioinformatique Université de Bordeaux - Projet de Programmation
- Licence – Université de Bordeaux - Python programming
11.1.2 Supervision
- Clémence Frioux - Internship of Mathieu Bolteau, Master 2 Bioinformatique, Université de Bordeaux (6 months)
11.2 Popularization
11.2.1 Interventions
- Clémence Frioux - Chiche! at Lycée Jean Renou (La Réole), March 2021. Presenting research in computer science to high school student to promote the digital sciences section while deconstructing gender stereotypes. 3 classes.
- Clémence Frioux - Fête de la Science "Circuit Bordelais 'Hors les Murs' 2021". October 2021. Presenting research in computer science to high school student to promote the digital sciences section while deconstructing gender stereotypes. 4 classes.
12 Scientific production
12.1 Major publications
- 1 articleA Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum..Nature Communications52014, 4044
- 2 articleModeling acclimatization by hybrid systems: Condition changes alter biological system behavior models.BioSystems121June 2014, 43-53
- 3 articleMetagenomic assessment of the global distribution of bacteria and fungi.Environmental MicrobiologyNovember 2020
- 4 articleMetage2Metabo, microbiota-scale metabolic complementarity for the identification of key species.eLife9December 2020
- 5 articleMetabolic Complementarity Between a Brown Alga and Associated Cultivable Bacteria Provide Indications of Beneficial Interactions.Frontiers in Marine Science7February 2020, 1-11
- 6 articleThe genome of Ectocarpus subulatus – A highly stress-tolerant brown alga.Marine Genomics52January 2020, 100740
- 7 articleUsing automated reasoning to explore the metabolism of unconventional organisms: a first step to explore host–microbial interactions.Biochemical Society Transactions483May 2020, 901-913
- 8 articleFrom bag-of-genes to bag-of-genomes: metabolic modelling of communities in the era of metagenome-assembled genomes.Computational and Structural Biotechnology JournalJune 2020
- 9 incollectionWhy We Need Sustainable Networks Bridging Countries, Disciplines, Cultures and Generations for Aquatic Biomonitoring 2.0: A Perspective Derived From the DNAqua-Net COST Action.Next Generation Biomonitoring: Part 158Elsevier2018, 63-99
- 10 articleExact or approximate inference in graphical models: why the choice is dictated by the treewidth, and how variable elimination can be exploited.Australian and New Zealand Journal of Statistics612to appearJune 2019, 89-133
- 11 articleGenolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes..Nucleic Acids Research372009, D550-D554
12.2 Publications of the year
International journals
Conferences without proceedings
Scientific book chapters
Reports & preprints
Other scientific publications
12.3 Cited publications
- 33 articleSIGPLAN Notices.Generating Embedded Software from Hierarchical Hybrid Models3872003, 171--82
- 34 articleRADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling.Mol. Ecol.22112013, 3179--90
- 35 articleReusing and composing models of cell fate regulation of human bone precursor cells.BioSystems1081-3April 2012, 63-72
- 36 articleImplementing biological hybrid systems: Allowing composition and avoiding stiffness.Applied Mathematics and Computation223August 2013, 167--79
- 37 inproceedingsReconciling competing models: a case study of wine fermentation kinetics.Algebraic and Numeric Biology 20106479Research Institute for Symbolic Computation, Johannes Kepler University of LinzHagenberg, AustriaSpringerJuly 2010, 68--83
- 38 articleThe Euclidean Distance Matrix Completion Problem.SIAM J. Matrix Anal. App.1621995, 646-654
- 39 techreportA geometric view of Biodiversity: scaling to metagenomics.RR-9144https://arxiv.org/abs/1803.02272INRIA ; INRAJanuary 2018, 1-16
- 40 articleExact Matrix Completion via Convex Optimization.Found. Comput. Math.92009, 717-772
- 41 articleReplacing fossil oil with fresh oil - with what and for what?Eur J Lipid Sci Technol11372011, 812-831
- 42 bookParasitism: The Ecology and Evolution of Intimate Interactions.University of Chicago Press2001
- 43 articleStructural analysis of expanding metabolic networks..Genome informatics. International Conference on Genome Informatics1512004, 35--45
- 44 articleNonlinear mapping and distance geometry.Optimization Letters142May 2019, 453-467
- 45 articleScalable and exhaustive screening of metabolic functions carried out by microbial consortia.Bioinformatics34172018, i934--i943
- 46 articleReference-free population genomics from next-generation transcriptome data and the vertebrate-invertebrate gap.PLoS Genetic94e10034572013
- 47 articleClingo = ASP + Control: Preliminary Report.CoRRabs/1405.36942014
- 48 inproceedingsAdvances in gringo Series 3.LPNMR6645Lecture Notes in Computer ScienceSpringer2011, 345---351
- 49 articleEuclidean Distance Geometry and Applications.SIAM review56(1)2014, 3-69
- 50 articleEstimation of Nucleotide Diversity, Disequilibrium Coefficients, and Mutation Rates from High-Coverage Genome-Sequencing Projects.Mol. Biol. Evol.25112008, 2409--19
- 51 articleImproving palm oil quality through identification and mapping of the lipase gene causing oil deterioration.Nat Commun42013, 2160URL: http://dx.doi.org/10.1038/ncomms3160
- 52 articleExact or approximate inference in graphical models: why the choice is dictated by the treewidth, and how variable elimination can be exploited.Australian and New Zealand Journal of Statistics612June 2019, 89-133
- 53 articleA comprehensive framework for global patterns in biodiversity.Ecology Letters712004, 1--15URL: http://dx.doi.org/10.1046/j.1461-0248.2003.00554.x
- 54 articleR-Syst::diatom: an open-access and curated barcode database for diatoms and freshwater monitoring.Database - The journal of Biological Databases and Curation2016February 2016
- 55 articleA unifying review of linear Gaussian Models.Neural Computation1121999, 305--45
- 56 articleThink globally, fit locally: unsupervised learning of low dimensional manifolds.Journal of Machine Learning Research42003, 119--55
- 57 bookOn Growth and Form.Cambridge University Press1917
- 58 bookGeometric structure of high-dimensional data and dimensionality reduction.Springer & Higher Education Press2012

