2024Activity reportProject-TeamCARMEN
RNSR: 201121001J- Research center Inria Centre at the University of Bordeaux
- In partnership with:Université de Bordeaux
- Team name: Modélisation et calculs pour l'électrophysiologie cardiaque
- In collaboration with:Institut de Mathématiques de Bordeaux (IMB)
- Domain:Digital Health, Biology and Earth
- Theme:Modeling and Control for Life Sciences
Keywords
Computer Science and Digital Science
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.1.4. Multiscale modeling
- A6.2.1. Numerical analysis of PDE and ODE
- A6.2.6. Optimization
- A6.2.7. High performance computing
- A6.2.8. Computational geometry and meshes
- A6.3.1. Inverse problems
- A6.3.3. Data processing
- A6.3.5. Uncertainty Quantification
Other Research Topics and Application Domains
- B1.1.2. Molecular and cellular biology
- B1.1.8. Mathematical biology
- B2.2.1. Cardiovascular and respiratory diseases
- B2.2.6. Neurodegenerative diseases
- B2.4.3. Surgery
- B2.6.1. Brain imaging
- B2.6.2. Cardiac imaging
1 Team members, visitors, external collaborators
Research Scientists
- Jacques Henry [INRIA, Emeritus]
- Peter Langfield [INRIA, Researcher]
- Michael Leguebe [INRIA, Researcher]
- Nejib Zemzemi [INRIA, Researcher]
Faculty Members
- Yves Coudière [Team leader, UNIV BORDEAUX, Professor Delegation]
- Mostafa Bendahmane [UNIV BORDEAUX, Associate Professor, HDR]
- Mark Potse [UNIV BORDEAUX, HDR]
- Lisl Weynans [UNIV BORDEAUX, Professor, from Sep 2024, HDR]
- Lisl Weynans [UNIV BORDEAUX, Associate Professor Delegation, until Aug 2024]
Post-Doctoral Fellows
- Delphine Deshors [UNIV BORDEAUX, Post-Doctoral Fellow, from Sep 2024]
- Chitaranjan Mahapatra [INRIA, Post-Doctoral Fellow, from Jul 2024 until Jul 2024]
- Laetitia Mottet [UNIV BORDEAUX, Post-Doctoral Fellow, until Aug 2024]
PhD Students
- Zeina Chehade [UNIV BORDEAUX]
- Sylvain Fourcade [INRIA, from Oct 2024]
- Narimane Gassa [UNIV BORDEAUX, until Mar 2024]
- Emma Lagracie [UNIV BORDEAUX]
- Niami Nasr [UNIV BORDEAUX, ATER, until Mar 2024]
- Valentin Pannetier [UNIV BORDEAUX]
Technical Staff
- Wissam Bouymedj [UNIV BORDEAUX, Engineer, until Jul 2024]
- Loïc Calvez [UNIV BORDEAUX, Engineer, from Jul 2024]
- Gengis Lourenco [UNIV BORDEAUX, Engineer, from Nov 2024]
- Xavier Muller [UNIV BORDEAUX, Engineer, from Jun 2024]
- Corentin Prigent [UNIV BORDEAUX, Engineer, until Sep 2024]
- Gwladys Ravon [INRIA, Engineer]
Interns and Apprentices
- Yassine Ayoun [UNIV BORDEAUX, Intern, from Jul 2024 until Sep 2024]
- Younes Bouhoreira [INRIA, Intern, from May 2024 until Jun 2024]
- Selma El Badaoui [UNIV BORDEAUX, Intern, from May 2024 until Jul 2024]
- Sylvain Fourcade [UNIV BORDEAUX, Intern, from Feb 2024 until Aug 2024]
- Paul Fournet [UNIV BORDEAUX, Intern, from Jun 2024 until Sep 2024]
- Pierre Sinel–Boucher [INRIA, Intern, from Jun 2024 until Jul 2024]
- Ananth Venkatesh [UNIV BORDEAUX, Intern, from Jun 2024 until Nov 2024]
- Mehdi Yahiaoui [INRIA, Intern, from May 2024 until Jun 2024]
Administrative Assistant
- Flavie Blondel [INRIA]
Visiting Scientists
- Narjess Ben Abid [ENIT TUNIS, from Sep 2024 until Nov 2024]
- Kyoung Lee [University of Auckland, New Zealand, from Jul 2024 until Jul 2024]
- Beata Ondrusova [Institute of Measurement Science, Bratislava]
- Joshua Steyer [KIT - ALLEMAGNE, from Sep 2024]
- Jana Svehlikova [Institute of Measurement Science, Bratislava]
2 Overall objectives
The Carmen team develops and uses models and numerical methods to simulate the electrophysiology of the heart from the molecular to the whole-organ scale, and its relation to measurable signals inside the heart and on the body surface. It aims at:
- improving understanding of normal and pathological cardiac electrophysiology,
- improving the efficiency and accuracy of numerical models,
- exploiting all available electrical signals for diagnosis,
- improving understanding and guidance of ablative treatment of cardiac arrhythmia.
The numerical models developed, analyzed, and used by the team incorporate essentially the gating dynamics of the ion channels in the cardiac cell membranes and the heterogeneities of the cardiac tissue, coupling processes on the cellular scale into macroscopic reaction-diffusion models. The team also works on incorporating any new biological knowledge, at any scale, that helps to understand the mechanisms of arrythmias, their diagnosis or treatment. At the same time we use simpler or reduced models to solve the inverse problems related to non-invasive electrical imaging of the heart.
The fields involved in our research are: ordinary and partial differential equations (ODE & PDE), inverse problems, numerical analysis, high-performance computing, image segmentation, and mesh construction.
A main goal of the team is to contribute to the work packages defined in the project of IHU Liryc, an institute founded in 2011 that focuses on cardiac arrhythmia.
We cooperate with physiologists and cardiologists on several projects. The team is building new models and powerful simulation tools that will help to understand the mechanisms behind cardiac arrhythmias and to establish personalized and optimized treatments. A particular challenge consists in making the simulations reliable and accessible to the medical community.
3 Research program
3.1 Complex models for the propagation of cardiac action potentials
The contraction of the heart is coordinated by a complex electrical activation process which relies on about a million ion channels, pumps, and exchangers of various kinds in the membrane of each cardiac cell. Their interaction results in a periodic change in transmembrane potential called an action potential. Action potentials in the cardiac muscle propagate rapidly from cell to cell, synchronizing the contraction of the entire muscle to achieve an efficient pump function. The spatio-temporal pattern of this propagation is related both to the function of the cellular membrane and to the structural organization of the cells into tissues. Cardiac arrythmias originate from malfunctions in this process. The field of cardiac electrophysiology studies the multiscale organization of the cardiac activation process from the subcellular scale up to the scale of the body. It relates the molecular processes in the cell membranes to the propagation process through the multiscale structure of the tissue and organ, to measurable signals in the heart and to the electrocardiogram, an electrical signal on the torso surface.
Several improvements of current models of the propagation of action potentials are being developed in the Carmen team, based on previous work 43 and on the data available at IHU Liryc:
- Enrichment of the current monodomain and bidomain models 43, 54 by accounting for structural heterogeneities of the tissue at cellular and intermediate scales. Here we focus on multiscale analysis techniques applied to the various high-resolution structural data available at IHU Liryc.
- Coupling of the tissues from the different cardiac compartments and conduction systems. Here, we develop models that couple 1D, 2D and 3D phenomena described by reaction- diffusion PDEs.
These models are essential to improve our understanding of cardiac electrical dysfunction. To this aim, we use high-performance computing techniques in order to numerically explore the complexity of these models.
We use these model codes for applied studies in two important areas of cardiac electrophysiology: atrial fibrillation 45 and sudden-cardiac-death (SCD) syndromes 8, 748. This work is performed in collaboration with several physiologists and clinicians both at IHU Liryc and abroad.
3.2 Simplified models and inverse problems
The medical and clinical exploration of the cardiac electric signals is based on accurate reconstruction of the patterns of propagation of the action potential. The correct detection of these complex patterns by non-invasive electrical imaging techniques has to be developed. This involves solving inverse problems that cannot be addressed with the more complex models. We want both to develop simple and fast models of the propagation of cardiac action potentials and improve the solutions to the reconstruction questions of cardiac electrical imaging techniques.
These questions concern the reconstruction of diverse information, such as cardiac activation maps or, more generally, the whole cardiac electrical activity, from high-density body surface electrocardiograms. It is a possibly powerful diagnosis technique, which success would be considered as a breakthrough. Although widely studied during the last decade, the reconstructed activation maps, for instance, are highly inacurate and have a poor clinical interest. It remains a challenge for the scientific community to understand how body surface signals can better inform on the fine details of arrhythmic mechanisms.
The most usual method consists in solving a Laplace equation on the volume delimited by the body surface and the epicardial surface, for which we contribute by:
- studying in depth the dependance of the inverse problem on inhomogeneities in the torso, conductivity values, the geometry, electrode positions, etc., and
- improving the solution to the inverse problem by using new regularization strategies, factorization of boundary value problems, and the theory of optimal control.
In addition, we have started to explore many alternative approaches including:
- using complete propagation models in the inverse problem, like the bidomain or monodomain equations, for instance in order to localize electrical sources,
- constructing data-based models using e.g. statistical learning techniques, which would accurately represent some families of well-identified pathologies, or allow to combine physics and biology-informed models and clinical data, and
- constructing simpler models of the propagation of the activation front, based on eikonal or level-set equations.
3.3 Numerical techniques
We want our numerical simulations to be efficient, accurate, and reliable with respect to the needs of the medical community. Based on previous work on solving the monodomain and bidomain equations 5, 4, 9, 1, we will focus on:
- high-order numerical techniques with respect to the variables with physiological meaning, like velocity, AP duration and restitution properties and
- efficient, dedicated preconditioning techniques coupled with parallel computing.
Existing simulation tools used in our team rely, among others, on mixtures of explicit and implicit integration methods for ODEs, hybrid MPI-OpenMP parallellization, algebraic multigrid preconditioning, and Krylov solvers. New developments include high-order explicit integration methods and task-based dynamic parallellism.
