Keywords
Computer Science and Digital Science
- A3.1.2. Data management, quering and storage
- A3.1.3. Distributed data
- A3.1.7. Open data
- A3.1.8. Big data (production, storage, transfer)
- A3.2.4. Semantic Web
- A3.3.3. Big data analysis
- A3.4.1. Supervised learning
- A3.4.2. Unsupervised learning
- A3.4.3. Reinforcement learning
- A3.4.4. Optimization and learning
- A3.4.6. Neural networks
- A3.4.8. Deep learning
- A5.1.4. Brain-computer interfaces, physiological computing
- A5.2. Data visualization
- A5.3.2. Sparse modeling and image representation
- A5.3.3. Pattern recognition
- A5.3.4. Registration
- A5.4.1. Object recognition
- A5.4.6. Object localization
- A5.9.2. Estimation, modeling
- A5.9.4. Signal processing over graphs
- A6.2.3. Probabilistic methods
- A6.2.4. Statistical methods
- A6.3.3. Data processing
- A6.3.4. Model reduction
- A9.2. Machine learning
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1.2. Neuroscience and cognitive science
- B1.2.1. Understanding and simulation of the brain and the nervous system
- B1.2.2. Cognitive science
- B2.1. Well being
- B2.2.2. Nervous system and endocrinology
- B2.2.6. Neurodegenerative diseases
- B2.5.1. Sensorimotor disabilities
- B2.5.2. Cognitive disabilities
- B2.6.1. Brain imaging
1 Team members, visitors, external collaborators
Research Scientists
- Emmanuel Caruyer [CNRS, Researcher, HDR]
- Julie Coloigner [CNRS, Researcher]
- Benoit Combès [INRIA, Researcher]
- Olivier Commowick [INRIA, Researcher, HDR]
- Claire Cury [INRIA, Researcher]
- Fanny Dégeilh [INRIA, Starting Research Position]
- Camille Maumet [INRIA, Researcher]
Faculty Members
- Pierre Maurel [Team leader, UNIV RENNES I, Associate Professor, HDR]
- Isabelle Bonan [UNIV RENNES I, CHU, Professor, HDR]
- Gilles Edan [UNIV RENNES I, CHU, Professor, HDR]
- Jean-Christophe Ferré [UNIV RENNES I, CHU, Professor, HDR]
- Francesca Galassi [UNIV RENNES I, Associate Professor]
- Jean-Yves Gauvrit [UNIV RENNES I, CHU, Professor, HDR]
- Anne Kerbrat [UNIV RENNES I, CHU, Associate Professor, HDR]
- Gabriel Robert [UNIV RENNES I, CHGR, Professor, HDR]
Post-Doctoral Fellows
- Agustina Fragueiro [INRIA, from May 2022]
- Burhan Rashid Hussein [INRIA, from Jun 2022]
- Lou Scotto Di Covella [INRIA, until Jul 2022]
PhD Students
- Constance Bocquillon [UNIV RENNES I, from Oct 2022]
- Sebastien Dam [INRIA, from Oct 2022]
- Thomas Durantel [UNIV RENNES I]
- Elodie Germani [UNIV RENNES I]
- Carla Joud [UNIV RENNES I, from Nov 2022]
- Alix Lamouroux [IMT Atlantique Brest, from Oct 2022]
- Caroline Pinte [UNIV RENNES I]
- Xavier Rolland [Université de Rennes 1 , until Mar 2022]
- Jean-Charles Roy [UNIV RENNES I]
- Ricky Walsh [UNIV RENNES I, from Nov 2022]
Technical Staff
- Elise Bannier [CHU, Engineer]
- Boris Clenet [Inria, Engineer, from Nov 2022]
- Isabelle Corouge [UNIV RENNES I, Engineer]
- Pierre-Henri Dauvergne [Inria, Engineer, from Nov 2022]
- Rene-Paul Debroize [Inria, Engineer, from Oct 2022]
- Jean-Côme Douteau [INRIA, Engineer]
- Quentin Duché [UNIV RENNES I, Engineer]
- Malo Gaubert [CHU, Engineer]
- Renaud Hedouin [INRIA, Engineer]
- Nolwenn Jegou [UNIV RENNES I, Engineer]
- Michael Kain [Inria, Engineer]
- Florent Leray [Inria, Engineer]
- Julien Louis [Inria, Engineer]
- Arthur Masson [INRIA, Engineer]
- Cédric Meurée [INRIA, Engineer, from Apr 2022]
- Sandesh Patil [Inria, Engineer]
Interns and Apprentices
- Clément Bion [UNIV RENNES I, from Jun 2022 until Aug 2022]
- Constance Bocquillon [UNIV RENNES I, from Oct 2022]
- Sarah Bucquet [UNIV RENNES I, from Feb 2022 until Aug 2022]
- Assia Chahidi [UNIV COTE D'AZUR, from Mar 2022 until Aug 2022]
- Sebastien Dam [Inria, from Feb 2022 until Jul 2022]
- Carla Joud [UNIV RENNES I, from Apr 2022 until Sep 2022]
- Thomas Leguay [UNIV RENNES I, until Jul 2022]
- Quentin Monnier [UNIV RENNES I, from Mar 2022 until Aug 2022]
Administrative Assistant
- Armelle Mozziconacci [CNRS]
Visiting Scientist
- Demian Vera [National University of the Center of the Buenos Aires Province, from Nov 2022]
2 Overall objectives
The research team Empenn ("Brain" in Breton language) ERL U1228 is co-affiliated with Inria, Inserm (National Institute for Health and Scientific Research), CNRS (INS2I institute), and the University of Rennes I. It is a team of IRISA/UMR CNRS 6074. Empenn is located in Rennes, on the medical and scientific campus. It succeeded in 2019 to the "VisAGeS" team, created in 2006 by Inria. As for "VisAGeS", Empenn holds the accreditation number U1228, renewed by Inserm in 2022 and for a period of 6 years, after an evaluation conducted by the HCERES and Inserm.
Thanks to this unique partnership, Empenn's ambition is to establish a multidisciplinary team of researchers in information sciences and medicine. Our medium and long term objective is to introduce our fundamental research into clinical practice, while maintaining the excellence of our methodological research.
Our goal is to foster research in medical imaging, neuroinformatics and population cohorts. In particular, the Empenn team aims at the detection and development of imaging biomarkers for brain diseases and focuses its efforts on transferring this research to the clinic and clinical neuroscience in general. More specifically, the objective of Empenn is to propose new statistical and computational methods, and to measure and model morphological, structural and functional states of the brain to better diagnose, monitor and treat mental, neurological and substance use disorders. We propose to combine advanced instrumental devices and novel computational models to provide advanced diagnostic, therapeutic, and neurorehabilitation solutions for some of the major developing and aging brain disorders.
Generic and challenging research topics in this broad area include finding new ways to compare models and data, aid in decision making and interpretation, and develop feedback. These activities are carried out in close collaboration with the Neurinfo imaging platform in vivo, which is an essential environment for the experimental implementation of our research on ambitious clinical research projects and the development of new clinical applications.
3 Research program
3.1 Glossary
-
Magnetic Resonance Imaging
- MR - Magnetic Resonance
- MRI - Magnetic Resonance Imaging
- fMRI - Functional Magnetic Resonance Imaging
- DWI - Diffusion-Weighted Imaging
- ASL - Arterial Spin Labeling
-
Other modalities
- PET - Positron Emission Tomography
- EEG - Eletroencephalograpy
- NIRS - Near InfraRed Spectroscopy
-
Medical terminology
- MS - Multiple Sclerosis
- TBI - Traumatic Brain Injury
-
Methodological terminology
- GLM - General Linear Model
- MCM - Multi-compartment models
3.2 Scientific Foundations
The scientific foundations of our team concern the design and development of new computational solutions for biological images, signals and measurements. Our goal is to develop a better understanding of the normal and pathological brain, at different scales.
This includes imaging brain pathologies in order to better understand pathological behavior from the organ level to the cellular level, and even to the molecular level (PET-MR imaging), and modeling of large groups of normal and pathological individuals (cohorts) from image descriptors. It also addresses the challenge of the discovery of episodic findings (i.e. rare events in large volumes of images and data), data mining and knowledge discovery from image descriptors, validation and certification of new drugs from imaging features, and, more generally, the integration of neuroimaging into neuroinformatics by promoting and supporting virtual organizations of biomedical actors using e-health technologies.
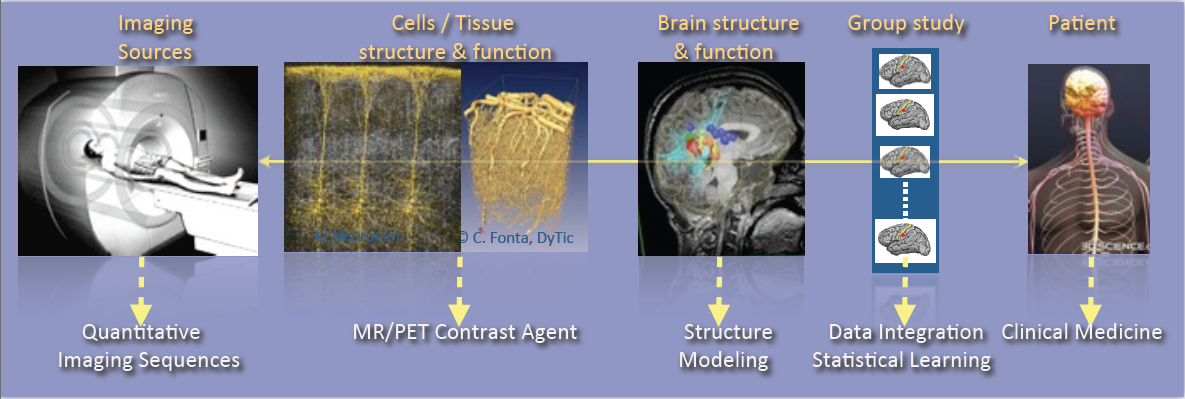
Illustration of the major scientific objectives of the Empenn team, including Imaging sources, Cells and tissue structure and function, Brain structure and function, Group study and Patient.
As shown in Figure 1, the research activities of the Empenn team closely link observations and models through the integration of clinical and multiscale data, and phenotypes (cellular, and later molecular, with structural or connectivity patterns in the first stage). Our ambition is to build personalized models of central nervous system organs and pathologies, and to compare these models with clinical research studies in order to establish a quantitative diagnosis, prevent the progression of diseases and provide new digital recovery strategies, while combining all these research areas with clinical validation. This approach is developed within a translational framework, where the data integration process to build the models is informed by specific clinical studies, and where the models are assessed regarding prospective clinical trials for diagnosis and therapy planning. All of these research activities are conducted in close collaboration with the Neurinfo platform, which benefited in 2018 from a new high-end 3T MRI system dedicated to research (3T Prisma™ system from Siemens), and through the development in the coming years of multimodal hybrid imaging (from the currently available EEG-MRI, to EEG-NIRS and PET-MRI in the future).
In this context, some of our major developments and newly arising issues and challenges include:
- The generation of new descriptors to study brain structure and function (e.g. the combination of variations in brain perfusion with and without a contrast agent; changes in brain structure in relation to normal, pathological, functional or connectivity patterns; or the modeling of brain state during cognitive stimulation using neurofeedback).
- The integration of additional spatiotemporal and hybrid imaging sequences covering a larger range of observations, from the molecular level to the organ level, via the cellular level (arterial spin labeling, diffusion MRI, MR relaxometry, MR cell labeling imaging, EEG-MRI functional imaging, EEG-NIRS-MRI).
- The creation of computational models through the data fusion of multimodal MR images, structural and functional image descriptors from group studies of normal and/or pathological subjects.
- The evaluation of these models in relation to acute pathologies, especially for the study of degenerative, psychiatric, traumatic or developmental brain diseases (primarily multiple sclerosis, stroke, traumatic brain injury (TBI) and depression, but applicable with a potential additional impact to epilepsy, Parkinson’s disease, dementia, post-traumatic stress disorder, etc.) within a translational framework.
In terms of new major methodological challenges, we address the development of models and algorithms to reconstruct, analyze and transform the images, and to manage the mass of data to store, distribute and “semanticize” (i.e. provide a logical division of the model’s components according to their meaning). As such, we expect to make methodological contributions in the fields of model inference; statistical analysis and modeling; the application of sparse representation (compressed sensing and dictionary learning) and machine learning (supervised/unsupervised classification and discrete model learning); data fusion (multimodal integration, registration, patch analysis, etc.); high-dimensional optimization; data integration; and brain-computer interfaces. As a team at the frontier between the digital sciences and clinical research in neuroscience, we do not claim to provide theoretical breakthroughs in these domains but rather to provide significant advances in using these algorithms through to the advanced applications we intend to address. In addition, we believe that by providing these significant advances using this set of algorithms, we will also contribute to exhibiting new theoretical problems that will fuel the domains of theoretical computer sciences and applied mathematics.
In summary, we expect to address the following major challenges:
- Developing new information processing methods able to detect imaging biomarkers in the context of mental, neurological, and substance use disorders.
- Providing new computational solutions for our target applications, allowing a more appropriate representation of data for image analysis and the detection of biomarkers specific to a form or grade of pathology, or specific to a population of subjects.
- Providing, for our target applications, new patient-adapted connectivity atlases for the study and characterization of diseases from quantitative MRI.
- Providing, for our target applications, new analytical models of dynamic regional perfusion, and deriving indices of dynamic brain local perfusion from normal and pathological populations.
- Investigating whether the theragnostics paradigm of rehabilitation from hybrid neurofeedback can be effective in some behavioral and disability pathologies.
These major advances are primarily developed and validated in the context of several priority applications in which we expect to play a leading role: multiple sclerosis, stroke rehabilitation, and the study and treatment of depression.
4 Application domains
Figure 2 summarizes the scientific organization of the research team through three basic research topics in information sciences (Population Imaging, Detection and Learning, and Quantitative Imaging) and three translational axes on central nervous system diseases (Behavior, Neuro-inflammation and Recovery).