3.4 Cardiac electrophysiology at the microscopic scale
Traditional numerical models of whole-heart physiology are based on the approximation of a perfect muscle using homogenisation methods. However, due to aging and cardiomyopathies, the cellular structure of the tissue changes. These modifications can give rise to life-threatening arrhythmias, the mechanisms of which we are investigating in collaboration with cardiologists at the IHU Liryc. For this research we are building models that describe the strong heterogeneity of the tissue at the cellular level.
The literature on this type of model is still very limited 60. Existing models are two-dimensional 49 or limited to idealized geometries, and use a linear (purely resistive) behaviour of the gap-juction channels that connect the cells. We propose a three-dimensional approach using realistic cellular geometry (Fig. 1), nonlinear gap-junction behaviour, and a numerical approach that can scale to hundreds of cells while maintaining a sub-micrometer spatial resolution (10 to 100 times smaller than the size of a cardiomyocyte). Following preliminary work in this area by us 39, 38, 37 and by others 60 we proposed a European project with 10 partner institutes and a 5.8M€ budget to develop software that can simulate such models, with micrometer resolution, on the scale of millions of cells, using future exascale supercomputers (microcard.eu). This project ran from April 2021 to October 2024, and involves also the Inria teams CAMUS, STORM and CARDAMOM as well as the Inria-led MMG Consortium.
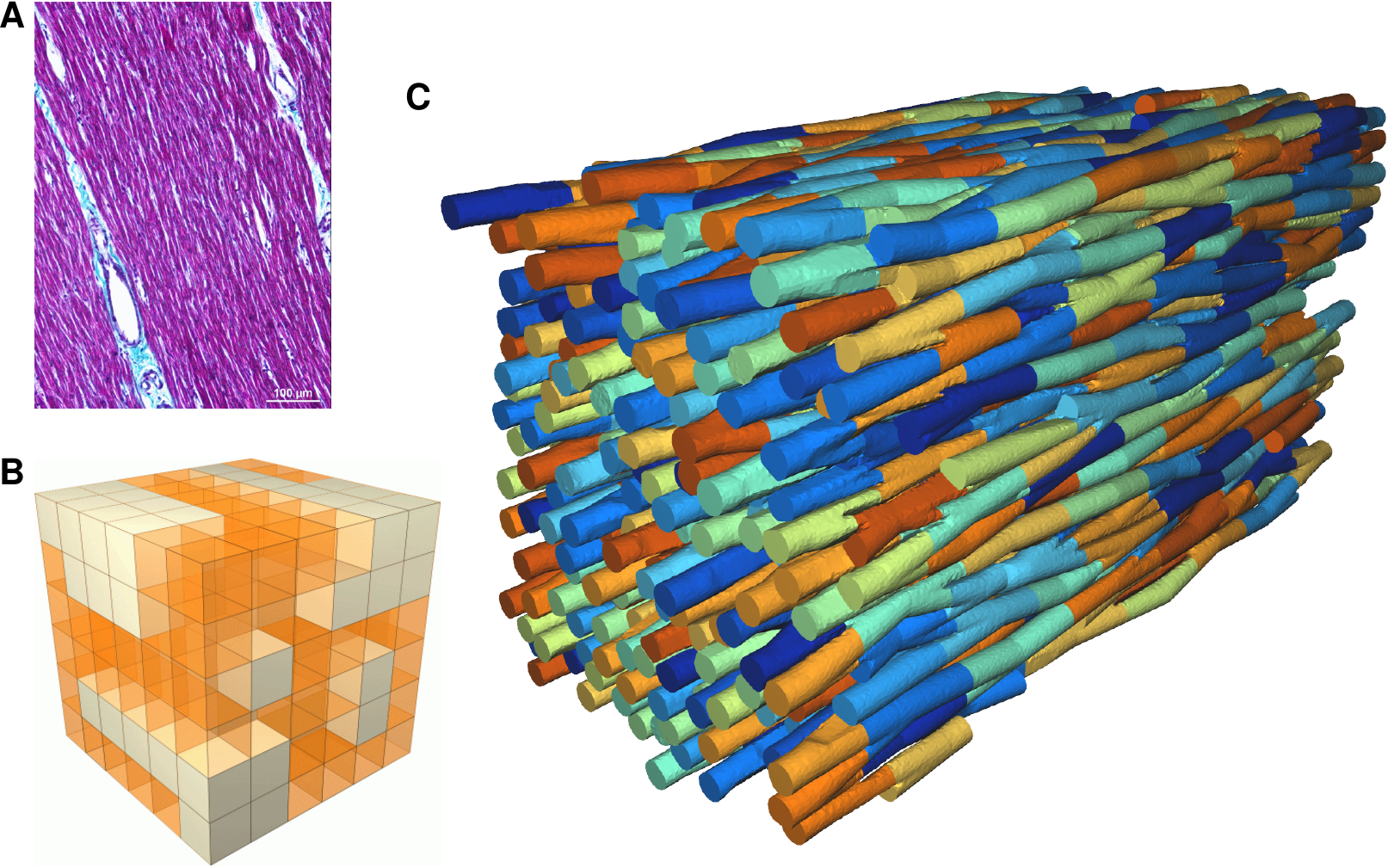
Image of the microstructure from histology, current, insufficient, representation of microstructural defects, and foreseen geometry to be used in microscopic models.
The cell-by-cell bidomain model presents numerous mathematical and computational challenges. First, mathematically, its unusual formulation providing time dynamics as an ordinary differential equation (ODE) at the cell- to-cell connections and cell-to-extracellular matrix interfaces. Second, the ionic model coupled to the nonstandard transmission conditions, introduce stiff non linear dynamics. Third, the simulation would be performed for billions of myocytes, leading to extremely large systems. In the MICROCARD project, we simulate the micromodel using finite volumes, finite elements and boundary element methods. In November 2024 the project was succeeded by the Centre of Excellence MICROCARD-2, in which the Inria teams CARMEN, STORM, CAMUS and TADAAM are involved.
3.5 Models and tools for ablative therapies
Today, the most effective way to treat arrhythmias is to ablate selected regions of the cardiac tissue. The lesions are assumed electrically passive, and consequently create conduction blocks that stop the disorganized propagation of action potentials. The ablation procedure consists in placing a catheter in contact with the targeted site and deliver energy into the tissue. The energy can be overheating by radio-frequency current, electroporating electric pulses or temperature drop (cryotherapy). In practice, the choice of the ablation site is done by the clinician based on previous signal measurements and imagery, and is also guided during the procedure with real-time measurement of the electric signal.
Our team works on several subjects related to ablation techniques that may improve the success rate of the treatments:
- accurate computation of electric fields generated by catheters: complex catheter shapes, contact models, tissue heterogeneities;
- models of creation of the lesions, either through temperature rise (radio-frequency) or electroporation; and
- localization tools to help clinicians target the optimal ablation sites, based on both data of previously ablated patients and synthetic data.
4 Application domains
4.1 Scientific context: IHU Liryc
The University Hospital of Bordeaux (CHU de Bordeaux) is equipped with a specialized cardiology hospital, the Hôpital Cardiologique du Haut-Lévêque, where the group of Professor Michel Haïssaguerre has established itself as a global leader in the field of cardiac electrophysiology 47, 46, 41. Their discoveries in the area of atrial fibrillation and sudden cardiac death syndromes are widely acclaimed, and the group is a national and international referral center for treatment of cardiac arrhythmia. Thus the group also sees large numbers of patients with rare cardiac diseases.
In 2011 the group has won the competition for a 40 million euro Investissements d'Avenir grant for the establishment of IHU Liryc, an institute that combines clinical, experimental, and numerical research in the area of cardiac arrhythmia. The institute works in all areas of modern cardiac electrophysiology: atrial arrhythmias, sudden death due to ventricular fibrillation, heart failure related to ventricular dyssynchrony, and metabolic disorders. It is recognized worldwide as one of the most important centers in this area.
The Carmen team was founded as a part of IHU Liryc. We bring applied mathematics and scientific computing closer to experimental and clinical cardiac electrophysiology. In collaboration with experimental and clinical researchers at Liryc we work to enhance fundamental knowledge of the normal and abnormal cardiac electrical activity and of the patterns of the electrocardiogram, and we develop new simulation tools for training, biological, and clinical applications.
4.2 Basic experimental electrophysiology
Our modeling is carried out in coordination with the experimental teams from IHU Liryc. It helps to write new concepts concerning the multiscale organisation of the cardiac action potentials that will serve our understanding in many electrical pathologies. For example, we model the structural heterogeneities at the cellular scale 40 (the MICROCARD project), and at an intermediate scale between the cellular and tissue scales.
At the atrial level, we apply our models to understand the mechanisms of complex arrythmias and the relation with the heterogeneities at the insertion of the pulmonary veins. We will model the heterogeneities specific to the atria, like fibrosis or fatty infiltration 56, 45. These heterogeneities are thought to play a major role in the development of atrial fibrillation.
At the ventricular level, we focus on (1) modeling the complex coupling between the Purkinje network and the ventricles, which is supposed to play a major role in sudden cardiac death, and (2) modeling the heteogeneities related to the complex organization and disorganization of the myocytes and fibroblasts, which is important in the study of infarct scars for instance.
4.3 Clinical electrophysiology
Treatment of cardiac arrhythmia is possible by pharmacological means, by implantation of pacemakers and defibrillators, and by curative ablation of diseased tissue by local heating, freezing or electroporation. In particular the ablative therapies create challenges that can be addressed by numerical means. Cardiologists would like to know, preferably by noninvasive means, where an arrhythmia originates and by what mechanism it is sustained.
We address this issue in the first place using inverse models, which attempt to estimate the cardiac activity from a (high-density) electrocardiogram. A new project aims to perform this estimation on-site in the catheterization laboratory and presenting the results, together with the cardiac anatomy, on the screen that the cardiologist uses to monitor the catheter positions 50, 35.
4.4 Application in Deep Brain Stimulation
Since 2017, we have been working with neurosurgeons from the Bordeaux University Hospital (Pr Cuny and Dr. Engelhardt) on improving the planning technique for deep brain surgery (DBS) for Parkinson's and Essential tremor diseases. DBS is the last resort to treat the symptoms of Parkinson's disease after the drug Levodopa. The surgery consists in placing electrodes in very specific regions of the patient's brain. These regions are unfortunately not visible on the 1.5 Tesla MRI, the most widely available MRI machines in hospitals. The most effective solution to date is to introduce 5 micro-electrodes (MER) to record the activity of neurons in the patient's brain and to prospect by moving the electrodes in order to find the best location. However, this approach renders the surgery very cumbersome because the patient must be awake during the exploration phase. In addition, this phase takes at least 3 hours and mobilizes a neurologist with his staff. The total duration of the operation is between 7 and 8 hours. Many elderly patients do not tolerate this surgery. We have proposed an approach that avoids the prospecting phase and performs surgery under general anesthesia. The idea is to learn on pairs of clinical landmarks and the position of active electrodes in order to predict the optimal position of the DBS from a pre-operative image. This approach simplifies and standardizes surgery planning. We tested several approaches, 6. We continue to seek approaches to fully automate the targeting process. We carried out a proof of concept by learning on the clinical database of the Bordeaux University Hospital. The clinical validation of our approach is in progress through a clinical trial which includes patients from the University Hospitals of Bordeaux and Lyon. Pr Cuny has submitted a phase 3 national clinical research hospital project (PHRCN) including 11 CHUs in France which has been accepted by the General Directorate for Care Offers (DGOS). The aim is to compare our new approach to the ones used in the other centers. Inria Bordeaux is a partner in this project and we maintain the OptimDBS software and solve any technical problem related to the compatibility of the MRIs exported by our software and the surgical robots in the different centers.
5 Social and environmental responsibility
5.1 Footprint of research activities
We avoid flying whenever we can, and try to keep computers for 7 years, despite the fact that they are not supported by the DSI that long.
Lisl Weynans is involved in the creation of courses about environmental and social transition at Bordeaux University.
5.2 Impact of research results
The MICROCARD project and its successor MICROCARD-2, which we coordinate, has energy efficiency as one of its goals. To this end, our partners in the STORM and CAMUS teams are developing methods to increase the time- and energy-efficiency of cardiac simulation codes.
6 Highlights of the year
6.1 Awards
New professor at the University of Bordeaux: Lisl Weynans since September 2024
6.2 Major Funding
We obtained a 5M€ grant from the EuroHPC joint undertaking to establish the MICROCARD Centre of Excellence (administrative name: MICROCARD-2) for numerical modeling of cardiac electrophysiology at the cellular scale. Details on the project are given in section 10.
7 New software, platforms, open data
7.1 Evolution of existing software
-
CEPS Our cardiac electrophysiology software CEPS received the following updates:
- Thanks to the recruitment of Loïc Calvez, several optimization bottlenecks were removed, which already improved the performance of several runs by more than 30%.
- Addition of the FDA benchmark for cardiac electrophysiology problems, as given in 52.
- A new build system has been proposed by Xavier Muller, which should ease installation of CEPS, and be integrated in a release early 2025.
-
OpenCarp is an open cardiac electrophysiology simulator for in-silico experiments (described below):
- A command-line tool is now available to visualize images of cross sections of .mesh/.sol combinations to obtain fly-through movies.
7.2 New software
7.2.1 CEPS
-
Name:
Cardiac ElectroPhysiology Simulation
-
Keywords:
Simulation, Health, Mesh, Cardiac, 3D, Cardiac Electrophysiology
-
Scientific Description:
As compared to other existing softwares, CEPS aims at providing a more general framework of integration for new methods or models and a better efficiency in parallel. CEPS is designed to run on massively parallel architectures, and to make use of state-of-the-art and well known computing libraries to achieve realistic and complex heart simulations. CEPS also includes software engineering and validation tools.
-
Functional Description:
CEPS is a modular high-performance computing software for performing numerical simulations in cardiac electrophysiology. It is based on modules : - management of geometries represented by meshes in 3D, 2D or 1D (volumes, surfaces, trees), - model simulation of cellular electrophysiology, - calculating the tissue propagation of the action potentials in the cardiac geometries, - calculation of extracardiac potentials, - time approximation methods in order 2, 3 and 4 specific to electrocardiography.
- URL:
-
Contact:
Ceps Dev Team
-
Participants:
Yves Coudière, Michael Leguebe, Valentin Pannetier, Pierre-Elliott Bécue, Florian Caro, Andjela Davidovic, Charlie Douanla Lontsi, Marc Fuentes, Mehdi Juhoor, Pauline Migerditichan, Nejib Zemzemi
-
Partners:
Université de Bordeaux, Fondation Bordeaux Université, CHU de Bordeaux, Inria
7.2.2 CirCE
-
Name:
Circuits for Cardiac Electrophysiology
-
Keywords:
Cardiac Electrophysiology, Electrical circuit
-
Scientific Description:
CirCE meets the need for numerical simulation in mathematical modeling of pacemakers.
-
Functional Description:
Uses vision with electrical schematics to simulate cardiac electrophysiology problems.
- URL:
- Publication:
-
Contact:
Valentin Pannetier
-
Participants:
Valentin Pannetier, Michael Leguebe
7.2.3 OptimDBS
-
Name:
Optimizing the Deep Brain Stimulation
-
Keywords:
Image analysis, Deep brain stimulation, Statistical learning
-
Functional Description:
Targeting software for deep brain stimulation
- URL:
-
Contact:
Nejib Zemzemi
-
Participants:
Nejib Zemzemi, Louise-Amelie Schmitt, Emmanuel Cuny, Julien Engelhardt
-
Partner:
CHU de Bordeaux
7.2.4 MUSIC - Carmen plugins
-
Name:
Carmen plugins for multi-modality imaging in Cardiology
-
Keywords:
Image segmentation, Mesh generation, Image filter, Numerical simulations, Cardiac Electrophysiology, Inverse problem, Finite element modelling, Visualization, 3D interaction, Registration
-
Scientific Description:
Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
-
Functional Description:
Carmen plugins is a collection of toolboxes and pipelines allowing the following functionalities: - Segmenting and filtering the heart, the surrounding organs and the body surface. - Generate/optimize surface and volume of computational meshes for the segmented organs. - Generate fibers orientations for Atria and Ventricles. - Interactively annotate meshes. - Read map and visualize electrical information collected from medical devices. - Numerical simulation of the forward problem using finite elements method. - Method of fundamental solutions for solving the ECGI inverse problem combined with the regularization methods including CRESO,Zero Crossing GCV, RGCV, ADPC and Ucurve. - Landmark based mesh registration. - ECGI pipeline: including segmentation, mesh generation, identification of the vest electrodes.
- Publications:
-
Contact:
Nejib Zemzemi
-
Participants:
Hubert Cochet, Florent Collot, Mathilde Merle, Maxime Sermesant, Julien Castelneau, Mehdi Juhoor, Pauline Migerditichan, Nejib Zemzemi, Yves Coudière
-
Partners:
Université de Bordeaux, IHU - LIRYC
7.3 New platforms
Participants: Mark Potse, Andony Arrieula, Laetitia Mottet, Corentin Prigent, Wissam Bouymedj, Yves Coudière, Gengis Lourenco, Xavier Muller.
OpenCARP
OpenCARP is an open cardiac electrophysiology simulator for in-silico experiments. Its source code is public and the software is freely available for academic purposes. OpenCARP is easy to use and offers single cell as well as multiscale simulations from ion channel to organ level. Additionally, openCARP includes a wide variety of functions for pre- and post-processing of data as well as visualization. The python-based CARPutils framework enables the user to develop and share simulation pipelines, i.e. automating in-silico experiments including all modeling/simulation steps.
KIT and the company Numericor, the main creators of openCARP, with the University of Graz, are partners of the MICROCARD project. The openCARP simulator is developed within the MICROCARD project targeting the creation of a microscopic cardiac model.
We have been contributing by managing the MICROCARD consortium and its contributions to openCARP, and by adapting the partitionning scheme (based on parMETIS) to the context of the microscopic model.
Mmg
Mmg is an open source software suite for simplicial remeshing and an open source Consortium, which participates in the MICROCARD project.
In this context we have been active developers of the Mmg software. Laetitia Mottet (postdoc) worked on ParMmg, the parallel version of Mmg, where she implemented level-set discretization and worked on the robustification of the software. Corentin Prigent (engineer) worked on the serial code mmg3d for 1) its robustification so that it can deal with the extremely complicated topology of the meshes created using segmented data provided by partners in the MICROCARD project, and 2) for the purposes of the MICROCARD project, Mark Potse uses mmg3d for the challenging task of creating artificial models of cardiac muscle fibers (Figure 1C).
In the MICROCARD project, tetrahedral meshes are created using both segmented data and synthetic models. Several improvements made this year in mmg now allow us to 1) use these segmented data to create valid meshes with much less elements than in the initial data, 2) improve the synthetic generation of cardiac tissue. Gengis Lourenco , hired to work on the MICROCARD project, has been working on postprocessing tools related to these meshing activities, notably the ability to construct images of cut planes of the 3D domain for better visualisation.
CEMPACK
CEMPACK is a collection of software that was previously archived in different places. It includes the high-performance simulation code Propag and a suite of software to create geometric models, prepare inputs for Propag, and analyse its outputs. In 2017 the code was collected in an archive on Inria's GitLab platform. The main components of CEMPACK are the following.