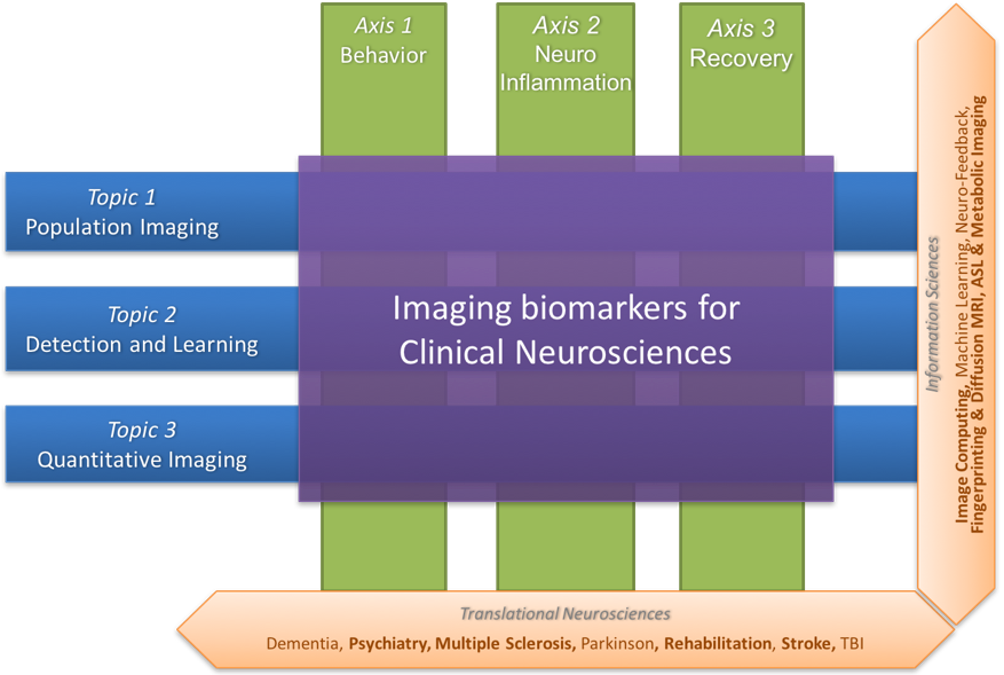
Illustration of the research topics and research axes of the Empenn team. The former (Population imaging, detection and learning and quantitative imaging) concern information sciences, and these topics intersect with the latter (Behavior, neuro-inflammation and recovery), which are translational neurosciences axes.
4.1 Basic research
4.1.1 Population imaging
One major objective of neuroimaging researchers and clinicians is to be able to stratify brain imaging data in order to derive new and more specific population models. In practice, this requires to set up large-scale experiments that, due to the lack of resources and capabilities to recruit locally subjects who meet specific inclusion criteria, motivates the need for sharing the load.
However, building and using multi-site large-scale resources pose specific challenges to deal with the huge quantity of data produced and their diversity. Empenn focuses on two challenges in particular:
- Providing computational environments for the computation and use of imaging biomarkers in the targeted brain diseases, a solution to be used by radiologists and neurologists/psychiatrists for the clinical follow-up of a large patient population.
- Modeling analytic variability of image processing pipelines to better understand and predict the behaviour of imaging biomarker detection solutions and improve reproducibility and productivity in clinical neuroimaging research.
4.1.2 Detection and learning
We intend to make significant contributions with major impacts in learning coupling models between functional recordings during neurofeedback procedures. These advances will provide a breakthrough in brain-computer interfaces for rehabilitation protocols. Our aim is to:
- Provide a computational environment that combines data-driven (machine learning) and Bayesian solutions to improve the detection of abnormal patterns in images through decision or evidence theory data fusion strategies. The major initial application is multiple sclerosis. Over the longer term, we also expect to adapt these methods to address a wider range of neurological diseases (epilepsy, stroke, tumors, etc.) in neonate and adult brains.
- Develop solutions for combining brain state measurements from multimodal sensors or sequences (e.g. fMRI, ASL, EEG, NIRS, etc.) with applications in the spatiotemporal reconstruction of brain activity from MRI-EEG or the combined detection of the endogenous hemodynamic and resting state network of the brain from ASL and NIRS. Over the longer term, the advent of new hybrid brain imaging sensors (e.g. PET-MRI) will require these methods to be extended to a larger spectrum of information combining structural, morphological, metabolic, electrophysiological and cellular/molecular information (e.g. through the use of specific ligands/nanocarriers).
4.1.3 Quantitative imaging
The Empenn research group focuses on the development of several quantitative techniques in magnetic resonance imaging of the brain. These methods allow for characterization of both the function and the structure of the brain with high precision. Arterial spin labelling (ASL) is a contrast agent-free imaging technique which labels arterial blood water as an endogenous tracer for perfusion and can measure resting-state cerebral blood flow. We are interested in estimating multiparametric hemodynamics using ASL, such as combined cerebral blood flow and arterial transit times, and derive statistical descriptors to represent significant differences between groups. In addition to quantitative perfusion parameters, our contributions on tissue compartment imaging aim at delineating neural circuits and characterize their microstructure properties, using both diffusion MRI and relaxometry. In diffusion MRI, arbitrary gradient waveforms were shown to exhibit higher sensitivity to microstructure parameters than standard pulsed gradients. We work on the optimization of sampling protocols in this domain, with the objective to propose sequences compatible with in vivo acquisition. Complementary to diffusion MRI, we develop methods for the reconstruction of myelin-bound, extra-axonal and cerebrospinal fluid water using multi-compartment modelling of the T2-relaxometry signal. We combine these techniques with tractography to identify trajectories of pathologies associated to the evolution of these microstructural parameters along specific fiber bundles in the brain white matter.
4.2 Translational research
4.2.1 Behavior
Advances in the field of in vivo imaging offer new opportunities for addressing the management of resistant affective disorders and their consequences (suicide risk and socio-professional impact), and the management of spatial cognition disorders after stroke and their consequences (postural perturbations and the loss of autonomy). Our objective, and the main challenge in this context, is to introduce medical image computing methods to the multidisciplinary field of behavioral disorders (cognitive disorders, particularly spatial and postural control disorders or anterograde memory impairment, mood disorders, notably resistant depression, schizophrenic disorders, pervasive developmental disorders, attention disorders, etc.) in order to gain a better understanding of the pathology and devise innovative therapeutic approaches.
We also expect to become a major player in the future and make important contributions with significant impacts, primarily in drug-resistant depression in young and old populations. In particular, we expect to provide new image-related metrics combining perfusion, metabolism and microstructural information regarding the brain in order to better characterize pathologies, provide prospective evolution values and potentially provide new brain stimulation targets that could be used in neurofeedback rehabilitation protocols or other types of brain stimulation procedures.
We aim to provide new imaging markers of mental diseases, especially in the context of mood disorders. The new biomarkers are derived from the metabolic (ASL and later ASL+PET) point of view as well as from the microstructural point of view (multicompartment diffusion MRI and relaxometry). Similarly, we expect to exhibit imaging biomarker regularities combining metabolic and structural information. Over the longer term, we expect these biomarkers to be the target of neurofeedback rehabilitation procedures. Also, over the longer term, we expect to supplement the MRI markers with molecular markers coming from new PET tracers, especially those associated with serotonin intake, at one time point or during a rehabilitation protocol under hybrid PET-EEG-MRI neurofeedback procedures.
4.2.2 Neuroinflammation
Some of the major ongoing research issues regarding neuroimaging of neuro-inflammatory diseases concern the definition of new biomarkers to track the development of the pathology using high- dimensional data (e.g. nD+t MRI). This includes the use of white matter-specific imaging, such as magnetization transfer MRI, relaxometry and diffusion-weighted imaging (DW-MRI). Our objective is (1) to develop information-processing tools to tag the spatiotemporal evolutions of Multiple Sclerosis patterns at the brain parenchyma and spinal cord levels from their different signatures (inflammatory cells visible with USPIO or Gd contrast agents on MRI, persistent black holes, eloquent regional atrophy and microstructure signatures); and (2) to test these new tools on new imaging cohorts. In this respect, we for instance conduct studies on brain and spinal cord imaging, continuing on from the PHRC multicentric EMISEP project (PI: G. Edan), as it is very likely that lesions in the spine will directly affect the ambulatory ability of the patient (and thereby the clinical scores). In order to extend this experiment to a larger MS population, based on our expertise from the OFSEP cohort, we also plan to improve the MS therapeutic decision process through the MUSIC project (Multiple Sclerosis Imaging Check out, a public/private project) and, notably, the RHU PRIMUS project. Our goal is to develop and assess a standardized monitoring tool that provides a robust, long-term computerized MRI follow-up that will become the gold standard in clinical practice for therapeutic decisions in MS treatment. As part of this project, Empenn will share its expertise in data management systems (Shanoir and FLI-IAM) and automatic processing tools (through the medInria and Anima software repositories) to extract quantitative indices from the images.
4.2.3 Recovery
Mental and neurological disorders are the leading cause of years lived with a disability. Treatment-resistant depression affects approximately 2% of the European population. Meanwhile, in the case of brain disorders, almost 1.5 million Europeans (15 million people worldwide) suffer a stroke event each year. Current recovery methods for brain disorders and traumatic brain injuries remain limited, preventing many from achieving full recuperation. We propose to address the issue of brain recovery by introducing new advances from recent breakthroughs in computational medical imaging, data processing and human-machine interfaces, and demonstrate how these new concepts can be used, in particular for the treatment of stroke and major depressive disorders.
We ambition to combine advanced instrumental devices (hybrid EEG, NIRS and MRI platforms), with new hybrid brain computer interface paradigms and new computational models to provide neurofeedback-based therapeutic and neuro-rehabilitation paradigms in some of the major mental and neurological disorders of the developmental and aging brain.
Neurofeedback involves using a brain-computer interface that provides an individual with real-time biofeedback about his or her brain activity in the form of sensory feedback. It enables individuals to learn to better control their brain activity, which can be measured in real time using various non-invasive sensors as described above. Although EEG is currently the only modality used by clinical practitioners in that context, it lacks specificity due to its low spatial resolution. Dynamic research into fMRI-neurofeedback has held promise for treating depression, chronic pain and stroke, since it offers the prospect of real-time imagery of the activity in deep brain structures with high spatial resolution. However, the low temporal resolution and high cost of fMRI-neurofeedback has hampered the development of many applications. We believe that the future belongs to hybrid responses that combine multimodal sensors and intend to demonstrate this in the Empenn project.
5 Social and environmental responsibility
- Francesca Galassi: member of the Women in MICCAI (WiM) board - to strengthen and widen the representation of female scientists in the MICCAI community.
- Francesca Galassi and Elise Bannier: members of the Groupe Développement Durable de l'Inria RBA - for the assessment and mitigation of the impact of our research activities on the environment.
- Camille Maumet: co-chair of the women-men equality group at Inria Rennes / IRISA.
- Elise Bannier: member of the Matching Committee for the Inria mentoring program.
6 Highlights of the year
- Starting of the RHU PRIMUS (Projection dans la sclérose en plaques) project: a 5-year project with the objective of developing and validating a CE-marked data-driven clinical decision support system (CDSS) for multiple sclerosis (MS).
- In the context of the project MMINCARAV (Multimodal Microstructure-Informed Neuronal Connectivity: Acquisition, Reconstruction, Analysis and Validation), we organised and received a 3-day visit from our associate team LTS5 in EPFL, Lausanne.
- Near-Infrared Spectroscopy (NIRS) Workshop: Empenn and Bionic organized a 2-day event at Inria RBA on the acquisition and processing of multimodal data e.g. EEG-NIRS, fMRI-NIRS.
7 New software and platforms
7.1 New software
7.1.1 Anima
-
Keywords:
Filtering, Medical imaging, Diffusion imaging, Registration, Relaxometry
-
Scientific Description:
Anima is a set of libraries and tools developed by the team as a common repository of research algorithms. As of now, it contains tools for image registration, statistical analysis (group comparison, patient to group comparison), diffusion imaging (model estimation, tractography, etc.), quantitative MRI processing (quantitative relaxation times estimation, MR simulation), image denoising and filtering, and segmentation tools. All of these tools are based on stable libraries (ITK, VTK), making it simple to maintain.
-
Functional Description:
Anima is a set of libraries and tools in command line mode for processing and analysing medical images.
- URL:
-
Contact:
Olivier Commowick
-
Participants:
Aymeric Stamm, Fang Cao, Florent Leray, Guillaume Pasquier, Laurence Catanese, Olivier Commowick, Renaud Hedouin, René-Paul Debroize
7.1.2 MedINRIA
-
Keywords:
Visualization, DWI, Health, Segmentation, Medical imaging
-
Scientific Description:
MedInria aims at creating an easily extensible platform for the distribution of research algorithms developed at Inria for medical image processing. This project has been funded by the D2T (ADT MedInria-NT) in 2010, renewed in 2012. A fast-track ADT was awarded in 2017 to transition the software core to more recent dependencies and study the possibility of a consortium creation.The Empenn team leads this Inria national project and participates in the development of the common core architecture and features of the software as well as in the development of specific plugins for the team's algorithm.
-
Functional Description:
MedInria is a free software platform dedicated to medical data visualization and processing.
- URL:
-
Contact:
Olivier Commowick
-
Participants:
Maxime Sermesant, Olivier Commowick, Théodore Papadopoulo
-
Partners:
HARVARD Medical School, IHU - LIRYC, NIH
7.1.3 autoMRI
-
Keywords:
FMRI, MRI, ASL, FASL, SPM, Automation
-
Scientific Description:
This software is highly configurable in order to fit a wide range of needs. Pre-processing includes segmentation of anatomical data, as well as co-registration, spatial normalization and atlas building of all data types. The analysis pipelines perform either within-group analysis or between-group or one subject-versus-group comparison, and produce statistical maps of regions with significant differences. These pipelines can be applied to structural data to exhibit patterns of atrophy or lesions, to ASL (both pulsed or pseudo-continuous sequences) data to detect perfusion abnormalities, to functional data - either BOLD or ASL - to outline brain activations related to block or event-related paradigms. New functionalities have been implemented to facilitate the management and processing of data coming from complex projects.
-
Functional Description:
AutoMRI is based on MATLAB and the SPM12 toolbox and provides complete pipelines to pre-process and analyze various types of images (anatomical, functional, perfusion).
- URL:
-
Contact:
Isabelle Corouge
-
Participants:
Camille Maumet, Elise Bannier, Isabelle Corouge, Pierre Maurel, Quentin Duché, Julie Coloigner
7.1.4 ShanoirUploader
-
Name:
ShanoirUploader (SHAring NeurOImaging Resources Uploader)
-
Keywords:
Webservices, PACS, Medical imaging, Neuroimaging, DICOM, Health, Biology, Java, Shanoir
-
Scientific Description:
ShanoirUploader is a desktop application on base of JavaWebStart (JWS). The application can be downloaded and installed using an internet browser. It interacts with a PACS to query and retrieve the data stored on it. After this ShanoirUploader sends the data to a Shanoir server instance in order to import these data. This application bypasses the situation, that in most of the clinical network infrastructures a server to server connection is complicated to set up between the PACS and a Shanoir server instance.
-
Functional Description:
ShanoirUploader is a Java desktop application that transfers data securely between a PACS and a Shanoir server instance (e.g., within a hospital). It uses either a DICOM query/retrieve connection or a local CD/DVD access to search and access images from a local PACS or the local CD/DVD. After having retrieved the data, the DICOM files are locally anonymized and then uploaded to the Shanoir server. A possible integration of a hash creation application for patient identifiers is provided as well. The primary goals of that application are to enable mass data transfers between different remote server instances and therefore reduce the waiting time of the users, when importing data into Shanoir. Most of the time during import is spent with data transfers.