-
Propag-5.1
Applied modeling studies performed by the Carmen team in collaboration with IHU Liryc and foreign partners 857, 45, 44, 42 rely on high-performance computations on the national supercomputers Irene, Zay, and Adastra. The Propag-5 code is optimized for these systems. It is the result of a decades-long development first at the Université de Montréal in Canada, then at Maastricht University in the Netherlands, and finally at the Institute of Computational Science of the Università della Svizzera italiana in Lugano, Switzerland. Since 2016 most of the development on Propag has been done by M. Potse at the Carmen team 59. The code scales excellently to large core counts 58 and, as it is controlled completely with command-line flags and configuration files, it can be used by non-programmers. It also features:
- a plugin system for membrane models,
- a completely parallel workflow, including the initial anatomy input and mesh partitioning, which allows it to work with meshes of more than nodes,
- a flexible output scheme allowing hundreds of different state variables and transient variables to be output to file, when desired, using any spatial and temporal subsampling,
- a configurable, LUSTRE-aware parallel output system in which groups of processes write HDF5/netCDF files, and
- CWEB documentation of the entire code base.
The code has been stable and reliable for many years. It can be considered the workhorse for our HPC work until CEPS takes over.
-
Gepetto
The Gepetto suite transforms a surface mesh of the heart into a set of (semi-)structured meshes for use by the Propag software or others. It creates the different fiber orientations in the model, including the transmurally rotating ventricular fibers and the various bundle structures in the atria (figure 2), and creates layers with possibly different electrophysiological properties across the wall. A practically important function is that it automatically builds the matching heart and torso meshes that Propag uses to simulate potentials in the torso (at a resolution of 1 mm) after projecting simulation results from the heart model (at 0.1 to 0.2 mm) on the coarser torso mesh 55. Like Propag, the Gepetto software results from a long-term development that started in Montreal, Canada, around 2002. The code for atrial fiber structure was developed by our team.
-
Blender plugins
Blender is a free software package for the production of 3-D models, renderings, and animations, comparable to commercial software such as Cinema4D. CEMPACK includes a set of plugins for Blender that facilitate the production of anatomical models and the visualization of measured and simulated data. It uses the MMG remeshing library, which is developed by the CARDAMOM team at Inria Bordeaux. Currently our segmentation work is mostly done with the MUSICardio software, but we still use Blender for finishing touches and high-quality visualization.
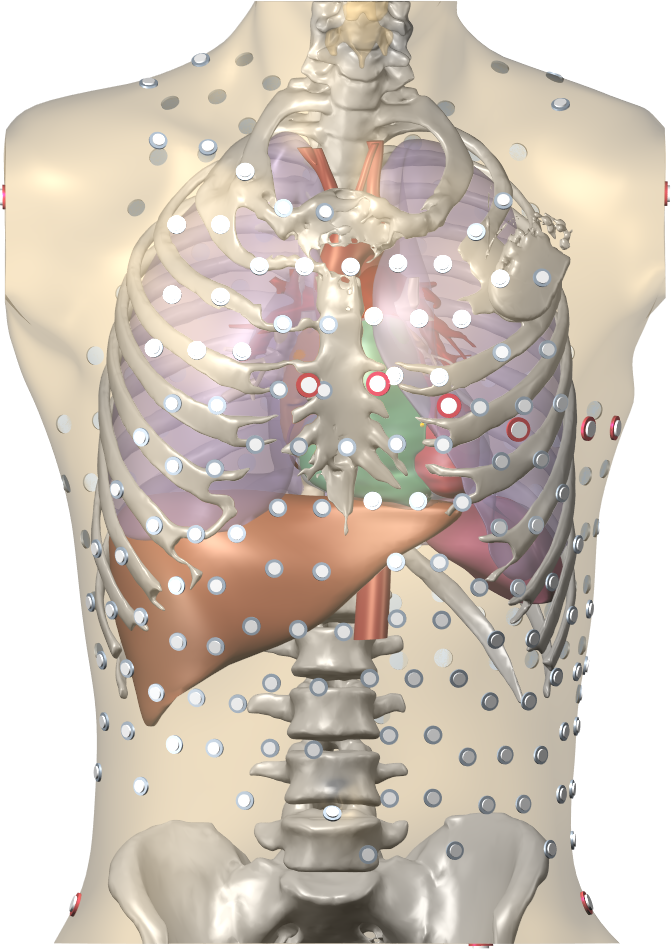


An image of a torso model including the heart, the ribs, the lungs, the livers, and the location of torso electrodes, and a detailed image of an atrial anatomical model including fiber directions.
8 New results
Participants: Mostafa Bendahmane, Yves Coudière, Jacques Henry, Peter Langfield, Michael Leguèbe, Mark Potse, Lisl Weynans, Nejib Zemzemi.
8.1 Analysis of partial differential equations
- Analysis of an extended bidomain model New results from the PhD thesis of Valentin Pannetier (still under embargo) en 2024 include the proof of the existence of solutions to the complete 3D model of pacemakers coupled to bidomain-with-bath equations, which was presented at CANUM 21, see also below about modeling.
- Analysis of a 1D/3D coupled problem in cardiac electrophysiology modeling In this work we analyze a coupled problem that describes the coupling two systems of reaction diffusion equation one on a three-dimensional domain representing the heart myocardium and the other on a one-dimensional tree representing the Purkinje network. Each system of PDEs is itself coupled to ordinary differential equations that describe the electrical activity at the cellular level. We establish the existence of a unique solution, utilizing a fixed-point approach with a judicious and non-conventional choice of functional spaces and contraction. 11
8.2 Numerical analysis and development of numerical methods
- Immersed boundary method for EIT In collaboration with Jérémi Dardé (IMT, Toulouse) and Niami Nasr (St Etienne) we studied the convergence of the gradient of the immersed boundary method for EIT that we previously developed during Niami Nasr's PhD thesis 51.
- Very high order finite volume methods In collaboration with R. Turpault and K. Khadra, Yves Coudière kept developing a very high order discretization method for the cardiac monodomain equations that scales well when executed in parallel.
- convergence analysisZeina Chehade analyzed numerically the convergence rate of the error for the discretization of the cardiac EMI model with the finite volume method that was developed earlier. She presented her results at the Eccomas conference in Lisbon (Portugal) 14 and at the Canum 19. This work is part of the MICROCARD project.
- Mechanisms underlying geometrical features of isochrons These tools allow us to study a highly-detailed depiction of the phase response of an oscillating cell in response to a stimulus, as a function of the stimulus properties. The results relate to the ongoing study of the Hodgkin-Huxley model with Profs Bernd Krauskopf and Hinke Osinga, where we had hypothesized a mechnism that gives rise to a distinct formation of isochrons. Following discussions with Kyoung Lee from University of Auckland in July, this hypothesis has been strengthened via his related observations in the planar FHN model. From this exchange, a clear mechanism was identified and has now been generalized to high-dimensional settings. Furthermore, a new structural feature has been uncovered, which, through geometrical and topological reasoning, we have shown is a necessary feature of all models.
- Locking rhythms of coupled phase oscillators In a theoretical study with Profs Leon Glass and Bard Ermentrout into the dynamics of generic coupled oscillators where a forcing oscillator drives a forced oscillator, we showed how certain rhythms are not possible for particular forms of coupling. Specifically, that the forced cycle must undergo at least one cycle for each cycle of the forcing, which is a phenomenon that has roles to play in biology such as cell division. These results are documented in a manuscript that is currently under review.
8.3 Modeling and inverse problems
-
Modeling pacemakers Within the H2020 SimCardioTest European project, Valentin Pannetier
defended his PhD thesis on Dec 6th 2024. In addition to the theoretical proof (see above) concerning the 3D model, the 0D model that was proposed the previous year was reworked for modeling accuracy. We presented a global sensitivity analysis of this model at CinC 2024 17. We pursued our efforts of software quality assurance within the V&V40 framework 36, presented at VPH 2024, by including in CEPS a benchmark for cardiac problems that was approved by the FDA 52. Finally, the 0D model was used as a testbench to demonstrate that it is possible to design a trial on new pacemaker, and run it entirely in-silico, as was explained in 15.
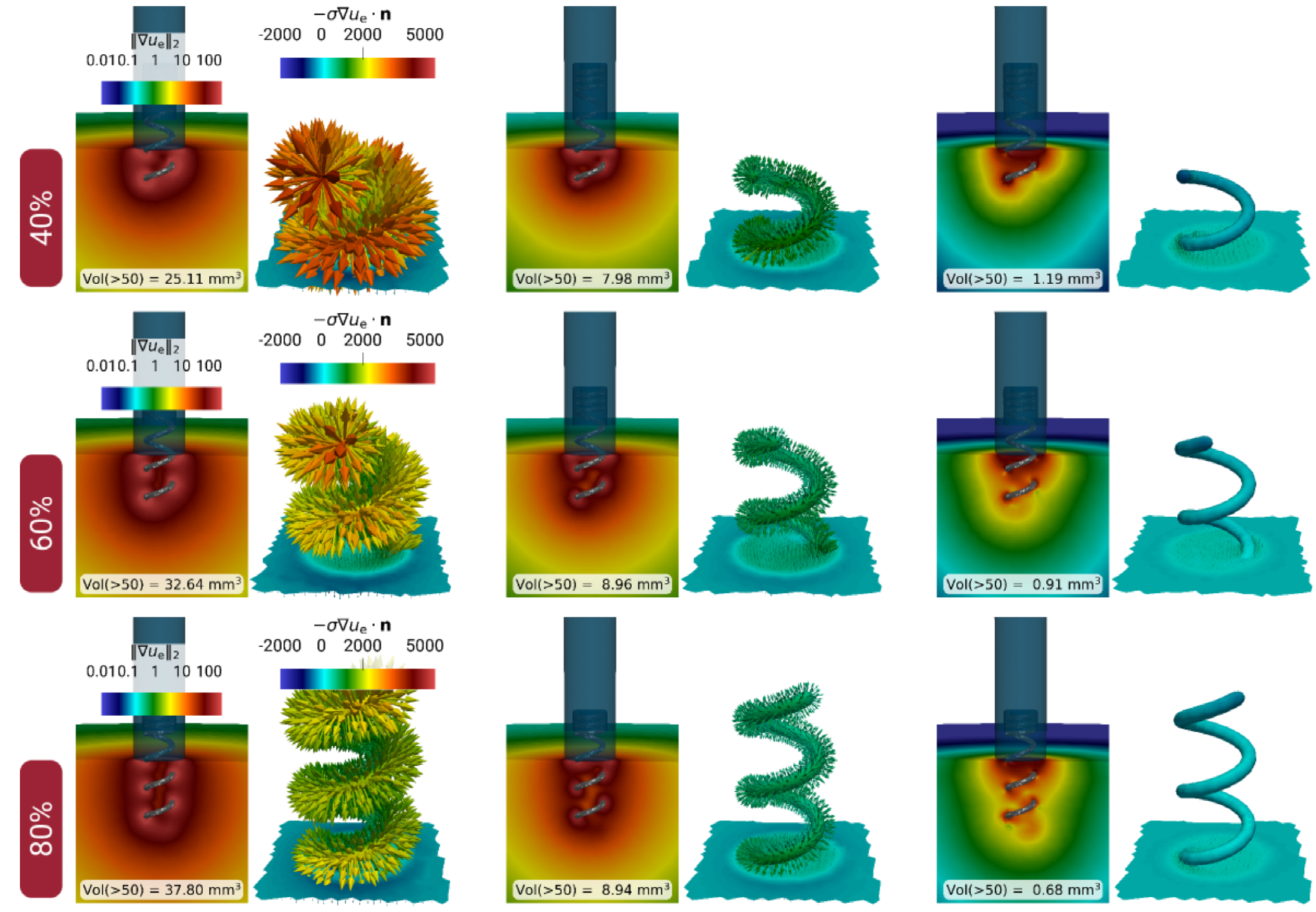
Repartition of current going thrhough the tip electrode of a pacemaker, which directly impacts the possibility of capture. Capture is effective when the current magnitude exceeds a given threshold. More surface of electrode within the tissue does not necessarily lead to more chances of activation. The image shows that the intensity of the current is larger when the electrode is inserted at 40% of the tissue, compared to 60 and 80% insertions.