- URL:
-
Contact:
Michael Kain
-
Participants:
Christian Barillot, Inès Fakhfakh, Justine Guillaumont, Michael Kain, Yao Chi
7.1.5 Shanoir-NG
-
Name:
Shanoir-NG (SHAring NeurOImaging Resources - Next Generation)
-
Keywords:
Neuroimaging, DICOM, Nifti
-
Functional Description:
Shanoir-NG (SHAring NeurOImaging Resources - Next Generation) is an open-source web platform designed to share, archive, search and visualize medical imaging data. "Next Generation" stands for a complete technological remake released in 2020 with a new technical architecture based on microservices. It provides a user-friendly secure web access and offers an intuitive workflow to facilitate the collecting and retrieving of imaging data from multiple sources. It supports the following formats: DICOM classic/enhanced (MR, CT, PT, NM), BIDS, processed datasets (NIfTI), Bruker, EEG(BrainVision/EDF).
Shanoir-NG comes along many features such as pseudonymization of data (based on standard profiles), support for multi-centric clinical studies on subjects or group of subjects. Shanoir-NG offers an ontology-based data organization (OntoNeuroLOG). Among other things, this facilitates the reuse of data and metadata, the integration of processed data and provides traceability trough an evolutionary approach. Shanoir-NG allows researchers, clinicians, PhD students and engineers to undertake quality research projects with an emphasis on remote collaboration.
-
News of the Year:
Shanoir-NG is a complete technological remake of the first version of the Shanoir application, but maintaining the key concepts of Shanoir.
- URL:
-
Contact:
Michael Kain
-
Participants:
Michael Kain, Anthony Baire, Julien Louis, Jean-Côme Douteau, Pierre-Henri Dauvergne, Arthur Masson
-
Partners:
CHU Grenoble, INSERM, CNRS, Université Grenoble Alpes, Université de Strasbourg
7.1.6 LongiSeg4MS
-
Name:
Longitudinal Segmentation For Multiple Sclerosis
-
Keywords:
3D, Brain MRI, Deep learning, Detection
-
Functional Description:
LongiSeg4MS is an automatic new multiple sclerosis (MS) lesion detection tool based on longitudinal data and using deep learning. The system uses FLAIR, T1 or T2 modalities, or a combination of those. The input is 2, 4 or 6 images (2 FLAIR, 2 FLAIR and 2 T1, etc.), a set of modalities for each time point, and outputs a segmentation map describing the location of new MS lesions.
- URL:
-
Authors:
Arthur Masson, Brandon Le Bon, Benoit Combes
-
Contact:
Arthur Masson
-
Partner:
OFSEP
7.1.7 Anima medInria plugins
-
Keywords:
IRM, Medical imaging, Diffusion imaging
-
Functional Description:
Plugins for the medInria software based on the open source software Anima developed in the Visages / Empenn team. These plugins are interfaces between anima and medinria allowing to use Anima functionalities within the clinical user interface provided by medInria. The current functionalities included in the plugins are right now: image registration, denoising, quantitative imaging (relaxometry), and model estimation and visualization from diffusion imaging.
- URL:
-
Contact:
Olivier Commowick
-
Participants:
Olivier Commowick, Florent Leray, René-Paul Debroize, Guillaume Pasquier
7.2 New platforms
7.2.1 The Neurinfo Platform
Participants: Elise Bannier, Emmanuel Caruyer, Isabelle Corouge, Quentin Duché, Jean-Christophe Ferré, Jean-Yves Gauvrit, Nolwenn Jégou.
Empenn is the founding actor of an experimental research platform which was installed in August 2009 at the University Hospital of Rennes. The University of Rennes 1, Inria, CNRS for the academic side, and the University Hospital of Rennes and the Cancer Institute “Eugene Marquis” for the clinical side, are partners of this neuroinformatics platform called Neurinfo (Neurinfo website). Concerning the Neurinfo Platform, the activity domain is a continuum between methodological and technological research built around specific clinical research projects. On the medical field, the translational research domain mainly concerns medical imaging and more specifically the clinical neurosciences. Among them are multiple sclerosis, epilepsy, neurodegenerative, neurodevelopmental and psychiatric diseases, surgical procedures of brain lesions, neuro-oncology and radiotherapy planning. Beyond these central nervous system applications, the platform is also open to alternative applications. Neurinfo ambitions to support the emergence of research projects based on their level of innovation, their pluri-disciplinarity and their ability to foster collaborations between different actors (public and private research entities, different medical specialties, different scientific profiles). In this context, a research 3T MRI system (Siemens Verio) was acquired in summer 2009 in order to develop the clinical research in the domain of morphological, functional, structural and cellular in-vivo imaging. A new 3T Siemens Prisma MRI scanner was installed at the Neuroinfo platform in February 2018. In 2014, an equipment for simultaneous recording of EEG and MRI images was acquired from Brain Product. In 2015, a mock scanner for experimental set-up was acquired as well as a High Performance Computing environment made of one large computing cluster and a data center that is shared and operated by the Inria center and IRISA (UMR CNRS 6074). The computation cluster (480 cores) and the data center (up to 150 TB) are dedicated to host and process imaging data produced by the Neurinfo platform, but also by other research partners that share their protocols on the Neurinfo neuroinformatics system (currently more than 60 sites). In 2019, an MRI and EEG-compatible fNIRS system was acquired through a co-funding from the INS2I institute of CNRS and FEDER. At the end of 2019, GIS IBISA awarded the Neurinfo platform with a complementary funding that will be dedicated to supplement the current system with additional sensors (from 8x8 optodes to 16x16 optodes). In 2022, the Regional Council of Britanny funding was renewed to provide engineer support for another year to develop and integrate this new imaging system.8 New results
8.1 Basic research
8.1.1 Population imaging
Population imaging is fundamental when it comes to evaluate clinical biomarkers. In this section we summarise our contributions over the last year to this theme. We studied how analytical variability can impact fMRI results and proposed recommendations and neuroinformatics models to describe the data. We also maintained our clinical interest regarding several pathologies by exploring brain function and connectivity. Also, technical recommendations regarding multicentric imaging protocols were proposed.
Towards efficient fMRI data re-use: can we run between-group analyses with datasets processed differently?
Participants: Pierre Maurel, Camille Maumet, Xavier Rolland.
In recent years, the lack of reproducibility of research findings has become an important source of concerns in many scientific fields, including functional Magnetic Resonance Imaging (fMRI). The low statistical power often observed in fMRI studies was identified as one of the leading causes of irreproducibility. The development of data sharing in the field of neuroimaging opens up new opportunities to achieve larger sample sizes by reusing existing data. FMRI studies use subject data processed with pipelines, and although most shared datasets currently include raw data, we may expect to see an increasing proportion of processed data among shared subject data in the future, for privacy and sustainability reasons. Pipelines consist of multiple steps, each with multiple possible methodological choices: this existing variability in terms of processing and analysis (analytical variability) has an impact on the results. We investigated the impact of analytical variability when combining subject data processed differently in between-group analyses. We created a set of pipelines for subject-level processing that we applied on data from the Human Connectome Project (n=1080). We then performed between-group analyses with subject data processed with different pipelines, under the null hypothesis (making any detection a false positive) using two different software packages: SPM and FSL. We compared the estimated false positive rates obtained to the nominal false positive rate. We found that the analytical variability induced by the studied parameters was found to be acceptable for some of these analyses and redhibitory for others. We concluded that different processed subject data cannot be combined without taking into account the processing applied on these data. Associated publication: 38.
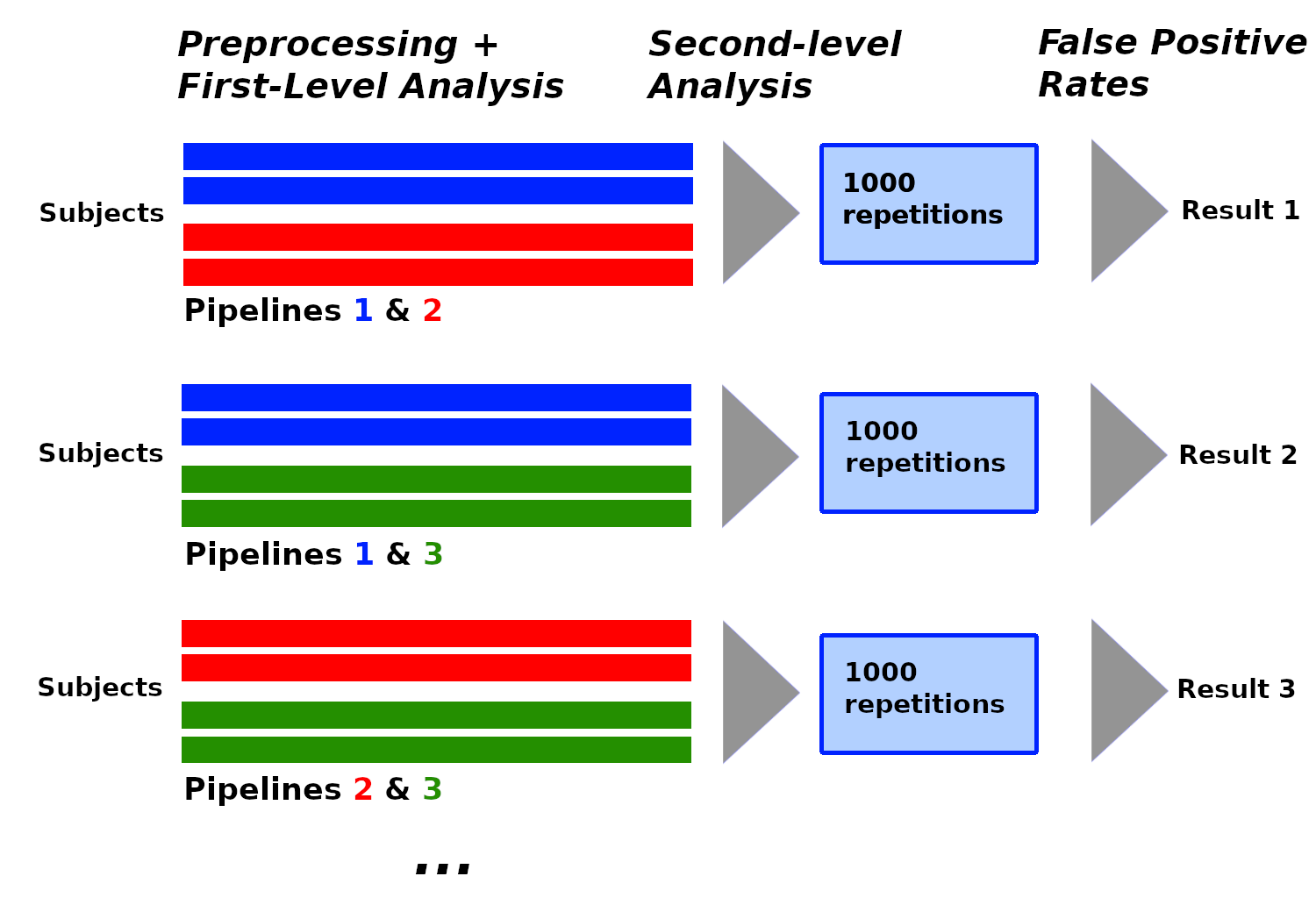
Figure from Rolland et al. ISBI
FMRI data analysis: How does analytical variability vary with sample size?
Participants: Elodie Germani, Camille Maumet.
Neuroimaging workflows are highly flexible leaving researchers with many possible choices at each step of their analysis. Recents studies have demonstrated how different analytical choices can substantially impact neuroimaging results, effectively leading to a “vibration of effects”. This observation is not limited to brain imaging and was also made across many scientific fields. In psychology, Klau et al., 2020 showed that the vibration of effects decreases and stabilizes as sample sizes increase. We built on the results of Botvinik-Nezer et al., 2020, in which a functional Magnetic Resonance Imaging (fMRI) “many analyst” study was conducted with 70 teams. In this study, each team used their favorite pipeline to analyze the same dataset and answer 9 pre-defined hypotheses. After reproducing the pipelines used by 4 teams, we observe the impact of varying sample sizes on the vibration of effects. Using descriptions provided in the original study, we reproduced the SPM pipelines of 4 teams and replicated the results of each pipeline with different subset of participants and with the full dataset. Our findings show that, in fMRI data analysis, the vibration of effects decreases with sample size. Our results also suggest that some variability remains even for large sample sizes. Further work will be needed in order to include more pipelines and investigate which part of the pipelines are the most impactful. Associated publication: 34.On the benefits of Self-Taught learning for Brain Decoding
Participants: Elodie Germani, Camille Maumet.
We study the benefits of using a large public neuroimaging database composed of fMRI statistic maps, in a self-taught learning framework, for improving brain decoding on new tasks. First, we leverage the NeuroVault database to train, on a selection of relevant statistic maps, a convolutional autoencoder to reconstruct these maps. Then, we use this trained encoder to initialize a supervised convolutional neural network to classify tasks or cognitive processes of unseen statistic maps from large collections of the NeuroVault database. We show that such a self-taught learning process always improves the performance of the classifiers but the magnitude of the benefits strongly depends on the number of samples available both for pre-training and finetuning the models and on the complexity of the targeted downstream task. This work was co-supervised by Prof. Elisa Fromont from the LACODAM team. Associated publication: 47.NARPS Open Pipelines project
Participants: Elodie Germani, Camille Maumet.
The goal of the NARPS Open Pipelines Project is to provide a public codebase that reproduces the 70 pipelines chosen by the 70 teams of the NARPS study. The project is public and the code hosted on GitHub at . This project initially emerged from the idea of creating an open repository of fMRI data analysis pipelines (as used by researchers in the field) with the broader goal to study and better understand the impact of analytical variability. NARPS – a many-analyst study in which 70 research teams were asked to analyze the same fMRI dataset with their favorite pipeline – was identified as an ideal usecase as it provides a large array of pipelines created by different labs. In addition, all teams in NARPS provided extensive (textual) description of their pipelines using the COBIDAS guidelines. All resulting statistic maps were shared on NeuroVault and can be used to assess the success of the reproductions. At the OHBM Brainhack 2022, our goal was to improve the accessibility and reusability of the database, to facilitate new contributions and to reproduce more pipelines. We focused our efforts on the first two goals. By trying to install the computing environment of the database, contributors provided feedback on the instructions and on specific issues they faced during the installation. Two major improvements were made for the download of the necessary data: the original fMRI dataset and the original results (statistic maps stored in NeuroVault) were added as submodules to the GitHub repository. Finally, propositions were made to facilitate contributions: the use of the Giraffe toolbox for people that are not familiar with NiPype and the creation of a standard template to reproduce a new pipeline. With these improvements, we hope that it will be easier for new people to contribute to reproduction of new pipelines and to grow the codebase.Successful reproduction of a large EEG study across software packages
Participants: Nina Forde, Camille Maumet.
As an active field of research and with the development of state-of-the-art algorithms to analyze EEG datasets, the parametrization of Electroencephalography (EEG) analysis workflows has become increasingly flexible and complex, with a great variety of methodological options and tools to be selected at each step. This high analytical flexibility can be problematic as it can yield to variability in research outcomes. Therefore, growing attention has been recently paid to understand the potential impact of different methodological decisions on the reproducibility of results. In this paper, we aim to examine how sensitive the results of EEG analyses are to variations in preprocessing with different software tools. We reanalyzed the shared EEG data (N=500) from (Williams et al. 2021) using three of the most commonly used EEG software tools: EEGLAB, Brainstorm and FieldTrip. After reproducing the same original preprocessing workflow in each software, the resulting evoked-related potentials (ERPs) were qualitatively and quantitatively compared in order to examine the degree of consistency/discrepancy between softwares. Our findings show a good degree of convergence in terms of the general profile of ERP waveforms, peak latencies and effect size estimates related to specific signal features. However, considerable variability was also observed in the magnitude of the absolute voltage observed with each software package as reflected by the similarity values and observed statistical differences at particular channels and time instants. In conclusion, we believe that this study provides valuable clues to better understand the impact of the software tool on the analysis of EEG results. This work was done in collaboration with Dr. Aya Kabbara and Dr. Mahmoud Hassan. Associated publication: 49.Open and reproducible neuroimaging: from study inception to publication
Participants: Camille Maumet.
Empirical observations of how labs conduct research indicate that the adoption rate of open practices for transparent, reproducible, and collaborative science remains in its infancy. This is at odds with the overwhelming evidence for the necessity of these practices and their benefits for individual researchers, scientific progress, and society in general. To date, information required for implementing open science practices throughout the different steps of a research project is scattered among many different sources. Even experienced researchers in the topic find it hard to navigate the ecosystem of tools and to make sustainable choices. Here, we provide an integrated overview of community-developed resources that can support collaborative, open, and reproducible neuroimaging throughout the entire research cycle from inception to publication and across different neuroimaging modalities. We review tools and practices supporting study inception and planning, data acquisition, research data management, data processing and analysis, and research dissemination. We believe it will prove helpful for researchers and institutions to make a successful and sustainable move towards open and reproducible science and to eventually take an active role in its future development. This work was done as part of an international collaboration led by Drs. Guiomar Niso, Rotem Botvinik-Nezer and Jochem Rieger.Review Paper: Reporting Practices for Task fMRI Studies
Participants: Camille Maumet.
What are the standards for the reporting methods and results of fMRI studies, and how have they evolved over the years? To answer this question we reviewed 160 papers published between 2004 and 2019. Reporting styles for methods and results of fMRI studies can differ greatly between published studies. However, adequate reporting is essential for the comprehension, replication and reuse of the study (for instance in a meta-analysis). To aid authors in reporting the methods and results of their task-based fMRI study the COBIDAS report was published in 2016, which provides researchers with clear guidelines on how to report the design, acquisition, preprocessing, statistical analysis and results (including data sharing) of fMRI studies (Nichols et al. in Best Practices in Data Analysis and Sharing in Neuroimaging using fMRI, 2016). In the past reviews have been published that evaluate how fMRI methods are reported based on the 2008 guidelines, but they did not focus on how task based fMRI results are reported. This review updates reporting practices of fMRI methods, and adds an extra focus on how fMRI results are reported. We discuss reporting practices about the design stage, specific participant characteristics, scanner characteristics, data processing methods, data analysis methods and reported results. This work was part as a collaboration during the PhD thesis of Freya Acar from Uni. Ghent, co-supervised by Drs. Ruth Seurinck and Beatrijs Moerkerke. Associated publication: 11.An Iterative Centroid Approach for Diffeomorphic Online Atlasing
Participants: Antoine Legouhy, Olivier Commowick.
Online atlasing, i.e., incrementing an atlas with new images as they are acquired, is key when performing studies on very large, or still being gathered, databases. Regular approaches to atlasing however do not focus on this aspect and impose a complete reconstruction of the atlas when adding images. We propose instead a diffeomorphic online atlasing method that allows gradual updates to an atlas. In this iterative centroid approach, we integrate new subjects in the atlas in an iterative manner, gradually moving the centroid of the images towards its final position. This leads to a computationally cheap approach since it only necessitates one additional registration per new subject added. We validate our approach on several experiments with three main goals: 1- to evaluate atlas image quality of the obtained atlases with sharpness and overlap measures, 2- to assess the deviation in terms of transformations with respect to a conventional atlasing method and 3- to compare its computational time with regular approaches of the literature. We demonstrate that the transformations divergence with respect to a state-of-the-art atlas construction method is small and reaches a plateau, that the two construction methods have the same ability to map subject homologous regions onto a common space and produce images of equivalent quality. The computational time of our approach is also drastically reduced for regular updates. Finally, we also present a direct extension of our method to update spatio-temporal atlases, especially useful for developmental studies. Associated publication: 23.Reproducibility of motor task-based fNIRS and comparison with functional MRI in healthy adults
Participants: Nolwenn Jegou, Elise Bannier, Emmanuel Caruyer, Isabelle Corouge.
We studied the ability and reproducibility of fNIRS to map the cortical motor areas. Simultaneously acquired fMRI was used as a reference and functional maps of both modalities were obtained from GLM analysis. NIRS results shows satisfactory reproducibility but partial agreement with fMRI 37.8.1.2 Detection and learning
In this section, we summarize our contributions that focus on information extraction from medical imaging data. Machine learning methods have been proposed to detect brain abnormalities, in order to assist and improve the quality of a diagnosis. We also contributed to the field of bimodal EEG-fMRI neurofeedback, crucial for a better understanding of brain mechanism.
A study on loss functions and decision thresholds for the segmentation of multiple sclerosis lesions on spinal cord MRI
Participants: Burhan Rashid Hussein, Cédric Meurée, Malo Gaubert, Arthur Masson, Anne Kerbrat, Benoit Combès, Francesca Galassi.
Multiple sclerosis (MS) patients often present hyper-intense T2-w lesions in the spinal cord. The severe imbalance between background and lesion classes poses a major challenge to Deep Learning segmentation approaches, requiring for ad hoc strategies. Careful selection of the loss function and adjustment of the conventional 0.5-thresholding may help mitigating this issue. Our results show the performance advantages of loss functions based on the Tversky Index and the benefits of threshold tuning over more standard settings and the state-of-the-art model for MS lesion segmentation on spinal cord MRI. Associated publication: 48.A deep learning solution for chronic stroke lesion segmentation in brain MRI
Participants: Lounès Meddahi, Arthur Masson, Elise Bannier, Stéphanie Leplaideur, Francesca Galassi.
Stroke is one of the leading causes of long-term adult disability worldwide. Post-stroke rehabilitation is crucial for long-term patient recovery. Determining the volume and location of lesions caused by stroke is essential to guide treatment and provide effective rehabilitation. Currently, the gold standard for chronic stroke lesion segmentation is manual tracing, a procedure that requires knowledge, is time consuming and prone to inter‐rater variability. Automatic segmentation algorithms have the potential to overcome these limitations. While a large number of solutions have been proposed for the automatic segmentation of lesions in the acute phase, tools for chronic stroke lesion segmentation are underdeveloped. Methods for acute stroke are not readily applicable to chronic stroke due to the different characteristics of the imaging protocol and of the lesion itself. In the context of an M1 internship, promising results have been obtained by adapting, training, and validating a fully convolutional neural network on a public dataset (ATLAS). Further validation and optimisation on a private annotated dataset is currently underway.Incomplete hippocampal inversion and hippocampal subfield volumes: implementation and inter-reliability of automatic segmentation
Participants: Agustina Fragueiro, Claire Cury.
The incomplete hippocampal inversion (IHI) is an atypical anatomical pattern of the hippocampus. However, the hippocampus is not a homogeneous structure, as it consists of segregated subfields with specific characteristics. While IHI is not related to whole hippocampal volume, higher IHI scores have been associated to smaller CA1 in aging. Although the segmentation of hippocampal subfields is challenging due to their small size, there are algorithms allowing their automatic segmentation. By using a Human Connectome Project dataset of healthy young adults, we first tested the inter-reliability of two methods for automatic segmentation of hippocampal subfields, and secondly, we explored the relationship between IHI and subfield volumes. Results evidenced strong correlations between volumes obtained thorough both segmentation methods. Furthermore, higher IHI scores were associated to bigger subiculum and smaller CA1 volumes. Here, we provide new insights regarding IHI subfields volumetry, and we offer support for automatic segmentation inter-method reliability.RNN-LSTM neural network for predicting fMRI neurofeedback scores from EEG signals
Participants: Claire Cury, Pierre Maurel, Caroline Pinte.
In the context of neurofeedback (NF), simultaneous acquisitions with electroencephalography (EEG) and functional magnetic resonance imaging (fMRI) provide more effective NF training due to their complementarity. However, the use of MRI is expensive and draining for a subject. Therefore, we would like to reduce its use. We propose a method based on a recurrent neural network that consists in learning a model from simultaneous EEG-fMRI acquisitions to predict NF-fMRI scores with EEG signals alone. Associated publication: 57.8.1.3 Quantitative imaging
Quantitative imaging is essential for an accurate and specific characterisation of tissue integrity or neural activity, among others. We contributed to methods in fiber tractography, and specifically tractograms evaluation measures.
A convolutional Wasserstein distance for tractography evaluation: complementary study to state-of-the-art measures.
Participants: Thomas Durantel, Julie Coloigner, Olivier Commowick.
Evaluation and comparison of tractograms are crucial and open problems that need to be solved in order to evaluate the false positives rate and variability of tractography algorithms. In this context, a lot of measures have been developed and are typically used to judge the quality of a tractogram. They however do not rely on the same quantities extracted from the tractograms to compare and may thus not evaluate the same aspects of tractogram quality. To evaluate this aspect and the measures redundancy or complementarity, we perform a quantitative analysis of the most common ones, both on simulated data and in real circumstances. We also propose a new evaluation measure based on optimal transport theory. We show that, when used in conjunction, these measures can provide a more in depth comparison of tractograms and thus a more complete evaluation. Associated publication: 32.8.2 Translational research
Our goal is also to provide new computational solutions for our target clinical applications (Alzheimer's disease, psychiatry, neurology or public health issues), allowing a more appropriate representation of the data for image analysis and detection of specific biomarkers. In this section, we present the contributions of the last year in the clinical applications of behavior and neuro-inflammation.
8.2.1 Behavior
Disrupted brain structural network connectivity among depressive patients and associations with treatment response
Participants: Julie Coloigner, Sebastien Dam, Pierre Maurel.
Mood depressive disorder (MDD) affects the emotional state as expressed as a persistent feeling of sadness and loss of interest. Antidepressant medications are first line treatment for depression. In this work, we propose to identify patterns of MDD via a cross-sectional cohort, with the assumption that alterations in brain connectivity may constitute a sensitive biomarker of depression and more specifically of poor outcome of a mood depressive episode. Using diffusion magnetic resonance imaging, we performed structural connectivity analyses using graph theory approach on a cohort of depressed patients and healthy volunteers. In order to study illness improvement, the MDD patients went through two clinical interviews at baseline and at 6 months follow-up, thus allowing us to classify them into “responders” (R) or “non-responders” (NR) based on the Clinical Global Impression-Improvement score. First, the threshold-free network-based statistics (TFNBS) was conducted to highlight the graph modifications between the different groups. Second, we performed a statistical analysis of topological metrics tests between depressed patients versus healthy controls and between R versus NR.Shape-based features of white matter fiber-tracts associated with outcome in Major Depression Disorder
Participants: Claire Cury, Julie Coloigner.
Major depression is a leading cause of disability due to its trend to recurrence and treatment resistance. Currently, there are no biomarkers which could potentially identify patients with risk of treatment resistance. In this original paper, we propose a two-level shape analysis of the white matter bundles based on the Large Diffeomorphic Deformation Metric Mapping framework, to study treatment resistant depression. Fiber bundles are characterised via the deformation of their center line from a centroid shape. We developed two statistical analyses at a global and a local level to identify the most relevant bundles related to treatment resistant depression. Using a prospective longitudinal cohort including 63 patients. We applied this approach at baseline on 50 white matter fiber-tracts, to predict the clinical improvement at 6 months. Our results show a strong association between three bundles and the clinical improvement 6 months after. More precisely, the rightsided thalamo-occipital fascicle and optic radiations are the most robust followed by the splenium. The present study shows the interest in considering white matter shape in the context of depression, contributing to improve our understanding of neurobiological process of treatment resistance depression. Associated publication: 29.Imaging biomarker discovery in major depressive disorder with diffusion MRI multi-compartment models
Participants: Renaud Hédouin, Olivier Commowick, Julie Coloigner, Gabriel Robert.
Major depressive disorder (MDD) is a disease widespread all over the world associated with a large and increasing economic, societal, and personal burden. Yet, the pathophysiology underlying MDD is not well understood, and rates of either non-response to treatment or MDD recurrence remain high. Many studies have already demonstrated impaired WM integrity, which corresponds to a FA decrease in the patient group compared to control subjects, with significant results found in several brain regions. Yet, the comparison between treatment-resistant and non-treatment-resistant populations has never been addressed. The FA at the voxel level provided by the classic diffusion tensor imaging (DTI) does not accurately describe the underlying microstructure. We propose a tract-based pipeline analysis using multi-compartment models (MCM) that allows to model water diffusion under the voxel resolution.Changes in emotional and behavior problems, and brain morphometry following mild traumatic brain injury in early adolescence: A pre-post study design
Participants: Fanny Dégeilh.
Studies comparing children with and without a traumatic brain injury (TBI) have shown that pediatric TBI is associated with difficulties in a large range of functional domains, including emotion and behavior (Catroppa et al., 2015), as well as with changes in brain morphometry (King et al., 2019). However, whether these differences already existed before the injury remains an unsolved question. The large population-based Adolescent Brain Cognitive Development (ABCD) Study (Casey et al., 2018) provides a rare opportunity to explore it. This pre-post design study aims to examine changes in emotional and behavioral problems, and brain morphometry following pediatric mTBI. Given its exploratory nature, no a priori hypothesis is formulated. The following baseline and 2-year follow-up data from the ABCD 4.0 curated data release will be used: 1) The Parent Ohio State TBI Screen-Short Modified report (Bogner et al., 2017) to identify children with no-TBI (n=6,394; baseline mean age = 9.9years; 3102 girls) and children who sustained a mild TBI between baseline and 2-year follow-up (n=132; baseline mean age = 9.9years; 58 girls); 2) The syndrome scales of the Child Behavior Checklist (Achenbach & Rescorla, 2000), and 3) Volumes and cortical thickness in 68 Desikan regions (Desikan et al., 2006) computed on T1-weighted images by the ABCD group. Scanner effects will be removed before analyses using longitudinal ComBat (Beer et al., 2020). Sex and parental education will be included as covariates. Multigroup latent change score models will be constructed with the lavaan 0.6-8 package (Rosseel, 2012) to estimate latent difference scores between baseline and follow-up for child behavior and brain structure. Group differences in 4 parameters of interest (mean of the baseline score, rate of change over time, and variances of the baseline and of the change) will be tested using chi-square difference tests (Kievit et al., 2018). Associated publication: 31.Using neuroimaging to assess the impact of alcohol ads on young drinkers
Participants: Quentin Duché, Elise Bannier.