Figure 3: Repartition of current going through the tip electrode of a pacemaker, which directly impacts the possibility of capture. Capture is effective when the current magnitude exceeds a given threshold. More surface of electrode within the tissue does not necessarily lead to better chances of activation. - A model for cardiac electroporation In the Dielectric project in collaboration with Inria Team MONC and IHU Liryc, Simon Bihoreau presented at CANUM 2024 30 and World Congress of Electroporation 2024 29 the first numerical results of the model we derived in 2023.
- Modeling the noise in EIT measurements During the internship of Sylvain Fourcade , in collaboration with Bénédicte Puig (LMAP, Pau), we developed a new formalism to study the inverse problem of EIT, based on the complete electrode model for EIT and the use of a Kohn-Vogelius functional to minimize, associated to the use of noise models to account for multiple measurements on the electrodes.
- A new source model for cardiac ECG imagingEmma Lagracie , Yves Coudière and Lisl Weynans , in collaboration with Yves Bourgault (Ottawa) proposed a new methodology for reconstructing activation maps from torso surface data, which incorporates information from the myocardial volume, while solving a surface problem on the heart. They formulated a 2D static forward model, derived from the bidomain model, by averaging equations in the heart. The averaged equations are coupled with the usual Laplace equations in the surrounding domains. For solving the inverse problem, this depth-averaged forward model is used as a constraint in an optimal control problem that allows to recover depth-averaged transmembrane voltage and extracellular potential in the heart, corresponding to observations on the body surface 12. This model was later extended to the epicardial surface in 3D (Epicardial Model), and used to study the effects of anisotropic conductivities in the source model on inverse problem reconstructions.
- Post processing methods for cardiac ECG imagingEmma Lagracie , Yves Coudière and Lisl Weynans compared several post-processing methods for computing activation maps from inverse ECGi reconstructions 16. In particular, using the Epicardial Model, they studied a Threshold Method on the transmembrane voltage, and showed that it tended to produce smooth maps, while the usual derivative-based methods produce artificial discontinuities.
8.4 Clinical applications
- Ananth Venkatesh was hired to work as part of a masters project on the ATLAS-RVA project together with Masimba Nemaire, in order to explore the annotation of cardiac signals via frequency decompositon. By combining these investigations with Ananth's prior experience with self-assisted learning, we were able to develop an annotation tool that can reliably detect the T-waves in both unipolar and ECG signals. The tool is adapted specifically to CARTO signals, requiring no prior processing, and achieves an accuracy beyond the current state-of-the-art. The general foundations of the method make it adaptable to different annotation problems and to different types of signal. A draft article presenting these results is currently in production.
- As part of the MICROCARD project, Joshua Steyer worked on how the cell-by-cell model can improve our understanding of cardiac electrograms, in particular in presence of a heterogeneous tissue 18, 34
- 27Narimane Gassa defended her PhD thesis in 2024. She presented her results on ???
8.5 High performance computing and Microscopic models
- More than a year of work of Corentin Prigent
and Algiane Froehly (SED) has resulted in a robustification of the mmg3d code that made it able to remesh the fiercely complex meshes of cardiac tissue generated by the MICROCARD project. This was a crucial step towards simulations with geometrically realistic cell-by-cell models of cardiac tissue. The improvements have been included in version 5.8.0 of Mmgtools, released in November 2024, and are expected so serve anyone dealing with numerous, tightly separated internal boundaries in volumetric meshes. This was notably discussed at the final project workshop, which took place as a satellite of the Cardiac Physiome workshop 31
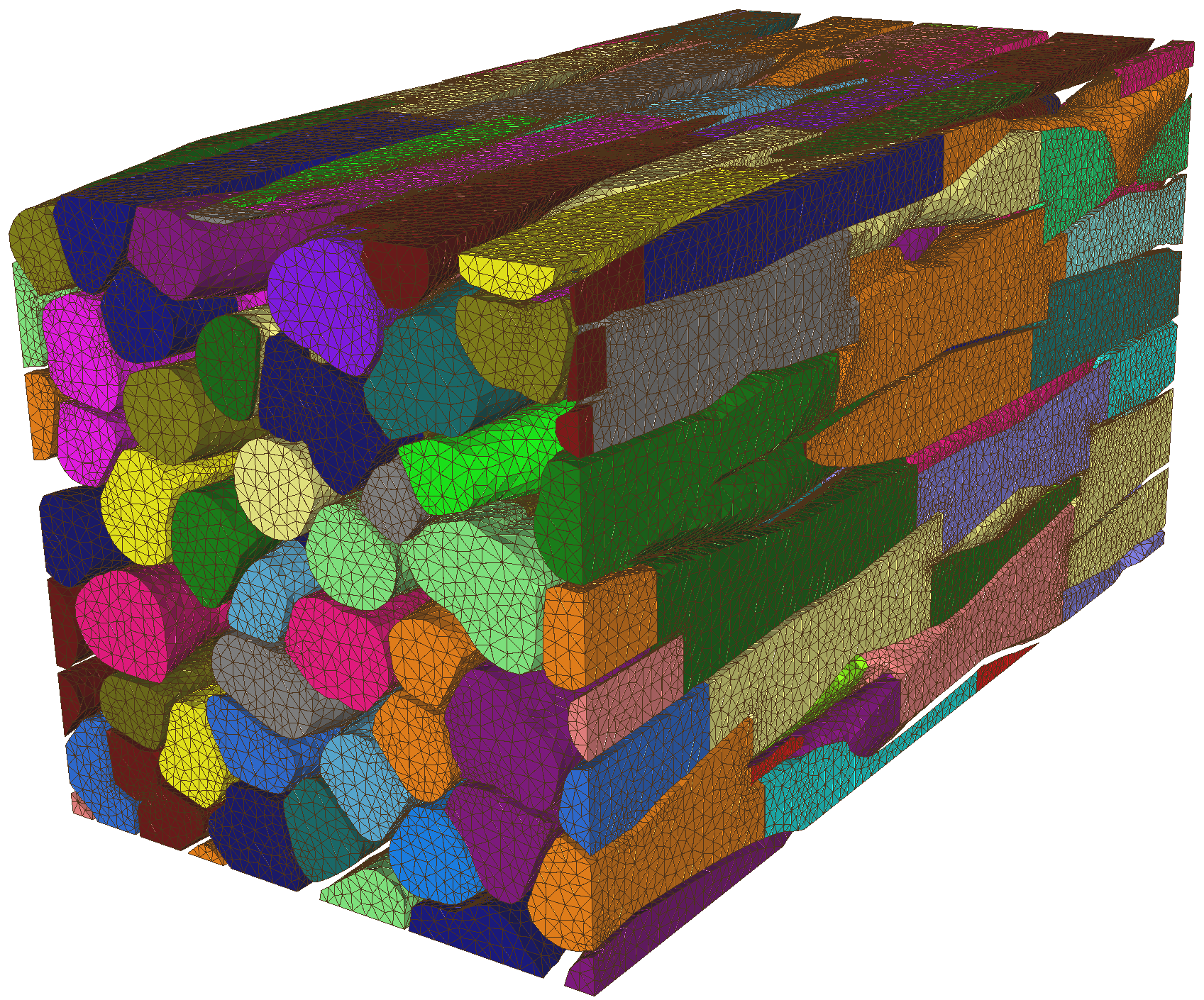
Small piece (0.4mm length) of a cardiac tissue model, produced by the MICROCARD project and remeshed with mmg3d. The figure illustrates the numerous internal boundaries in this volumetric mesh, and the small angles under which they meet. The ability to remesh under these conditions is a major breakthrough for the project.
Figure 4: Small piece (0.4mm length) of a cardiac tissue model, produced by the MICROCARD project and remeshed with mmg3d. The figure illustrates the numerous internal boundaries in this volumetric mesh, and the small angles under which they meet. The ability to remesh under these conditions is a major breakthrough for the project.
9 Bilateral contracts and grants with industry
Participants: Nejib Zemzemi, Lisl Weynans.
9.1 Bilateral Grants with Industry
- RebrAIn and Inria contracted an agreement allowing Nejib Zemzemi to pass 90 % of his time working at RebrAIn. The startup RebrAIn has been co-founded by N. Zemzemi and E. Cuny.
- Lisl Weynans obtained a grant from Fondation EDF to fund a research project about Electrical Impedance Tomography.