Although alcohol advertising content regulation has been adopted in many countries, its effects remain under-investigated. The IMAJ project is a multidisciplinary collaboration involving different research groups in Rennes. First results, focusing on the effect of ad content on the reward circuit showed significant increase in activation in the fusiform gyrus, the amygdala, the medial and lateral orbito frontal cortex, caudate nucleus and hippocampus when participants were exposed to attractive ad contents for alcohol brands compared to informative ones. Focusing the analysis on the effect of warnings show that image based warnings reduce activation in the reward circuit. The changes in activations in brain areas linked to emotions and the reward system plead for regulation of alcohol ad contents.Building memories on prior knowledge: behavioral and fMRI evidence of impairment in early Alzheimer’s disease
Participants: Pierre-Yves Jonin, Quentin Duché, Elise Bannier, Isabelle Corouge, Jean-Christophe Ferré.
Impaired memory is a hallmark of prodromal Alzheimer's disease (AD). Prior knowledge associated with the memoranda improves memory in healthy individuals, but we ignore whether the same occurs in early AD. We used functional MRI to investigate whether prior knowledge enhances memory encoding in early AD, and whether the nature of this prior knowledge matters. Patients with early AD and Controls underwent a task-based fMRI experiment where they learned face-scene associations. Famous faces carried pre-experimental knowledge (PEK), while unknown faces with which participants were familiarized prior to learning carried experimental knowledge (EK). Surprisingly, PEK strongly enhanced subsequent memory in healthy controls, but importantly not in patients. Partly nonoverlapping brain networks supported PEK vs. EK associative encoding in healthy controls. No such networks were identified in patients. In addition, patients displayed impaired activation in a right sub hippocampal region where activity predicted successful associative memory formation for PEK stimuli. Despite the limited sample sizes of this study, these findings suggest that the role prior knowledge in new learning might have been so far overlooked and underestimated in AD patients. Prior knowledge may drive critical differences in the way healthy elderly and early AD patients learn novel associations. Associated publication: 21.Multimodal MRI cerebral correlates of verbal fluency switching and its impairment in women with depression
Participants: Elise Bannier, Isabelle Corouge, Jean-Christophe Ferré, Gabriel Robert.
Background The search of biomarkers in the field of depression requires easy implementable tests that are biologically rooted. Qualitative analysis of verbal fluency tests (VFT) are good candidates, but its cerebral correlates are unknown. Methods We collected qualitative semantic and phonemic VFT scores along with grey and white matter anatomical MRI of depressed (n = 26) and healthy controls (HC, n = 25) women. Qualitative VFT variables are the “clustering score” (i.e. the ability to produce words within subcategories) and the “switching score” (i.e. the ability to switch between clusters). The clustering and switching scores were automatically calculated using a data-driven approach. Brain measures were cortical thickness (CT) and fractional anisotropy (FA). We tested for associations between CT, FA and qualitative VFT variables within each group. Results Patients had reduced switching VFT scores compared to HC. Thicker cortex was associated with better switching score in semantic VFT bilaterally in the frontal (superior, rostral middle and inferior gyri), parietal (inferior parietal lobule including the supramarginal gyri), temporal (transverse and fusiform gyri) and occipital (lingual gyri) lobes in the depressed group. Positive association between FA and the switching score in semantic VFT was retrieved in depressed patients within the corpus callosum, right inferior fronto-occipital fasciculus, right superior longitudinal fasciculus extending to the anterior thalamic radiation (all p < 0.05, corrected). Conclusion Together, these results suggest that automatic qualitative VFT scores are associated with brain anatomy and reinforce its potential use as a surrogate for depression cerebral bases. Associated publication: 16.Interactions between emotions and eating behaviors: Main issues, neuroimaging contributions, and innovative preventive or corrective strategies
Participants: Elise Bannier.
Emotional eating is commonly defined as the tendency to (over)eat in response to emotion. Insofar as it involves the (over) consumption of high-calorie palatable foods, emotional eating is a maladaptive behavior that can lead to eating disorders, and ultimately to metabolic disorders and obesity. Emotional eating is associated with eating disorder subtypes and with abnormalities in emotion processing at a behavioral level. However, not enough is known about the neural pathways involved in both emotion processing and food intake. In this review, we provide an overview of recent neuroimaging studies, highlighting the brain correlates between emotions and eating behavior that may be involved in emotional eating. Interaction between neural and neuro-endocrine pathways (HPA axis) may be involved. In addition to behavioral interventions, there is a need for a holistic approach encompassing both neural and physiological levels to prevent emotional eating. Based on recent imaging, this review indicates that more attention should be paid to prefrontal areas, the insular and orbitofrontal cortices, and reward pathways, in addition to regions that play a major role in both the cognitive control of emotions and eating behavior. Identifying these brain regions could allow for neuromodulation interventions, including neurofeedback training, which deserves further investigation. Associated publication: 19.Brain responses to food choices and decisions depend on individual hedonic profiles and eating habits in healthy young women
Participants: Elise Bannier.
The way different food consumption habits in healthy normal-weight individuals can shape their emotional and cognitive relationship with food and further disease susceptibility has been poorly investigated. Documenting the individual consumption of Western-type foods (i.e., high-calorie, sweet, fatty, and/or salty) in relation to psychological traits and brain responses to food-related situations can shed light on the early neurocognitive susceptibility to further diseases and disorders. We aimed to explore the relationship between eating habits, psychological components of eating, and brain responses as measured by blood oxygen level-dependent functional magnetic resonance imaging (fMRI) during a cognitive food choice task and using functional connectivity (FC) during resting-state fMRI (rsfMRI) in a population of 50 healthy normal-weight young women. A Food Consumption Frequency Questionnaire (FCFQ) was used to classify them on the basis of their eating habits and preferences by principal component analysis (PCA). Based on the PCA, we defined two eating habit profiles, namely, prudent-type consumers (PTc, N = 25) and Western-type consumers (WTc, N = 25), i.e., low and high consumers of western diet (WD) foods, respectively. The first two PCA dimensions, PCA1 and PCA2, were associated with different psychological components of eating and brain responses in regions involved in reward and motivation (striatum), hedonic evaluation (orbitofrontal cortex, OFC), decision conflict (anterior cingulate cortex, ACC), and cognitive control of eating (prefrontal cortex). PCA1 was inversely correlated with the FC between the right nucleus accumbens and the left lateral OFC, while PCA2 was inversely correlated with the FC between the right insula and the ACC. Our results suggest that, among a healthy population, distinct eating profiles can be detected, with specific correlates in the psychological components of eating behavior, which are also related to a modulation in the reward and motivation system during food choices. We could detect different patterns in brain functioning at rest, with reduced connectivity between the reward system and the frontal brain region in Western-type food consumers, which might be considered as an initial change toward ongoing modified cortico-striatal control. Associated publication: 15.8.2.2 Neuro-inflammation
Impact of lesion damages along the whole motor pathways on disability in multiple sclerosis
Participants: Malo Gaubert, Benoit Combès, Elise Bannier, Jean-Christophe Ferré, Anne Kerbrat.
The anatomical substrate of motor disability in MS patients is not fully understood. Studying the distribution of corticospinal tracts (CST) lesions per side, from the brain to the end of the thoracic spinal cord (SC) could provide a better association with patient motor deficits evaluated per limb. Objectives: i) To describe lesion preferential location along the CST; ii) To investigate the association between CST lesions and motor functional consequences, as measured using the EDSS, and the ASIA motor scores and electrophysiology (Central motor conduction time (CMCT)) per limb. Methods: 21 relapsing remitting MS (median EDSS=2.5) and 9 progressive MS patients (median EDSS=5.2) with clinical pyramidal symptoms were scanned on a 3T Siemens MRI scanner. White matter lesions were segmented on 3D FLAIR for the brain, on T2* for cervical SC and T2 for thoracic SC. For each patient, registration to an atlas was computed using Anima and SCT toolboxes. Lesion volume fraction along the CST (defined as "lesion volume along the CST"/"overall CST volume") was calculated separately for the both sides on 3 regions: brain including brainstem, C1 to C7 (C1C7) and T1 to T10 (T1T10). Finally, the relationships between lesion volume fraction and the associated lateralized disability scores were assessed using multiple linear models, adjusting for age and disease duration. Results: In MS patients, lesion volume fraction was higher in the C1C7 portion compared to the brain and T1T10 portion (all p’s<.001; mean=2%, 10% and 2%, for brain, C1C7 and T1T10, resp). No evidence of correlation was found between lateralized lesion volume fraction in each portion of the CST and EDSS score or ASIA score per limb, except for a mild correlation between EDSS and lesion volume fraction on the right CST (standardized beta=.39, p=.041). We observed strong positive associations between lesion volume fraction in C1C7 and CMCT for superior and inferior limbs on the right side and for superior limbs on the left side (all std-beta>.6; all p’s<.005). Finally, we observed a mild positive association between lesion volume fraction in T1T10 and CMCT for inferior limbs on the left side (std-beta=.53; p=.02). Conclusions: CST damage is not homogeneous along the tract and predominates in the cervical portion. It has clear consequences on motor conduction velocities measured using electrophysiology. Future work will include an assessment of lesion severity to better explain lesion consequences on motor disability. Associated publication: 51.Magnetization transfer imaging of the whole spinal cord in multiple sclerosis patients
Participants: Malo Gaubert, Elise Bannier, Jean-Christophe Ferré, Anne Kerbrat, Benoit Combès.
Magnetization transfer ratio (MTR) has shown promise to assess tissue microstructure modification in MS patients. To date, such exploration has been limited to the brain and the cervical spinal cord (SC) portions of the central nervous system (CNS). Studying the MTR abnormalities in the whole SC could provide a better association with ambulatory disability in MS patients. The objectives are i) to compare mean MTR values in MS patients and healthy controls (HC) according to the SC level; ii ) to describe the link between MTR measurements at the cervical and thoracic SC levels; iii) to evaluate the link between MTR measures and disability according to the spinal cord level. For that purpose, 21 relapsing remitting MS (RRMS; median EDSS=2.5), 10 progressive MS patients (PMS; median EDSS=5.25) and 13 HC were scanned on a 3T Siemens MRI scanner. The imaging protocol included 3 MT imaging acquisition slabs to cover the whole SC. For each subject, MTR maps and vertebra labeling were computed using the SCT toolbox. MTR means were computed in semi-automatic delineated SC for the following vertebral levels: C4 to C6, T4 to T6, T9 to T10. Group differences as well as correlations with lesions in the whole SC and EDSS were assessed controlling for age. Evidence of group difference was only found in the cervical SC (C4C6; mean MTR=41.7pu, 39.4pu, 35.4pu for HC, RRMS and PMS resp.; p<.001). No evidence for group difference was found in the thoracic SC. A positive association was found between the mean MTR in the cervical SC and in the thoracic SC (r=.45, p=.01 for T4T6 and r=.54, p=.002 for T9T10) in MS patients. We observed negative associations between mean MTR in the cervical SC and the EDSS score (r=-.51, p=.004) and between mean MTR in the cervical SC and the SC lesion load (r=-.6, p<.001), while no clear evidence of correlation was found between SC lesion load and EDSS score (r=.35; p=.084). No evidence of correlation was found between mean MTR in the thoracic cord and EDSS score. To conclude, the microstructural damage in the SC of MS patients seems to be predominant in the cervical SC and is linked to the lesion load and the disability. In our sample data, the added value of exploring thoracic SC in addition to cervical SC using MTR to explain disability in MS patients seems limited. Potential explanations could be the presence of higher variabilities in MTR measurement in the thoracic SC or the preferential location of MS lesions in the cervical SC. Associated publication: 52.9 Bilateral contracts and grants with industry
9.1 Bilateral contracts with industry
9.1.1 Siemens
Participants: Elise Bannier, Emmanuel Caruyer, Olivier Commowick, Isabelle Corouge, Jean-Christophe Ferré, Jean-Yves Gauvrit.
A collaboration between Siemens, Empenn and the Neurinfo platform is in place and formalized by a research contract. Thanks to this agreement, the Neurinfo platform has received the object code of MRI sequences under development at Siemens for evaluation in clinical research. In addition, the Neurinfo platform has received the source code of selected MRI sequences. As a result, MRI sequences can be developed on site by our team. For example, an MRI diffusion sequence was modified to load arbitrarly diffusion gradient waveforms for the FastMicroDiff project (led by E. Caruyer).
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
MMINCARAV
-
Title:
Multimodal Microstructure-Informed Neuronal Connectivity: Acquisition, Reconstruction, Analysis and Validation
-
Duration:
2019 ->
-
Coordinator:
Jean-Philippe Thiran (jean-philippe.thiran@epfl.ch)
-
Partners:
- Ecole Polytechnique Fédérale de Lausanne (Suisse)
-
Inria contact:
Emmanuel Caruyer
-
Summary:
The objectives of this associate team will be to address new scientific challenges related to the use of multimodal magnetic resonance imaging (MRI) to derive microstructure indices and apply them to the measure of brain connectivity. We will focus on 4 aspects of this: first we will develop novel sampling techniques, with the objective to reduce acquisition time for the accurate reconstruction of microstructure indices using diffusion MRI; next we will propose joint T2 relaxometry and diffusion models for the description of microstructure, to take advantage of the complementarity of both modalities in the estimation of microstructure indices; in continuation, we will propose new statistical and network analysis methods using the microstructure-informed connectome, and evaluate its potential to reduce bias and false positives; last we will develop a realistic simulation tool combining a fine macroscopic description of fiber bundles, with a fast and realistic simulator at the mesoscopic scale developed by LTS5.
10.1.2 Visits of international scientists
Prof. Tim Dyrby
-
Status
Associate Professor
-
Institution of origin:
Technical University of Denmark
-
Country:
Denmark
-
Dates:
Dec 9, 2022
-
Context of the visit:
HDR Emmanuel Caruyer
Prof. Gloria Menegaz
-
Status
Professor
-
Institution of origin:
Università di Verona
-
Country:
Italy
-
Dates:
Dec 9, 2022
-
Context of the visit:
HDR Emmanuel Caruyer
Prof. Derek Jones
-
Status
Professor
-
Institution of origin:
Cardiff University
-
Country:
UK
-
Dates:
Dec 9, 2022
-
Context of the visit:
HDR Emmanuel Caruyer
Prof. Jean-Baptiste Poline
-
Status
Associate Professor
-
Institution of origin:
University of McGill
-
Country:
Canada
-
Dates:
Dec 15, 2022
-
Context of the visit:
discussion of joint projects around analytical variability
Demian Vera
-
Status
PhD Candidate
-
Institution of origin:
National University of Central Buenos Aires
-
Country:
Argentina
-
Dates:
Since Oct 2022
-
Context of the visit:
implementation of a multilayer fNIRS model and experimentation at the Neurinfo platform
10.2 European initiatives
10.2.1 Other european programs/initiatives
European COST Action GLIMR
Participants: Elise Bannier, Camille Maumet.
The GLIMR COST Action (PI: Esther Warnert, Erasmus MC, Netherlands) aims to build a pan-European and multidisciplinary network of international experts in glioma research, patient organisations, data scientists, and MR imaging scientists by uniting the glioma imaging community within Europe and progressing the development and application of advanced MR imaging for improved decision making in diagnosis, patient monitoring, and assessment of treatment response in clinical trials and clinical practice. Camille Maumet leads the work package "WG2 - Multi-site data integration" with Cyril Pernet (University of Edinburgh, UK). GliMR’s first grant period ran from September 2019 to April 2020, during which several meetings were held and projects were initiated, such as reviewing the current knowledge on advanced MRI; developing a General Data Protection Regulation (GDPR) compliant consent form; and setting up the website. A publication led by Patricia Clement (Ghent University, Belgium) was published in 2021 describing the results of this GliMR’s first grant period 14. The Action overcomes the pre-existing limitations of glioma research and is funded until September 2023. New members will be accepted during its entire duration.
10.3 National initiatives
10.3.1 ANR-20-THIA-0018: programme Contrats doctoraux en intelligence artificielle
Participants: Francesca Galassi, Ricky Walsh, Benoît Combès, Olivier Commowick, Ricky Walsh.
-
Funding:
Co-funding for a PhD thesis in AI - Duration: 2022-2025.
-
Summary:
Co-funding (50% with UR1) for a doctoral program in Artificial Intelligence. The PhD concerns the automatic segmentation of MS lesions in spinal cord MRI by means of AI-based solutions.
10.3.2 RHU PRIMUS: Transforming the care of patients with Multiple Sclerosis using a multidimensional data-driven clinical decision support system
Participants: Elise Bannier, Benoît Combès, Olivier Commowick, Gilles Edan, Jean-Christophe Ferré, Francesca Galassi, Anne Kerbrat.
-
Funding:
RHU - Duration: 2022-2026 - Budget: 8272kC
-
Partners:
Observatoire Français de la Sclérose en Plaques (OFSEP), France Life Imaging (FLI), Pixyl.
-
Summary:
The overall objective of PRIMUS is to develop and validate a CE-marked data-driven clinical decision support system (CDSS) for multiple sclerosis (MS). The CDSS will support clinical decision-making by providing easily interpretable information on treatment options. MS is a complex disease, with different phenotypes and heterogeneous progression patterns. Over the past two decades, MS practice has been flooded with data and the number of available treatments has considerably increased. Although clinical, biological and imaging information is now being generated on a massive scale, it contributes to clinical decision-making in a rather haphazard, siloed and non-standardised fashion, so that selecting the most appropriate therapeutic option remains hard. PRIMUS contributes to data-driven homogenization of shared decision practices with and for patients with MS. To achieve this goal, the project will develop advanced artificial intelligence solutions, for a patient- and physician-centred CDSS.
10.3.3 EyeSkin-NF : Eye-tracking and skin conductance measures for neurofeedback analysis and validation
Participants: Claire Cury, Elise Bannier, Pierre Maurel, Hachim Bani, Rene-Paul Debroize, Agustina Fragueido.
-
Funding:
Exploratory action Inria - Duration: 2021 - 2024.
-
Summary:
Neurofeedback techniques (NF) or restorative brain computer interfaces (BCI) consist in providing a subject with real-time feedback about its own brain activity, in order to learn self-regulate specific brain regions during NF training. Brain activity can be measured by various techniques such as EEG and/or fMRI. However, analysis of NF sessions is limited due to the difficulty at identifying the origin of failed training. To enhance and monitor participant's motivation in real-time during EEG-fMRI recording, bio-signal can be measured via eye-tracking (ET) or skin conductance (SC) devices. For a precise evaluation of the motivation mental states of interest such as focus, arousal, mind wandering or mental load can be analysed. The main objective of this project is to investigate measures from eye-tracking and skin conductance signals to evaluate in real-time subject’s motivation during NF training.
10.3.4 GRASP: Generalizing Results Across Scientific Pipelines
Participants: Camille Maumet, Boris Clenet.
-
Funding:
Exploratory action Inria - Duration: 2022 - 2024.
-
Summary:
Scientific pipelines are at the heart of modern experimental sciences. But practionners face a highly complex pipeline landscape – different tools, algorithms, parameters – in which different pipelines can lead to contradictory research findings. GRASP will model pipeline-induced variability to derive valid and generalizable results in the field of brain imaging.
10.3.5 ANR-NODAL: Identification de biomarqueurs de maladies neurodégénératives par l'analyse de la connectivité multimodale.
Participants: Julie Coloigner.
-
Funding:
Appel à projets générique 2022 - Duration: 2022 - 2026.
-
Summary:
The neurodegenerative diseases like Alzheimer’s (AD) and Parkinson’s (PD) disease are the consequences of pathological processes that begin decades before the onset of the typical clinical symptoms. However, current diagnosis comes quite late in the course of the disease, while evidences underline the multiple benefits that would be associated with earlier diagnosis. An outstanding challenge for clinical neurosciences is therefore to provide reliable, non-invasive, affordable and easy-to-track biomarkers able to improve both the early detection and the monitoring of neurodegenerative diseases. Recent advances in non-invasive connectome mapping techniques offer great hope for significant progress in taking up this challenge by investigating cerebral organization. Mathematical modelling using graph theory has recently played a significant role in analyzing complex cerebral networks and exhibiting connectivity disruptions that are characteristic of specific brain disorders. However, it is well acknowledged that AD and PD display a progressive multifactorial disruption of functional and structural cerebral networks, all along the course of the diseases. A recent framework called Graph signal Processing is particular promising to shed new light on the complex interplay between brain function and structure, by jointly analyzing functional activity and the underlying structural connectome. For the first time, GSP will be extended to the development of more sensitive metric of AD and PD progression, taking into account the cerebral functional-structural coupling, contrary to the classical biomarkers using a single-modality data and clinical symptoms. In the PRESCO project, we will develop a new multimodal and multi-stage approach using innovative machine learning (ML) methods, adapted for GSP-based features, to provide non-invasive, reliable and easy-to-track candidate biomarkers for each stage of AD and PD diseases. We will apply this approach on two large patients’ cohorts and, then, assess the effectiveness of candidate disease-specific biomarkers on a new innovative local multimodal cohorts including patients with and without cognitive impairment, at various stages of the diseases.
10.3.6 Rapid Neocortical Declarative Learning in normal aging and memory disorders
Participants: Pierre-Yves Jonin, Julie Coloigner.
-
Funding:
Fondation de l'Avenir - Duration: 2020-2022 - Budget: 40k€.
-
Summary:
Our project aims at making the case for the existence of a rapid declarative learning system largely independent from the extended hippocampal system and characterizing its neural bases, by use of experimental psychology, cognitive neuropsychology and neuroimaging methods. This project is led in collaboration with Dr Audrey Noël, Assistant Prof., University of Rennes 2, France, with Dr Gabriel Besson, Associate Researcher, University of Coimbra, Portugal, with Dr Ann-Kathrin Zaiser, Associate Researcher, University of Heidelberg, Germany, with Dr Serge Belliard, PhD, MD, Rennes University Hospital, Neurology Dept., France, and with Dr Anca Pasnicu, MD, Rennes University Hospital, Neurology Dept., France.
10.3.7 Connectivity of the amygdala in depression
Participants: Olivier Commowick, Emmanuel Caruyer, Julie Coloigner, Claire Cury.
-
Funding:
Fondation de France + INCR – Institut des Neurosciences Cliniques de Rennes - Duration: 2019-2023 - Budget: 250k€
-
Summary:
The onset of depression in teenagers and young adults increases the risk to develop a drug-resistant depression in the adulthood. This project aims at evaluating the role of early changes in the microstructure and connectivity of the amygdala. Using a cohort of drug-resistant patients (N=30), non drug-resistant patients (N=30) and controls (N=30), the aim is to identify imaging biomarkers of the pathology and to compare these with emotional and cognitive phenotypes in this population, searching for early differences in the development of the amygdala connectivity. Inclusions are ongoing. This is a colloborative project with M.-L. Paillère Martinot from Paris-Descartes University, as Principal Investigator.
10.3.8 Hybrid EEG/MRI Neurofeedback for rehabilitation of brain pathologies
Participants: Elise Bannier, Isabelle Bonan, Isabelle Corouge, Jean-Christophe Ferré, Jean-Yves Gauvrit, Pierre Maurel, Mathis Fleury.
-
Funding:
Fondation pour la recherche médicale (FRM) - Duration: 2017-2022 - Budget: 370k€
-
Summary:
This project is a continuation of the HEMISFER project ("Hybrid Eeg-MrI and Simultaneous neuro-FEedback for brain Rehabilitation") conducted at Inria Rennes with the support of the Labex "CominLabs". The goal of this project is to make full use of neurofeedback (NF) paradigm in the context of brain rehabilitation. The major breakthrough will come from the coupling associating functional and metabolic information from Magnetic Resonance Imaging (fMRI) to Electro-encephalography (EEG) to “optimize” the neurofeedback protocol. We propose to combine advanced instrumental devices (Hybrid EEG and MRI platforms), with new hybrid Brain computer interface (BCI) paradigms and new computational models to provide novel therapeutic and neuro-rehabilitation paradigms in some of the major mental and neurological disorders of the developmental and the aging brain (e.g. stroke, language disorders, Mood Depressive Disorder (MDD)). Though the concept of using neurofeedback paradigms for brain therapy has somehow been experimented recently (mostly through case studies), performing neurofeedback through simultaneous fMRI and EEG has almost never been done before so far (two teams in the world including us within the HEMISFER CominLabs project). This project will be conducted through a very complementary set of competences over the different involved teams: Empenn U1228, HYBRID and PANAMA Teams from Inria/Irisa Rennes and EA 4712 team from University of Rennes 1.
10.3.9 Knowledge addition through Neuroimaging of Alcohol consumption in healthy young Volunteers, causes or consequences
Participants: Elise Bannier, Quentin Duché, Gabriel Robert.
-
Funding:
Funding: INCR - Duration: 2020-2023 - Budget: 45k€
-
Summary:
Alcohol consumption is responsible for 3 million annual deaths worldwide (5.1 percent of the global burden of disease). It causes disease (liver cirrhosis, cancers, etc.) and other social costs (injuries, road accidents, alcohol dependence, etc.). Excessive alcohol consumption grows through adolescence. This type of behavior has also been shown to have subtle but significant deleterious effects on cognitive function in adolescents. Advances in the field of neuroimaging make it possible to characterize anatomical changes and the evolution of neuropsychological deficits. Besides, focusing on the societal causes of alcohol abuse, a large body of studies show that exposure to alcohol advertising through media bootstraps early consumption initiation, greater desire to drink, increased alcohol use and binge drinking patterns among young people, especially minors. We aim to combine the analysis of the locally acquired IMAJ dataset (PI Karine Gallopel-Morvan, INCA Funding) and data from the european consortium IMAGEN datasets to determine whether there are functional characteristics and external factors that can explain behavior towards alcohol and to extract biomarkers capable of predicting excessive behavior. Relying on the IMAJ dataset, we will analyze whether, depending on warning formats displayed on ads (small and text-only vs. larger, shock-inducing and pictorial), health messages can influence brain activity by decreasing the effect of attractive alcohol content ads on the reward system area and on behavioral responses. Relying on the already effective collaboration of Dr Robert with Prof Schumann, we will explore the longitudinal anatomical and functional data from the IMAGEN cohort to extract biomarkers of alcohol consumption evolution and complement the analysis with the results obtained from the IMAJ dataset.
10.3.10 PHRC EMISEP: Evaluation of early spinal cord injury and late physical disability in Relapsing Remitting Multiple Sclerosis
Participants: Elise Bannier, Emmanuel Caruyer, Benoit Combès, Olivier Commowick, Gilles Edan, Jean-Christophe Ferré, Anne Kerbrat.
-
Funding:
PHRC - Duration: 2016-2023 - Budget: 200k€
-
Summary:
Multiple Sclerosis (MS) is the most frequent acquired neurological disease affecting young adults (1 over 1000 inhabitants in France) and leading to impairment. Early and well adapted treatment is essential for patients presenting aggressive forms of MS. This PHRC (Programme hospitalier de recherche clinique) project focuses on physical impairment and especially on the ability to walk. Several studies, whether epidemiologic or based on brain MRI, have shown that several factors are likely to announce aggressive development of the disease, such as age, number of focal lesions on baseline MRI, clinical activity. However, these factors only partially explain physical impairment progression, preventing their use at the individual level. Spinal cord is often affected in MS, as demonstrated in postmortem or imaging studies. Yet, early radiological depiction of spinal cord lesions is not always correlated with clinical symptoms. Preliminary data, on reduced number of patients, and only investigating the cervical spinal cord, have shown that diffuse spinal cord injury, observed via diffusion or magnetisation transfer imaging, would be correlated with physical impairment as evaluated by the (EDSS) Expanded Disability Status Scale score. Besides, the role of early spinal cord affection (first two years) in the evolution of physical impairment remains unknown. In this project, we propose to address these different issues and perform a longitudinal study on Relapsing Remitting Multiple Sclerosis (RRMS) patients, recruited in the first year of the disease. Our goal is to show that diffuse and focal lesions detected spinal cord MRI in the first two years can be used to predict disease evolution and physical impairment at 5 years. Twelve centers are involved in the study to include 80 patients. To date, all subjects have been included and the last visit of the last patient is scheduled early 2023. The EMISEP data consists of brain and spinal cord structural and quantitative MR images of early MS patients followed over 5 years. Four papers have been published so far on data acquired at baseline on healthy controls and patients. Three papers were co-authored in the context of international collaborations. Additional papers are in preparation.
10.3.11 Estimating the impact of multiple sclerosis lesions in motor and proprioceptive tracts, from the brain to the thoracic spinal cord, on their functions, assessed from clinical tests (MS-TRACTS and MAP-MS)
Participants: Elise Bannier, Benoit Combès, Malo Gaubert, Anne Kerbrat.
-
Funding:
ARSEP, COREC and INCR - Duration: 2020-2023 - Budget: 200k
-
Summary:
Previous studies, whether epidemiologic or based on brain MRI, have shown that several factors were likely to announce aggressive development of the disease, such as age, clinical relapses, number of focal lesions on baseline MRI. However, these factors only partially explain physical disability progression, preventing their use at the individual level. We hypothesize that a fine assessment of damage on specific networks, from the brain to the thoracic cord, offers a relevant biomarker of disability progression in MS. Such damage assessments must take into account both lesion location, assessed on structural brain and cord MR images and lesion severity, assessed using advanced brain and cord imaging through quantitative MRI.We propose to test this hypothesis by combining assessments of lesion location and severity on corticospinal and proprioceptive tracts from the brain to the thoracic cord with clinical and () electrophysiological measurements. The MS-TRACTS study involves two French centers (Rennes,Marseille) and includes a total of 60 relapsing remitting MS patients. The expected outcome is to obtain early biomarkers of physical impairment evolution in RRMS patients, first treated with immunomodulatory treatment. The long-term goal is to provide the clinician with biomarkers able to anticipate therapeutic decisions and support the switch to alternative more aggressive treatment. Inclusions are ongoing. The MAP-MS study involves the same two French centers and will nclude 40 progressive MS patients. The investigation will focus on motor asymetry in these more advanced patients. This study includes two French centers (Rennes, Marseille) and includes a total of 60 patients. The expected outcome is to obtain early biomarkers of physical impairment evolution in RRMS patients, first treated with immunomodulatory treatment. The long-term goal is to provide the clinician with biomarkers able to anticipate therapeutic decisions and support the switch to alternative more aggressive treatment. Inclusions are ongoing.