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
SPIMCY
-
Title:
Stochastic forward and inverse Problems In electrical and Mechanical Cardiac physiologY
-
Duration:
2024 ->
-
Coordinator:
Mahjoub Moncef (moncef.mahjoub@enit.utm.tn)
-
Partners:
- Université de Tunis El Manar Tunis (Tunisie)
-
Inria contact:
Mostafa Bendahmane
-
Summary:
The electrocardiography imaging inverse problem is frequently solved using the deterministic quasi-static models. These models don't take into account the heart dynamic in time, channel noise and external random perturbations acting in the torso. Recent numerical studies in the direct problem have shown that such randomness cannot be suppressed. Occasionally deterministic equations give qualitatively incorrect results. Therefore, it is important to quantify the nature of the noise and choose an appropriate model incorporating randomness. In our project, we study the inverse problem constrained by the stochastic monodomain or bidomain equations in electrocardiology. The state equations consist in a coupled stochastic reaction-diffusion system modelling the propagation of the intracelullar and extracellular electrical potentials, and stochastic ionic currents in the heart. These equations are coupled to the stochastic quasi-static elliptic equation in the torso. Thus, we will demonstrate that the novel concept of applying the stochastic model will be useful to improve noninvasive reconstruction of electrical and mechanical heart activity. Moreover, we will study the stability result for the conductivities and numerically solve the parameters estimations problem in the stochastic model.
-
Achievments of the year 2024
In this year, Narjess Benabid with Moncef Mahjoub and Mostafa Bendahmane submitted the first work on nonlocal diffusion control problem in electrocardiology. In this project, they establish a new Carleman estimate related to nonlocal diffusion. Compared to the classical inverse problem in electro-cardiology, the goal is to study the effect of the various diffusion terms on the construction of electrical potential on the heart. Moreover, they almost finished the project concerning the theoretical and numerical study of the optimal control of the stochastic monodomain model in cardiac electrophysiology modelling. The purpose of this project is to introduce a “stochastically forced” version of the inverse problem to the monodomain model that accounts for various random effects. The main goal of this work is to study the effect of the ionic model parameters in the genesis and the sustainability of arrhythmia using a stochastic bidomain model. The third topic is in preparation and it is related to the question of the convergence of the numerical solution to the optimal control of stochastic electromechanical monodomain model. To this, we establish the existence of the finite element scheme, and convergence of the unique weak solution of the stochastic electromechanical model. The convergence proof is based on deriving a series of a priori estimates and using a general L2-compactness criterion. This work is in progress and we expect to finish it next year. Two new doctoral Phd-students from FST of Marrakech Fatima Miya and Amine Ahamada Kari (supervisor: Mostafa Bendahmane and Noureddine Alaa) joined the SPIMCY project. Miya will use numerical VEM (Virtual element method) applied to optimal control and inverse problems in electro-cardiology. Kari will study the mechanical tri-domain with gap-junctions. Nejib Zemzemi Khouloud Kordoghli and Saloua Aouadi have been finalizing a work on the Mathematical analysis of a 1D-3D coupled problem in cardiac electrophysiology modelling.The objective of this work iis to analyze a coupled problem that describes the propagation of the electric wave in the heart. The problem comprises coupled partial differential equations posed on a three-dimensional domain representing the heart and on a one-dimensional tree representing the Purkinje network. Each system of PDEs is itself coupled to ordinary differential equations that describe the electrical activity at the cellular level. We establish the existence of a unique solution, utilizing a fixed-point approach with a judicious and non-conventional choice of functional spaces and contraction. Hamza Ammar, Moncef Mahjoub and Nejib Zemzemi have been finalizing a work on How does the optimal constant of the Carleman stability estimate depends on the size of the observation boundary? In this work, we elucidate the question of the Carleman estimate constants dependancy on the measurement boundary size. This allows to quantify the effect of measurement uncertainty on the stability of parameters identification inverse problem. We deliberately construct a class of space dependent weight functions that depend on the measurement boundary size. Then we identify the optimal weight function that allows us to minimize the stability constant. Using our approach, we found that when the measurement boundary covers 80% of the domain boundary, we can explicitly provide the formula of the optimal constant. When the measurement boundary is less than 80%, we are not able to find the explicit formula of the optimal expression, but we are able to numerically approximate the optimal constant.
10.1.2 Participation in other International Programs
ECOS Nord
Participants: Jacques Henry, Yves Coudière.
-
Title:
Qualitative and numerical analysis of inverse problems in cardiology
-
Partner Institution(s):
Cuerpo Academico de Ecuaciones Diferenciales y Modelacion Matematica, Facultad de Ciencias Fisico Matematicas
- Benemerita Universidad Autonoma de Puebla, Mexico
-
Date/Duration:
2021-2025 (prolonged due to Covid)
ECOS Sud
Participants: Mostafa Bendahmane, Yves Coudière.
-
Title:
Virtual Element Methods for Different Bidomain Models for Cardiac Electrphysiology
-
Partner Institution(s):
Departamento de Matemática, Facultad de Ciencias
- Universidad del Bio-Bio, Conception, Chile
-
Date/Duration:
2021-2024
10.2 International research visitors
10.2.1 Visits of international scientists
Joshua Steyer
-
Status
PhD student
-
Institution of origin:
Karlsruher Institut für Technologie
-
Country:
Allemagne
-
Dates:
9/2024 – 12-2024
-
Context of the visit:
The MICROCARD project
-
Mobility program/type of mobility:
research stay
Beata Ondrusova
-
Status
PhD student
-
Institution of origin:
Institute of Measurement Science, Slovak Academy of Sciences
-
Country:
Slovakia
-
Dates:
05/2024
-
Context of the visit:
scientific collaboration
-
Mobility program/type of mobility:
research stay
Jana Svehlikova
-
Status
Researcher
-
Institution of origin:
Institute of Measurement Science, Slovak Academy of Sciences
-
Country:
Slovakia
-
Dates:
05/2024
-
Context of the visit:
scientific collaboration
-
Mobility program/type of mobility:
research stay
10.3 European initiatives
10.3.1 Horizon Europe – EuroHPC
MICROCARD-2
Participants: Mark Potse, Yves Coudière.
MICROCARD-2 on the EuroHPC website
-
Title:
MICROCARD-2: numerical modeling of cardiac electrophysiology at the cellular scale
-
Duration:
from November 1, 2024 to April 30, 2027
-
Partners:
- Inria, France
- Karlsruher Institut Für Technologie, Germany
- Megware, Germany
- Simula Research Laboratory (Simula), Norway
- Technical University München (TUM), Germany
- Università degli Studi di Pavia, Italy
- Università di Trento (UTrento), Italy
- Université de Bordeaux, France
- Université de Strasbourg, France
-
Inria contact:
Olivier Aumage (Storm)
-
Coordinator:
Mark Potse, Université de Bordeaux
-
Summary:
The MICROCARD-2 project is coordinated by Université de Bordeaux and involves the Inria teams Carmen, Storm, and Tadaam in Bordeaux and CAMUS in Strasbourg, among a total of ten partner institutions in France, Germany, Italy, and Norway. This Centre of Excellence for numerical modeling of cardiac electrophysiology at the cellular scale builds on the MICROCARD project (2021–2024) and has the same website.
The modelling of cardiac electrophysiology at the cellular scale requires thousands of model elements per cell, of which there are billions in a human heart. Even for small tissue samples such models require at least exascale supercomputers. In addition the production of meshes of the complex tissue structure is extremely challenging, even more so at this scale. MICROCARD-2 works, in concert, on every aspect of this problem: tailored numerical schemes, linear-system solvers, and preconditioners; dedicated compilers to produce efficient system code for different CPU and GPU architectures (including the EPI and other ARM architectures); mitigation of energy usage; mesh production and partitioning; simulation workflows; and benchmarking.
10.3.2 H2020 projects
MICROCARD
Participants: Mark Potse, Yves Coudière, Corentin Prigent, Gengis Lourenco, Joshua Steyer, Laetitia Mottet, Wissam Bouymedj, Xavier Muller, Zeina Chehade.
MICROCARD project on cordis.europa.eu
-
Title:
Numerical modeling of cardiac electrophysiology at the cellular scale
-
Duration:
From April 1, 2021 to September 30, 2024
-
Partners:
- UNIVERSITE DE BORDEAUX (UBx), France
- MEGWARE COMPUTER VERTRIEB UND SERVICE GMBH, Germany
- SIMULA RESEARCH LABORATORY AS, Norway
- UNIVERSITE DE STRASBOURG (UNISTRA), France
- ZUSE-INSTITUT BERLIN (ZUSE INSTITUTE BERLIN), Germany
- UNIVERSITA DELLA SVIZZERA ITALIANA (USI), Switzerland
- KARLSRUHER INSTITUT FUER TECHNOLOGIE (KIT), Germany
- UNIVERSITA DEGLI STUDI DI PAVIA (UNIPV), Italy
- NUMERICOR GMBH, Austria
- OROBIX SRL (OROBIX), Italy
-
Linked third parties:
- INSTITUT NATIONAL DE RECHERCHE EN INFORMATIQUE ET AUTOMATIQUE (INRIA), France
- INSTITUT POLYTECHNIQUE DE BORDEAUX (Bordeaux INP), France
-
Inria contact:
Mark POTSE
-
Coordinator:
Université de Bordeaux
-
Summary:
Cardiovascular diseases are the most frequent cause of death worldwide and half of these deaths are due to cardiac arrhythmia, a disorder of the heart's electrical synchronization system. Numerical models of this complex system are highly sophisticated and widely used, but to match observations in aging and diseased hearts they need to move from a continuum approach to a representation of individual cells and their interconnections. This implies a different, harder numerical problem and a 10,000-fold increase in problem size. Exascale computers will be needed to run such models.
We propose to develop an exascale application platform for cardiac electrophysiology simulations that is usable for cell-by-cell simulations. The platform will be co-designed by HPC experts, numerical scientists, biomedical engineers, and biomedical scientists, from academia and industry. We will develop, in concert, numerical schemes suitable for exascale parallelism, problem-tailored linear-system solvers and preconditioners, and a compiler to translate high-level model descriptions into optimized, energy-efficient system code for heterogeneous computing systems. The code will be parallelized with a recently developed runtime system that is resilient to hardware failures and will use an energy-aware task placement strategy.