10.3.12 France Life Imaging (FLI)
Participants: Olivier Commowick, Michael Kain, Camille Maumet, Jean-Christophe Ferré.
-
Funding:
Funding: FLI - Duration: 2012-2023 - Total budget: 2000k€ (phase 1) + 1200k€ (phase 2) + 800k€ (phase 3)
-
Summary:
France Life Imaging (FLI) is a large-scale research infrastructure project to establish a coordinated and harmonized network of biomedical imaging in France. This project was selected by the call “Investissements d’Avenir - Infrastructure en Biologie et Santé”. One node of this project is the node Information Analysis and Management (IAM), a transversal node built by a consortium of teams that contribute to the construction of a network for data storage and information processing. Instead of building yet other dedicated facilities, the IAM node use already existing data storage and information processing facilities (LaTIM Brest; CREATIS Lyon; CIC-IT Nancy; Empenn U1228 Inria Rennes; CATI CEA Saclay; ICube Strasbourg) that increase their capacities for the FLI infrastructure. Inter-connections and access to services are achieved through a dedicated software platform that is developed based on the expertise gained through successful existing developments. The IAM node has several goals. It is building a versatile facility for data management that inter-connects the data production sites and data processing for which state-of-the-art solutions, hardware and software, are available to infrastructure users. Modular solutions are preferred to accommodate the large variety of modalities acquisitions, scientific problems, data size, and to be adapted for future challenges. Second, it offers the latest development that are made available to image processing research teams. The team Empenn fulfills multiple roles in this nation-wide project. Olivier Commowick is participating in the working group workflow and image processing and Michael Kain is the technical manager. Apart from the team members, software solutions like MedInria and Shanoir are part of the software platform.
10.3.13 OFSEP: French Multiple Sclerosis Observatory
Participants: Elise Bannier, Olivier Commowick, Gilles Edan, Jean-Christophe Ferré, Francesca Galassi, Arthur Masson, Benoît Combès, Anne Kerbrat.
-
Funding:
ANR-PIA - Duration: since 2017 - Budget: 175k€
-
Summary:
The French Observatory of Multiple Sclerosis (OFSEP) is one of ten projects selected in January 2011 in response to the call for proposal in the "Investissements d’Avenir - Cohorts 2010" program aunched by the French Government. It allows support from the National Agency for Research (ANR) of approximately 10 million € for 10 years. It is coordinated by the Department of Neurology at the Neurological Hospital Pierre Wertheimer in Lyon (Professor Christian Confavreux), and it is supported by the EDMUS Foundation against multiple sclerosis, the University Claude Bernard Lyon 1 and the Hospices Civils de Lyon. OFSEP is based on a network of neurologists and radiologists distributed throughout the French territory and linked to 61 centers. OFSEP national cohort includes more than 50,000 people with Multiple Sclerosis, approximately half of the patients residing in France. The generalization of longitudinal monitoring and systematic association of clinical data and neuroimaging data is one of the objectives of OFSEP in order to improve the quality, efficiency and safety of care and promote clinical, basic and translational research in MS. For the concern of data management, the Shanoir platform of Inria has been retained to manage the imaging data of the National OFSEP cohort in multiple sclerosis. One long term objective of the OFSEP project is to identify prognostic factors of the evolution of Multiple Sclerosis. The HD Cohort is an enhanced cohort specifically designed for this purpose in which some patients are followed-up on a yearly basis. Additional clinical, quality of life and other patient-reported data is also collected. This study aims at developing personalized predictive tools to improve patient care management, and help in making decision to start, maintain or adapt medical care. Collected data will be processed to extract valuable information enabling to determine specific biomarkers of the evolution of the disease. Multiple Sclerosis brain lesions are of particular interest, hence the need for a careful comparison of lesion segmentation methods. A litterature review enabled to gather most promising cross-sectionnal methods, designed to identify and localize lesions with precise measurement of the lesion load at one particular point in time ; and longitudinal methods which gives more insight on the evolution of those lesions over the different time points. Those later methods are particularly interesting for clinicians for whom the type of lesion evolution is of foremost importance. A cross-sectionnal method and a longitudinal method were trained and evaluated to select the ones which will be used to analyze the entire HD Cohort dataset. Moreover, an experimental and a statistical design to compare the accuracy, sensitivity and specificity of the active/inactive classification of MS patients based on brain MRI as assessed using the analysis of brain was proposed. These designs will allow to assess the interest of re-analyzing the MRI data to improve the quality of the standardized reports used in most epidemiologic studies from the OFSEP cohorts.
10.4 Regional initiatives
10.4.1 PEPERONI : Portable and Personalized Neurofeedback for Stroke Rehabilitation
Participants: Elise Bannier, Julie Coloigner, Isabelle Corouge, Claire Cury, Pierre Maurel, Caroline Pinte.
-
Funding:
Labex CominLabs : from Sept. 2022 to end of 2024 - Budget: 290k€
-
Summary:
Neurofeedback (NF) consists in presenting a person with a stimulus directly related to his or her ongoing brain activity. NF can be used to teach subjects how to regulate their own brain functions by providing real-time sensory feedback of the brain “in action”. Recent studies showed that NF is promising for the treatment of various neuronal pathologies. Electroencephalography (EEG), which has historically been the preferred modality for NF, suffers from a lack of specificity, preventing the transfer of this treatment to clinical use. On the other hand functional Magnetic Resonance Imaging (fMRI) has a good specificity, but it is a cumbersome and expensive modality, making it difficult to develop personalized protocols. In this project, we aim to develop a methodological and experimental framework opening the door to a more portable and personalized NF, for easier and effective clinical use, with a focus on post-stroke motor rehabilitation. We propose to organize the project in four work packages, grouped in two axes. The following figure summarizes the organization of the project.
10.4.2 Région Bretagne: SAD
Participants: Francesca Galassi, Benoit Combès, Burhan Rashid Hussein.
-
Funding:
SAD 2022 - PRIMUS : RHU PRIMUS - Duration: 48 months (start: December 2022) - Budget: 75k€
-
Summary:
Complementary funding for a post-doctoral position (Burhan Rashid Hussein) within the RHU PRIMUS project.
10.4.3 Région Bretagne: ARED MAPPIS
Participants: Camille Maumet, Elodie Germani.
-
Funding:
Budget: 0.5 PhD thesis
-
Summary:
The goal of this thesis is to build a "map" of the neuroimaging pipeline space that will be used to study one or more of the following challenges: 1) Debugging pipelines; 2) Identifying cases where pipelines suffer from violations of their underlying assumptions; 3) Eliminating unsuitable pipelines based on the input dataset. In all of the above cases, a systematic exploration of the processing chain space is impossible. Furthermore, there is generally no ground truth that can be used to compare pipeline results in the neuroimaging setting. In this thesis, we propose to provide a representation of fMRI statistics in a lower dimensional space and use this space to estimate the proximity or distance of different pipelines. By using this map of distances between pipelines in different contexts, we will find a solution to the problems outlined above.
10.4.4 Région Bretagne: Boost Europe MIND
Participants: Camille Maumet.
-
Funding:
Budget: 20k EUR
-
Summary:
The goal of the Boost'Europe MIND project is to implement and share an open dataset including several different treatments of the same neuroimaging study.
10.4.5 Région Bretagne: Boost Europe
Participants: Fanny Dégeilh.
-
Funding:
Budget: 20k EUR
-
Summary:
The main objective of this action is to conduct a pilot study allowing to initiate the implementation of the specific infrastructure for acquiring non-sedated MRI in young children with and without traumatic brain injury at the Neurinfo platform.
11 Dissemination
Participants: Hachim Bani, Elise Bannier, Emmanuel Caruyer, Julie Coloigner, Benoit Combès, Olivier Commowick, Isabelle Corouge, Claire Cury, Sébastien Dam, Fanny Dégeilh, Quentin Duché, Jean-Christophe Ferré, Francesca Galassi, Malo Gaubert, Elodie Germani, Renaud Hédouin, Burhan Rashid Hussein, Nolwenn Jégou, Anne Kerbrat, Arthur Masson, Camille Maumet, Pierre Maurel, Cédric Meurée, Caroline Pinte, Lou Scotto Di Covella.
11.1 Promoting scientific activities
11.1.1 Scientific events: organisation
Member of the organizing committees
- Francesca Galassi and Claire Cury: special track on "Artificial Intelligence in Medical Imaging: from the research lab to the clinical practice", IEEE International Symposium on Computer-Based Medical Systems (CBMS) 2023.
National workshop
- Empenn co-organized, with the Bionic France company, the 2022 national workshop on NIRS, "Mulitmodal acquisition and data analysis", gathering 50 attendants both at Inria-Rennes Center and remote, Dec 15-16.
11.1.2 Scientific events: selection
Chair of conference program committees
- Julie Coloigner: communication chair of IEEE International Symposium on Biomedical Imaging (ISBI) 2023.
- Claire Cury and Pierre Maurel: scientific committee of IABM23, first edition of the French Conference on Artificial Intelligence in Biomedical Imaging
Reviewer
- Fanny Dégeilh: International Neuropsychology Society (INS) Annual Meeting
- Francesca Galassi, Burhan Rashid Hussein, Cédric Meurée, Julie Coloigner: International Symposium on Biomedical Imaging (ISBI)
- Francesca Galassi, Cédric Meurée: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
- Julie Coloigner: International Symposium on Medical Information Processing and Analysis(SIPAIM)
11.1.3 Journal
Member of the editorial boards
- Olivier Commowick and Benoit Combès, editorial board member, Frontiers resarch topic : "Automatic methods for multiple sclerosis new lesions detection and segmentation"
- Camille Maumet, editorial board member, Scientific Data (Nature).
- Camille Maumet, editorial board member, Communications Biology (Nature).
- Camille Maumet, editorial board member, Neuroinformatics (Springer).
Reviewer - reviewing activities
- Elise Bannier, Scientific Data (Nature)
- Julie Coloigner, Brain Connectivity
- Claire Cury and Olivier Commowick, international conference of Medical Image Computing and Computer Assisted Intervention - MICCAI 2022
- Claire Cury, Pierre Maurel, NeuroImage
- Claire Cury, international conference of Human-Computer Interaction 2023
- Fanny Dégeilh, BRAIN
- Fanny Dégeilh, Cortex
- Fanny Dégeilh, Infant Behavior and Development
- Francesca Galassi, Frontiers in Neuroinformatics
- Malo Gaubert, Benoit Combès: American Journal of Neuroradiology
- Malo Gaubert, Neurobiology of Aging
- Renaud Hedouin: Frontiers in Neuroimaging
- Burhan Rashid Hussein, Computers in Biology and Medicine
- Burhan Rashid Hussein, Expert Systems With Applications
- Burhan Rashid Hussein, MDPI
- Burhan Rashid Hussein, Engineering Applications of Artificial Intelligence
- Burhan Rashid Hussein, Applied Soft Computing
- Burhan Rashid Hussein, International Journal of Computers and Applications
- Burhan Rashid Hussein, Cédric Meurée: PlosOne
- Burhan Rashid Hussein, Computer and Electronics in Agriculture
- Camille Maumet: Nature Methods
- Camille Maumet, Julie Coloigner: Nature Communications
- Pierre Maurel, French Conference on Artificial Intelligence in Biomedical Imaging
- Cédric Meurée, Pierre Maurel: Frontiers in Neuroscience
11.1.4 Invited talks
- Elise Bannier, SOFMER 2022, congress dedicated to physical medecine and rehabilitation : "BOLD functional MRI".
- Isabelle Corouge, "Arterial Spin Labeling (ASL): data processing and quantification", REMI network, January 2022.
- Benoit Combès, Arthur Masson: Assise de l’OFSEP 2022: Challenge nouvelles lésions : perspectives d’évaluation.
- Elodie Germani, "L Codent, L Créent : s'initier à l'informatique par la programmation créative", Colloque Femmes & Sciences, November 2022.
- Nolwenn Jégou, "Reproductibilité d'une tâche motrice en NIRS et comparaison avec l'IRM sur des sujets sains", Workshop Acquisition multimodale et analyse des données - 15-16/12/2022, Rennes.
- Camille Maumet, "An introduction to neuroinformatics" Joint meeting of R Ladies Gaborone and PyData Botswana, Online, May 2022.
- Camille Maumet, "Sharing research products FAIRly", OHBM 2022 educational course: Cultivating open science practices in academic research and culture, Glasgow, UK, June 2022.
- Camille Maumet, "Reviewing neuroimaging flexibility: Components and records of provenance". OHBM 2022: Symposium: What should we do with neuroimaging analytical flexibility?, Glasgow, UK, June 2022.
- Camille Maumet, "Open science: A journey from sharing research artefacts to collaborative research". Rigor and Reproducibility Seminar Series, UF Interdisciplinary T32 in Movement Disorders and Neurorestoration, Gainsville (Online), US, July 2022.
- Camille Maumet, "MRI Post-processing Platforms and How they Compare", MRI Together Global workshop on open, reproducible, and inclusive MR research, session Juggling with data, December 2022.
11.1.5 Leadership within the scientific community
- Camille Maumet is member (by selection) of the national committee on Open Science, Working group "open software" led by Roberto Di Cosmo and François Pellegrini and oversees the activities of the working group "national and international" with Dr. Nicolas Rougier.
- Elise Bannier is member of the organization committee of the ARSEP MRI Workshop
- Elise Bannier is board member of the REMI Network and took part in the organization of 2 virtual meetings (January 20th and November 7th) and one physical meeting (May 30th at ICM) on methodological and pratical aspects of MRI in clinical research
- Elise Bannier is board member (by election) of the French society for MRI (SFRMBM) in Biology and Medecine
11.1.6 Research administration
- Anne Kerbrat, Francesca Galassi and Benoit Combès are involved in the research administration (Accord de Consortium, Analyse de risque) of the RHU Primus project.
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
- ESIR, École Supérieure d’Ingénieur de Rennes:
- Pierre Maurel is the co-head of the Master program "imagerie numérique" (two last year of the Engineering School)
- Pierre Maurel, "General image processing" (30h).
- Pierre Maurel, "Algorithmique et complexité" (30h).
- Pierre Maurel, "Imagerie médicale" (30h).
- Julie Coloigner, "Analyse Multimodale de Signaux Biomédicaux" (12h).
- Claire Cury, "Traitement avancé des images" (Plenary : 12h).
- Claire Cury, "Statistiques" (TD : 8h).
- Claire Cury, "Base de données" (TP : 32h).
- Francesca Galassi, co-responsable of the "Apprentissage artificiel" course, ESIR2 INFO (30h).
- Francesca Galassi, responsable of the "Algoritmique des graphes" course, ESIR1 (24h).
- Francesca Galassi, responsable of the "Base de données" course, ESIR2 SNR (24h).
- Francesca Galassi, "Imagerie médicale" (TP: 15h).
- Cédric Meurée, "Algorithmique et complexité" (TD: 14h, TP: 14h)
- Caroline Pinte, "Mathématiques appliquées au traitement d'images" (TP: 18h)
- Caroline Pinte, "Projets d'imagerie médicale" (TP: 10h)
- Caroline Pinte, "Neurofeedback (NF) et Brain Computer Interface (BCI) : Une Introduction" (Plenary: 4h)
- Sébastien Dam, "Algorithmique et complexité" (TP: 28h).
- Master SIBM, M2, University of Angers-Brest-Rennes:
- Jean-Christophe Ferré is head of the master.
- Benoît Combès is co-head of the UE “Modélisation et Apprentissage Automatique pour le Traitement des Images Médicales”.
- Camille Maumet is co-head of the UE “Gestion de données massives et complexes”.
- Emmanuel Caruyer, "Méthodes d'analyse d'IRM de diffusion" (Plenary: 3h).
- Julie Coloigner, "Méthode d'analyse de la connectivité cérébrale" (Plenary: 3h).
- Benoit Combès, "Méthodes de segmentation pour l'imagerie médicale" (Plenary: 3h).
- Benoit Combès, “Méthodes de recalage linéaire et non-linéaires des images médicales” (Plenary: 6h).
- Benoit Combès, “Applications des méthodes de traitement des images médicales” (Plenary: 3h).
- Benoit Combès, “Eléments de statistiques pour l’induction scientifique” (Plenary: 4.5h).
- Benoit Combès, Camille Maumet, “Soutenance de présentations critiques d'articles scientifiques” (TD: 3h).
- Isabelle Corouge, "IRM de perfusion par Arterial Spin Labeling (ASL)" (Plenary: 3h).
- Camille Maumet, "Workflows de traitements d'images" (Plenary: 3h).
- Elise Bannier, "Imagerie fonctionnelle cérébrale" (Plenary: 1h).
- Quentin Duché, "Traitement des données d'IRM fonctionnelle" (Plenary: 1h).
- Elise Bannier, “Utilisation et réutilisation des données d'imagerie” (Plenary: 1h).
- ENS Rennes/UR1:
- Emmanuel Caruyer, "Méthodes numériques pour le traitement d'images", L3 SIF (Plenary: 20h).
- Pierre Maurel, "Introduction to image processing" (20h).
- Master Informatique, ISTIC, UR1:
- Julie Coloigner, "Computer Vision" (Plenary: 10h)parcours Science informatique (SIF), M2.
- Elodie Germani, "Option Machine Learning" (TP: 10h), specialités IL (Ingenierie Logiciel) et CCN (Competences Complémentaires Numériques).
- Licence BECV (Biologie, Environnement, Chimie du Vivant), University of Rennes 1: Elodie Germani, "Mathématiques pour la biologie" (TD: 27h).
- Bachelor for speech therapy, L3, University of Rennes: Elise Bannier, "Imagerie fonctionnelle cérébrale du langage" (Plenary: 2h).
- Diplome Universitaire MERC (Manipulateur en Recherche Clinique), University of Montpellier: Elise Bannier, "Spécificités de la recherche clinique en imagerie" (Plenary: 7h).
- Master Physique Médicale, M2, University of Rennes:
- Elise Bannier, "Imagerie par Résonance Magnétique " (TD: 2h).
- L2, MR Technologists, University Hospital of Rennes:
- Elise Bannier, "Imagerie par Résonance Magnétique " (Plenary: 6h).
- Master Neuropsychologie, M2, University of Savoie: Pierre-Yves Jonin, "Limites méthodologiques du bilan neuropsychologique à visée diagnostique" (Plenary: 3h30).
- Master Psychologie et neuropsychologie de l’enfant et de l’adulte : langage, cognition et apprentissage, M2, University of Poitiers: Pierre-Yves Jonin, "Méthodologie de l'étude de cas" (Plenary: 3h).
- Master Biologie et Santé, M1, University of Bretagne Occidentale: Pierre-Yves Jonin, "Explorations neuropsychologiques des maladies neurologiques et psychiatriques" (Plenary: 4h).
- Master Psychologie Clinique, Psychopathologie et Psychologie de la Santé, M2, University of Rennes 2:
- Pierre-Yves Jonin, "Neuropsychologie clinique des pathologies neurodégénératives" (Plenary: 4h).
- Pierre-Yves Jonin, "Méthodologie de l'étude de cas" (Plenary: 4h).
- Licence Psychologie, L3, University of Rennes 2:
- Pierre-Yves Jonin, "Les syndromes neuropsychologiques" (TD: 16h).
- Pierre-Yves Jonin, "Approche neuropsychologique du handicap" (TD: 4h).
- Master Neurosciences Cliniques, M2, University of Rennes 1: Pierre-Yves Jonin, "Neurosciences cognitives et cliniques de la mémoire humaine" (Plenary: 3h).
11.2.2 Supervision
PhD
- PhD in progress: Ricky Walsh, “Accurate and robust segmentation of MS lesions in spinal cord MRI”, UR1, from Nov 2022, Francesca Galassi, Benoit Combes, Olivier Commowick (Anne Kerbrat from 2023).
- PhD in progress: Xavier Rolland, “Modeling analytic variability in brain imaging”, CNRS, from Oct 2018 to May 2022, Camille Maumet and Pierre Maurel.
- PhD in progress: Elodie Germani, “Mapping the fMRI pipeline-space towards more robust pipelines”, University of Rennes 1, from Oct 2021, Camille Maumet and Elisa Fromont (Lacodam).
- PhD in progress: Caroline Pinte, “Methodology for enhanced and adapted Neurofeedback training”, Univ. Rennes, from Oct 2021, Claire Cury and Pierre Maurel.
- PhD in progress: Carla Joud, "Analyse conjointe de données multimodales en épilepsie", University of Rennes1, from Nov 2022, Julie Coloigner.
- PhD in progress: Sébastien Dam, "Structural Brain Connectivity and Treatment Response in Mood Depressive Disorder", Inria, from Oct 2022, Julie Coloigner and Pierre Maurel.
- PhD in progress: Jean-Charles Roy, "Apathy in Late Life Depression: New Biomarkers Using Actimetry and Magnetic Resonance Imaging (ACTIDEP)", UR1, from 2021, Julie Coloigner and Garbriel Robert.
- PhD in progress: Constance Bocquillon, "Optimisation of acquisition parameters in diffusion MRI for reconstruction of brain connectivity", UR1, from Oct 2022, Emmanuel Caruyer and Isabelle Corouge.
- PhD in progress: Lisa Hemforth "Methodology for automatic scoring of Incomplete Hippocampal Inversion", Sorbonne University, from Oct. 2021, Claire Cury, Baptiste Couvy-Duchesne (Aramis) and Olivier Colliot (Aramis).
- PhD in progress; Thomas Durantel, "Anatomy and microstructure informed tractography for connectivity evaluation in neurological pathologies", from Nov 2020, Olivier Commowick and Julie Coloigner.
- PhD in progress; Alix Lamouroux, "Connectivity and Neurofeedback", at IMT Atlantique Brest, from Oct. 2022, co-supervised by Julie Coloigner and Pierre Maurel, with Giulia Lioi (Brain team, IMT Atlantique) and Nicolas Farrugia (Brain team, IMT Atlantique).
M2 Internship
- Benoit Combès and Francesca Galassi: Thomas Leguay, "Segmentation of MS lesions in spinal cord MRI", M2 SIF UR1.
- Julie Coloigner and Renaud Hédouin: Carla Joud, "Approche multimodale d'apprentissage automatique pour le diagnostic et la caractérisation de la dépression précoce", M2 calcul scientifique et modélisation, Univ Rennes 1.
- Julie Coloigner and Pierre Maurel: Sébastien Dam, "Méthode d’apprentissage pour le traitement de la connectivité cérébrale dans le cadre de la dépression", M2 UTC.
- Elise Bannier and Julie Coloigner: Sarah Bucquet, "Effect of prenatal exposures to neurotoxicants on metabolism and structural connectivity of the metabolism and structural connectivity as assessed by imaging", M2 IMT.
- Elise Bannier (and colleagues from Inrae): Brieuc Léger, "Cognitive behavioural therapies (CBT) usage in the prevention and treatment of eating disorders".
- Emmanuel Caruyer and Isabelle Corouge: Constance Bocquillon, "Tractography in diffusion MRI: influence of acquisition parameters", M2 SIF.
- Isabelle Corouge and Pierre Maurel: Quentin Monnier, "Evaluation of inter-machine reproducibility in MRI", ESIR
M1 Internship
- Francesca Galassi and Elise Bannier: Lounès Meddahi co-supervised with Stéphanie Leplaideur, "Segmentation of chronic stroke lesions", M1 SIF UR1/ENS.
- Benoit Combès and Francesca Galassi: Clément Bion, "Développement et déploiement d’outils pour la visualisation et l'évaluation de segmentations automatiques de lésions dans des images médicales 3D", M1 ESIR.
- Camille Maumet: Nina Forde co-supervised with Mahmoud Hassan, "Variability in EEG preprocessing", M1 UTC.
Other projects
- Cédric Meurée and Burhan Rashid Hussein: Olivier Dufour and Jaime Morillo, "Fine-tuning of Deep Learning Models for Segmentation of Multiple Sclerosis Lesions on Spinal Cord MRI", Master EIT Data Science, M2, University of Rennes.
- Camille Maumet, co-supervised project: Freya Acar, "Review Paper: Reporting Practices for Task fMRI Studies", in collaboration with Ruth Seurinck, Beatrijs Moerkerke (Ghent University).
- Francesca Galassi: supervisions of Projet Industriel and Contrat Pro, ESIR3.
11.2.3 Juries
- Francesca Galassi: member of Ghiles Reguig's PhD thesis committee, "Contrôle qualité rétrospectif pour l'IRM quantitative", Institut du Cerveau, Sorbonne Université.
- Camille Maumet: member of Sahar Allouch's PhD thesis committee, "La variabilité analytique dans l’estimation des réseaux cérébraux fonctionnels à partir de l’électroencéphalographie", Université Libanaise et Laboratoire Traitement du Signal et de l’Image (LTSI – UMR 1099)
- Elise Bannier was panel expert for two HCERES evaluations
- Julie Coloigner was in the jury of Moyen incitatif Inria 2022, Rennes.
- Claire Cury and Julie Coloigner were in the jury of Ecole Doctorale Biologie Santé Rennes.
11.3 Popularization
11.3.1 Internal or external Inria responsibilities
- Claire Cury, Scientific mediation Officer of the Inria Rennes Scientific mediation team.
- Camille Maumet is a co-organizer of the local version of the program "L codent L créent", an outreach program to send PhD students to teach Python to middle school students in 8 sessions of 45 minutes. It was initiated in Lille, with Anne-Cécile Orgerie and Tassadit Bouadi. The program is currently supported by: Fondation Blaise Pascal, ED MathSTIC, Inria and Fondation Rennes 1.
11.3.2 Articles and contents
- Book "Le corps en images : les nouvelles imagerie pour la santé", CNRS Editions, may 2022. Co-directed by Laure Blanc-Féraud, Emmanuel Caruyer, Christian Jutten, Hervé Liebgott.
- Olivier Commowick: "L'imagerie cérébrale néonatale".
- Elise Bannier, Camille Maumet with Anne Hespel: "L'imagerie au service de la santé : questions éthiques et sociétales".
- Claire Cury, Isabelle Bonan, with Anatole Lécuyer (Inria), Giulia Lioi (IMT): "L'imagerie cérébrale au service de la rééducation" 41.
11.3.3 Education
- Elodie Germani, Caroline Pinte. L codent L créent - Teaching middle schoolers at Collège Le Landry and Les Gayeulles, Rennes. Animation of practical session that aim to initiate young girls to programming and to show them that careers in computer science are accessible.
- Camille Maumet, "L codent, L créent, Rennes Initiation à l’informatique via la programmation créative", Training for teachers in secondary and high schools of the Colloque Femmes & Sciences, November 2022.
11.3.4 Interventions
- Quentin Duché, Hachim Bani, Elise Bannier, Isabelle Corouge, Emmanuel Caruyer, Pierre Maurel. Semaine du cerveau 2022, 14 mars 2022 : Visite de la plateforme Neurinfo.
- Calire Cury, Quentin Duché, Fanny Dégeilh, Elodie Germani, Malo Gaubert, Nolwenn Jégou, Caroline Pinte. Semaine du cerveau 2022, 16 mars 2022 : présentation du quiz grand public "À la découverte des métiers de la recherche en neuroimagerie".
- Claire Cury, Caroline Pinte, Lou Scotto Di Covella, "Le neurofeedback, ou comment peut-on rééduquer notre cerveau ?". Animation, Espace des sciences de Rennes, 14 juin 2022.
- Elise Bannier, Pierre Maurel, "Numérique et Cerveau : ce que nous apporte l'imagerie IRM". Animation, Espace des sciences de Rennes, 21 juin 2022.
- Pierre Maurel, "1 scientifique, 1 classe : Chiche !". Présentation des thématiques et du métier à une classe de terminale et 3 classes de seconde. 17 novembre 2022.
- Elise Bannier, Claire Cury, Fanny Dégeilh, Elodie Germani, Pierre Maurel, participating to the "Speed searching" activity for the Festival des Sciences at Les Champs Libres, October 2022
- Quentin Duché, Elise Bannier - October 2022 : Formation des salariés et bénévoles de l’association Nozambule à Rennes. Présentation des résultats de l'étude sur le marketing de l'alcool.
12 Scientific production
12.1 Major publications
- 1 articleUnsupervised Domain Adaptation With Optimal Transport in Multi-Site Segmentation of Multiple Sclerosis Lesions From MRI Data.Frontiers in Computational Neuroscience14March 2020, 1-13
- 2 articleIsolating the Sources of Pipeline-Variability in Group-Level Task-fMRI results.Human Brain MappingNovember 2021
- 3 articleFocal and diffuse cervical spinal cord damage in patients with early relapsing--remitting MS: A multicentre magnetisation transfer ratio study.Multiple Sclerosis Journal258February 2019, 1113-1123
- 4 articleObjective Evaluation of Multiple Sclerosis Lesion Segmentation using a Data Management and Processing Infrastructure.Scientific Reports81December 2018, 13650
- 5 articleA sparse EEG-informed fMRI model for hybrid EEG-fMRI neurofeedback prediction.Frontiers in Neuroscience13January 2020
- 6 articleMultiple sclerosis lesions in motor tracts from brain to cervical cord: spatial distribution and correlation with disability.Brain - A Journal of Neurology 1437July 2020, 2089-2105
- 7 articleAn iterative centroid approach for diffeomorphic online atlasing.IEEE Transactions on Medical Imaging4192022, 2521-2531
- 8 articleThe impact of Neurofeedback on effective connectivity networks in chronic stroke patients: an exploratory study.Journal of Neural Engineering185September 2021, 056052
- 9 articlePatch-Based Super-Resolution of Arterial Spin Labeling Magnetic Resonance Images.NeuroImage189January 2019, 85-94
- 10 articleAssociation of Gray Matter and Personality Development With Increased Drunkenness Frequency During Adolescence.JAMA Psychiatry774April 2020, 409-419
12.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Scientific books
Scientific book chapters
Doctoral dissertations and habilitation theses
Reports & preprints
Other scientific publications
12.3 Other
Scientific popularization
Educational activities