The platform will be applied in real-life use cases with high impact in the biomedical domain and will showcase HPC in this area where it is painfully underused. It will be made accessible for a wide range of users both as code and through a web interface.
We will further employ our HPC and biomedical expertise to accelerate the development of parallel segmentation and (re)meshing software, necessary to create the extremely large and complex meshes needed from available large volumes of microscopy data.
The platform will be adaptable to similar biological systems such as nerves, and components of the platform will be reusable in a wide range of applications.
SimCardioTest
Participants: Yves Coudière, Michael Leguebe, Valentin Pannetier, Delphine Deshors, Loïc Calvez.
SimCardioTest project on cordis.europa.eu
-
Title:
Simulation of Cardiac Devices & Drugs for in-silico Testing and Certification
-
Duration:
From January 1, 2021 to June 30, 2025
-
Partners:
- UNIVERSITE DE BORDEAUX, France
- UNIVERSIDAD POMPEU FABRA, Spain
- UNIVERSITAT POLITECNICA DE VALENCIA, Spain
- SIMULA RESEARCH LABORATORY AS, Norway
- INSILICOTRIALS TECHNOLOGIES S.P.A., Italy
- SORIN CRM SAS, France
- EXACTCURE, France
- VIRTUAL PHYSIOLOGICAL HUMAN INSTITUTE FOR INTEGRATIVE BIOMEDICAL RESEARCH VZW, Belgium
- INSTITUT NATIONAL DE RECHERCHE EN INFORMATIQUE ET AUTOMATIQUE (INRIA), France
-
Inria contact:
Maxime Sermesant
-
Coordinator:
Inria
-
Summary:
Computer modelling and simulation have the power to increase speed and reduce costs in most product development pipelines. The EU-funded SimCardioTest project aims to implement computer modelling, simulation and artificial intelligence to design and test cardiac drugs and medical devices. Scientists will establish a platform for running in silico trials and obtaining scientific evidence based on controlled investigations. The simulation of disease conditions and cohort characteristics has the potential to overcome clinical trial limitations, such as under-representation of groups. It also reduces the size and duration of human clinical trials as well as animal testing, and offers robust, personalised information. Leveraging in silico technology in healthcare will expedite product and drug certification and offer patients the best possible care.
10.4 National initiatives
GENCI
Exceptionally, we have no GENCI allocation this year.
Dielectric project
The Dielectric project, co-funded by the Federation Française de Cardiologie and Inria, started in January 2023, and is co-piloted by Pr. Pierre Jaïs (IHU Liryc) and Clair Poignard (Inria MONC). It aims at a better understanding of cardiac ablation by electroporation. Both Inria teams MONC and CARMEN are involved, with the PhD of Simon Bihoreau, co-directed by Annabelle Collin (MONC) and Michael Leguèbe (CARMEN).
ANR Mire4VTach
PI Annabelle Collin (Inria MONC), started late 2023. Michael Leguèbe contributes in Mire4VTach, another project on cardiac electroporation which is more focused on the application and confrontation with data than the work in the Dielectric project. Mire4Tach also involves people from Inria MONC, CARMEN and IHU Liryc.
10.5 Regional initiatives
EITCardio, from September 2023 to September 2027
- Participants: Laura Bear, Yves Coudière , Charles Pierre, Bénédicte Puig, Lisl Weynans .
- Partners: Centre Inria de l'université de Bordeaux, IHU Liryc, Université de Pau et des Pays de l'Adour
- Coordinated by Lisl Weynans
- Summary: The objective of this project is to develop mathematical methods for solving Electrical Impedance Tomography (EIT) to enhance the resolution of the ECGi (Electrocardiographic Imaging) problem and validate them experimentally. Specifically, the project consists of two parts:
- Development of mathematical and numerical methods to solve the inverse problem of EIT in the torso and identify influential parameters for the propagation of the electric field, such as conductivities and organ movements.
- Experimental validation of the ECGi + EIT coupling. This experimental validation will be conducted first within the experimental setup, the torso tank, currently available at Liryc, which allows measurements for ECGi in a controlled environment. Subsequently, it will be conducted as in-vivo experiments, meaning a context closer to clinical reality.
10.6 Other national collaborations
ATLAS-RVA
The ATLAS-RVA project was funded by IHU Liryc. It is coordinated by Peter Langfield and Karim Benali (IHU Liryc), and aims to investigate repolarization patterns in the cardiac ventricles via in-vivo clinical. The main goals are to describe typical repolarization patterns in humans, from which localized variablility of such patterns can be assessed, and in turn, how particular pathologies manifest.
REALPRIOREIT
A project (exploratory action) funded by Inria, coordinated by Lisl Weynans and Jing-Rebecca Li (Saclay). This project aims at develop bayesian inference tools for EIT.
11 Dissemination
11.1 Promoting scientific activities
11.1.1 Scientific events: selection
Member of the conference program committees
- Lisl Weynans was a member of scientific board of the conference CANUM 2024.
Reviewer
- Mark Potse was a reviewer for the Computing in Cardiology conference 2024.
- Yves Coudière was a reviewer for the Computing in Cardiology conference 2024, and a jury member for the Rosana Delonghi Young Investigator award distributed at the conference.
11.1.2 Journal
Member of the editorial boards
- Mark Potse is associate editor for Frontiers in Cardiac Electrophysiology and Journal of Electrocardiology.
Reviewer - reviewing activities
All permanent researchers are involved in a lot of review for various journals. Let's mention some of them
- In applied mathematics and scientific computing: JCP, M2AN...
- In biomedical engineering: Journal of Electrocardiology...
- Others:
11.1.3 Invited talks
- Lisl Weynans was invited to the seminar of the IRMAR (Rennes) and ICJ (Lyon).
- Mark Potse gave a keynote at CinC 2024, 25
- Mark Potse gave an invited presentation at the INdAM 2024 Workshop of Mathematical and Numerical Modeling of the Cardiovascular System in Roma, Italy 26
- Yves Coudière gave an invited presentation at the French annual journée Math Bio Santé in Nantes, France 22
- Yves Coudière gave an invited presentation at the French annual 48ème colloque AFSTAL (Association Française des Sciences et Techniques de l'Animal de Laboratoire) in Lille, France 23
11.1.4 Scientific expertise
- Lisl Weynans was a member of the hiring committee for CR Inria at Bordeaux
- Yves Coudière has been an expert for: CIFRE PhD theses, a call for fundings (large amounts) from Inserm, and the annual French ANR call
11.1.5 Research administration
- Lisl Weynans was a member of the board of CNU 26 as "assesseure".
- Yves Coudière is a member of the Comité de Direction of IHU Liryc
- Yves Coudière is in charge of International relations for the math laboratory, Institut de Mathématiques de Bordeaux
- Peter Langfield is a member of the scientfic animation sessions comittee at IHU Liryc.
- Peter Langfield was the organizer of the modeling journal club at Liryc until September.
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
The 2 assistant professors and 1 professor, changing to 1 assistant professor and 2 professors since September 2024, teach at several levels of the Bordeaux University programs in Mathematics, Neurosciences, and Medicine (respectively, 192, 192 and 96 h/year on average). The researchers also have a regular teaching activity, contributing to several courses in the Applied Mathematics at the Bachelor and Master levels (between 16 and 72 h/year).
The PhD students who ask for it are used to teach between 32 and 64 h/year, usually courses of general mathematics in L1 or mathematics for biologists in L1 or L2.
Typical courses taught by team members (L for Bachelor level, M for Master level):
- Numerical analysis (L2)
- Programming for scientific computing with C++ (L3) and Modern Fortran (M1)
- Solving sparse linear systems (L3)
- Differential calculus and ordinary differential equations
- Numerical approximation of PDEs: Finite Differences, Finite Elements, Finite Volumes (M1, M2)
- Supervision of programming projects (L3, M1)
- Analysis, L2
- Computational Neurosciences, M2
- Neuropsychology and Psychophysiology, L3
- Physics for students in medicine, one lecture on cardiac modelling
In addition to the recurrent activities, for 2024:
- Mark Potse and Corentin Prigent gave lectures in a summer school organized by the MICROCARD project in collaboration with SIMULA in Oslo, June 2024.
- Peter Langfield gave a lecture at ISEN Ouest École d'ingénieurs in January on the subject of digital twins in biology. He also gave a lecture at IHU Liryc as part of the Ecole Santé Sciences Master winter workshop on the topic of mathematical modelling in cardiology.
11.2.2 Teaching administration
- Lisl Weynans: in charge of the Bachelor track Licence Mathématique parcours ingénierie mathématique,
- Yves Coudière is an elected member of the Conseil du Collège Sciences et Techniques of the University of Bordeaux
- Yves Coudière is an elected member of the Conseil de l'UF Mathématiques et Interactions of the University of Bordeaux
11.2.3 Supervision
We have been supervising several internship students at various levels (L3, M1, M2), as can be seen on the list of team members.
- Yassine Ayoun, Intern, from Jul 2024 until Sep 2024, supervised by Lisl Weynans
- Younes Bouhoreira Intern, from May 2024 until Jun 2024, supervised by Yves Coudière , Michael Leguebe
- Selma El Badaoui, Intern, from May 2024 until Jul 2024, supervised by Yves Coudière
- Sylvain Fourcade, Intern, from Feb 2024 until Aug 2024, supervised by Lisl Weynans
- Paul Fournet, Intern, from Jun 2024 until Sep 2024, supervised by Yves Coudière and Masimba Nemaire, a collaborator from Liryc
- Pierre Sinel–Boucher, Intern, from Jun 2024 until Jul 2024, supervised by Yves Coudière and Rodolphe Turpault, a collaborator from IMB
- Ananth Venkatesh, Intern, from Jun 2024 until Nov 2024, supervised by Peter Langfield and Masimba Nemaire
- Mehdi Yahiaoui, Intern, from May 2024 until Jun 2024, supervised by Michael Leguebe and Yves Coudière
11.2.4 Juries
- Mark Potse was examiner in the PhD jury of Arun Thangamani at the University of Strasbourg in September 2024.
-
Lisl Weynans
was
- president of the PhD jury of Simone Nati Poltri (Bordeaux),
- reviewer of the PhD thesis of Haibo Liu (Sorbonnes Université)
- reviewer of the PhD thesis of Benjamin Sulis (Reims)
- an examiner in the PhD jury of Bastien Jouy (EDF).
-
Nejib Zemzemi
was
- Jury member of the PhD thesis of Haibo Liu (Sorbonnes Université)
- Jury member of the PhD jury of Narimane Gassa (Bordeaux).
-
Yves Coudière
was
- reviewer of the PhD thesis of Paul Paragot (Nice)
- president of the HDR jury of Mark Potse (Bordeaux)
- an examiner in the PhD jury of Narimane Gassa (Bordeaux).
11.3 Popularization
11.3.1 Productions (articles, videos, podcasts, serious games, ...)
- Article Dealer de Science Yves Coudière
11.3.2 Participation in Live events
- Fête de la Science 2024 : Xavier Muller participated
- 20 ans d'accompagnement de startups par Inria et ses partenaires en Nouvelle-Aquitaine: Table ronde sur le financement de la R&D dans la startup Rebrain. Nejib Zemzemi avec Natalia Araujo (BPI France) et Vincent Prêtet (M Capital) 4 décembre 2024.
- DeepTech Tour Université de Bordeaux: Table ronde sur l'éxperience de création d'entreprise et son évolution: équipe, écosystème, défits reglémentaires,... Nejib Zemzemi avec deux autre intervenant de deux stratups.
- Nuit des chercheurs 2024 à Cap Sciences : Yves Coudière
- Opération Chiche : Yves Coudière (5-10 classes), Laetitia Mottet
- Semaine des Maths : Yves Coudière
- Moi Informaticienne, Moi Mathématicienne: Lisl Weynans , Emma Lagracie
- Recieving schoolchildren visits (stages de 3e)
12 Scientific production
12.1 Major publications
- 1 articleConvergence of discrete duality finite volume schemes for the cardiac bidomain model.Networks and Heterogeneous Media622011, 195-240HALback to text
- 2 articleA mathematical model of Purkinje-Muscle Junctions.Mathematical Biosciences and Engineering842011, 915-930
- 3 articleExistence And Uniqueness Of The Solution For The Bidomain Model Used In Cardiac Electrophysiology.Nonlinear Anal. Real World Appl.1012009, 458-482URL: http://hal.archives-ouvertes.fr/hal-00101458/fr
- 4 articleA 2D/3D Discrete Duality Finite Volume Scheme. Application to ECG simulation.International Journal on Finite Volumes612009, URL: http://hal.archives-ouvertes.fr/hal-00328251/frback to text
- 5 articleStability And Convergence Of A Finite Volume Method For Two Systems Of Reaction-Diffusion Equations In Electro-Cardiology.Nonlinear Anal. Real World Appl.742006, 916--935URL: http://hal.archives-ouvertes.fr/hal-00016816/frback to text
- 6 articlePrediction of Clinical Deep Brain Stimulation Target for Essential Tremor From 1.5 Tesla MRI Anatomical Landmarks.Frontiers in Neurology12October 2021HALDOIback to text
- 7 articleThe Early Repolarization Pattern; A Consensus Paper.Journal of the American College of Cardiology66resulting from the Symposium on J Wave Patterns and a J Wave Syndrome, Glasgow, August 2013. Defines terminology for the J point: Jo, Jp, Jt. Provisionally accepted 19 May 2015.2015, 470-477URL: http://dx.doi.org/10.1016/j.jacc.2015.05.033back to text
- 8 articleReduced Sodium Current in the Lateral Ventricular Wall Induces Inferolateral J-Waves.Front Physiol7365August 2016HALDOIback to textback to text
- 9 articlePreconditioning the bidomain model with almost linear complexity.Journal of Computational Physics2311January 2012, 82--97URL: http://www.sciencedirect.com/science/article/pii/S0021999111005122DOIback to text
- 10 articleMethodological and Mechanistic Considerations in Local Repolarization Mapping.JACC: Clinical Electrophysiology102February 2024, 376-377HALDOI
12.2 Publications of the year
International journals
International peer-reviewed conferences
National peer-reviewed Conferences
Conferences without proceedings
Doctoral dissertations and habilitation theses
Other scientific publications
12.3 Cited publications
- 35 inproceedingsAn Improved Iterative Pace-Mapping Algorithm to Detect the Origin of Premature Ventricular Contractions.Computing in Cardiology47abstract nr 62, really early. Oral presentation.RiminiComputing in Cardiology2020, 62HALDOIback to text
- 36 bookAssessing the credibility of computational modeling through verification and validation: Application to medical devices.2018back to text
- 37 inproceedingsModélisation et simulation de l'électrophysiologie cardiaque à l'échelle microscopique.43e Congrès National d'Analyse Numérique (CANUM)oral presentationSMAIObernai, Alsace, FranceMay 2016, URL: http://smai.emath.fr/canum2016/resumesPDF/peb/Abstract.pdfback to text
- 38 miscTheoretical and Numerical Study of Cardiac Electrophysiology Problems at the Microscopic Scale..PosterJuly 2016HALback to text
- 39 inproceedingsA Three-Dimensional Computational Model of Action Potential Propagation Through a Network of Individual Cells.Computing in Cardiology 2017Rennes, FranceSeptember 2017, 1-4HALback to text
- 40 inproceedingsMicroscopic Simulation of the Cardiac Electrophysiology: A Study of the Influence of Different Gap Junctions Models.Computing in CardiologyMaastricht, NetherlandsSeptember 2018HALback to text
- 41 articleImpact of Septal Radiofrequency Ventricular Tachycardia Ablation; Insights From Magnetic Resonance Imaging.Circulation1302014, 716-718URL: https://www.ahajournals.org/doi/10.1161/CIRCULATIONAHA.114.010175back to text
- 42 inproceedingsDo we need to enforce the homogeneous Neuman condition on the Torso for solving the inverse electrocardiographic problem by using the method of fundamental solution ?Computing in Cardiology 201643Computing in Cardiology 2016Vancouver, CanadaSeptember 2016, 425-428HALback to text
- 43 articleOptimal monodomain approximations of the bidomain equations used in cardiac electrophysiology.Mathematical Models and Methods in Applied Sciences246February 2014, 1115-1140HALback to textback to text
- 44 articleSpatially Coherent Activation Maps for Electrocardiographic Imaging.IEEE Transactions on Biomedical Engineering64May 2017, 1149-1156HALDOIback to text
- 45 inproceedingsEpicardial Fibrosis Explains Increased Transmural Conduction in a Computer Model of Atrial Fibrillation .Computing in CardiologyVancouver, CanadaSeptember 2016HALback to textback to textback to text
- 46 articleSudden cardiac arrest associated with early repolarization.N. Engl. J. Med.3582008, 2016--2023back to text
- 47 articleSpontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins.N. Engl. J. Med.3391998, 659-666URL: https://www.nejm.org/doi/full/10.1056/NEJM199809033391003back to text
- 48 articleMechanism of Right Precordial ST-Segment Elevation in Structural Heart Disease: Excitation Failure by Current-to-Load Mismatch.Heart Rhythm72010, 238-248URL: http://dx.doi.org/10.1016/j.hrthm.2009.10.007back to text
- 49 articleA microstructural model of reentry arising from focal breakthrough at sites of source-load mismatch in a central region of slow conduction.Am. J. Physiol. Heart Circ. Physiol.3062014, H1341-1352back to text
- 50 inproceedingsA new ECG-based method to guide catheter ablation of ventricular tachycardia.iMAging and eLectrical TechnologiesUppsala, SwedenApril 2018HALback to text
- 51 phdthesisMéthodes numériques pour la tomographie par impédance électrique dans le cadre de l'électrocardiographie.Université de BordeauxDecember 2023HALback to text
- 52 articleVerification of computational models of cardiac electro-physiology.Int. J. Numer. Method. Biomed. Eng.305May 2014, 525--544back to textback to text
- 53 inproceedingsA practical algorithm to build geometric models of cardiac muscle structure.ECCOMAS 2022 - The 8th European Congress on Computational Methods in Applied Sciences and EngineeringOslo, NorwayJune 2022HALback to text
- 54 articleA Comparison of monodomain and bidomain reaction-diffusion.IEEE Transactions on Biomedical Engineering53122006, 2425-2435URL: http://dx.doi.org/10.1109/TBME.2006.880875back to text
- 55 articleCardiac Anisotropy in Boundary-Element Models for the Electrocardiogram.Medical and Biological Engineering and Computing472009, 719--729URL: http://dx.doi.org/10.1007/s11517-009-0472-xback to text
- 56 inproceedingsAnatomically-induced Fibrillation in a 3D model of the Human Atria.Computing in CardiologyMaastricht, NetherlandsSeptember 2018HALback to text
- 57 miscRegional conduction slowing can explain inferolateral J waves and their attenuation by sodium channel blockers.PosterSeptember 2016HALback to text
- 58 inproceedingsFeasibility of Whole-Heart Electrophysiological Models With Near-Cellular Resolution.CinC 2020 - Computing in CardiologyRimini / Virtual, ItalySeptember 2020HALDOIback to text
- 59 articleScalable and Accurate ECG Simulation for Reaction-Diffusion Models of the Human Heart.Frontiers in Physiology9April 2018, 370HALDOIback to text
- 60 articleA Cell-Based Framework for Numerical Modeling of Electrical Conduction in Cardiac Tissue.Front. Phys.5This is very very similar to what we were doing with PEB... Looks like we're scooped at least for the approach, but we do have a few abstracts ([becue:cinc17], [becue16a], MMCE meeting in Ottawa November 2017). Note it's in Frontiers in (Biomedical) Physics, not Physiology.2017, 48back to textback to text

