2024Activity reportProject-TeamMORPHEO
RNSR: 201120981M- Research center Inria Centre at Université Grenoble Alpes
- In partnership with:CNRS, Université de Grenoble Alpes
- Team name: Capture and Analysis of Shapes in Motion
- In collaboration with:Laboratoire Jean Kuntzmann (LJK)
- Domain:Perception, Cognition and Interaction
- Theme:Vision, perception and multimedia interpretation
Keywords
Computer Science and Digital Science
- A5.1.8. 3D User Interfaces
- A5.3.4. Registration
- A5.4.4. 3D and spatio-temporal reconstruction
- A5.4.5. Object tracking and motion analysis
- A5.5.1. Geometrical modeling
- A5.5.4. Animation
- A5.6. Virtual reality, augmented reality
- A6.2.8. Computational geometry and meshes
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B2.7.2. Health monitoring systems
- B2.8. Sports, performance, motor skills
- B9.2.2. Cinema, Television
- B9.2.3. Video games
- B9.4. Sports
- B9.5.1. Computer science
- B9.5.2. Mathematics
- B9.7. Knowledge dissemination
- B9.7.1. Open access
- B9.7.2. Open data
1 Team members, visitors, external collaborators
Research Scientists
- Jean Franco [Team leader, INRIA, Senior Researcher, from Oct 2024]
- Stefanie Wuhrer [INRIA, Researcher]
Faculty Members
- Jean Franco [Team leader, INP, Associate Professor, until Sep 2024]
- Sergi PUJADES [UGA, Associate Professor]
PhD Students
- Youssef Ben Cheikh [INRIA, from May 2024]
- Nicolas Comte [ANATOSCOP, CIFRE, until Mar 2024]
- Antoine Dumoulin [INRIA]
- Anilkumar Erappanakoppal Swamy [NAVER LABS, CIFRE, from Jul 2024 until Nov 2024]
- Anilkumar Erappanakoppal Swamy [NAVER LABS, until Jul 2024]
- Vicente Estopier Castillo [INSERM]
- Samara Ghrer [INRIA, from Dec 2024]
- Aymen Merrouche [INRIA]
- Laura Neschen [INRIA, from Oct 2024]
- Gabriel Ravelomanana [INRIA, from Nov 2024]
- Rim Rekik Dit Nekhili [INRIA]
- Briac Toussaint [INRIA, from Oct 2024]
- Briac Toussaint [UGA, until Sep 2024]
- Quentin Zoppis [UGA, from Oct 2024]
Technical Staff
- Vaibhav Arora [INRIA, Engineer]
- Léo Challier [LJK, Engineer, until Sep 2024]
- Mohammed Chekroun [INRIA, Engineer]
- Abdelmouttaleb Dakri [INRIA, Engineer]
- Samara Ghrer [INRIA, Engineer, from Sep 2024 until Nov 2024]
- Samara Ghrer [UGA, Engineer, until Aug 2024]
- Julien Pansiot [INRIA, Engineer]
Interns and Apprentices
- Felix Kuper [INRIA, Intern, from Mar 2024 until Sep 2024]
- Leon Lepers [INRIA, Intern, from May 2024 until Jul 2024]
- Leon Lepers [INRIA, Intern, from Mar 2024 until Apr 2024]
- Thibault Penning [ENS RENNES, Intern, from Feb 2024 until Jul 2024]
Administrative Assistant
- Nathalie Gillot [INRIA]
Visiting Scientist
- David Bojanic [UNIV ZAGREB, from Feb 2024 until Jul 2024]
2 Overall objectives

Dynamic Geometry Modeling: a human is captured while running. The different temporal acquisition instances are shown on the same figure.
MORPHEO's ambition is to perceive and interpret shapes that move using multiple camera systems. Departing from standard motion capture systems, based on markers, that provide only sparse information on moving shapes, multiple camera systems allow dense information on both shapes and their motion to be recovered from visual cues. Such ability to perceive shapes in motion brings a rich domain for research investigations on how to model, understand and animate real dynamic shapes, and finds applications, for instance, in gait analysis, bio-metric and bio-mechanical analysis, animation, games and, more insistently in recent years, in the virtual and augmented reality domain. The MORPHEO team particularly focuses on four different axes within the overall theme of 3D dynamic scene vision or 4D vision:
- Shape and appearance models: how to build precise geometric and photometric models of shapes, including human bodies but not limited to, given temporal sequences.
- Dynamic shape vision: how to register and track moving shapes, build pose spaces and animate captured shapes.
- Inside shape vision: how to capture and model inside parts of moving shapes using combined color and X-ray imaging.
- Shape animation: Morpheo is actively investigating animation acquisition and parameterization methodologies for efficient representation and manipulability of acquired 4D data.
The strategy developed by Morpheo to address the mentioned challenges is based on methodological tools that include in particular learning-based approaches, geometry, Bayesian inference and numerical optimization. In recent years, and following many successes in the team and the computer vision community as a whole, a particular effort is ongoing in the team to investigate the use of machine learning and neural learning tools in 4D vision.
3 Research program
3.1 Shape and Appearance Modeling
Standard acquisition platforms, including commercial solutions proposed by companies such as Microsoft, 3dMD or 4DViews, now give access to precise 3D models with geometry, e.g. meshes, and appearance information, e.g. textures. Still, state-of-the-art solutions are limited in many respects: They generally consider limited contexts and close setups with typically at most a few meter side lengths. As a result, many dynamic scenes, even a body running sequence, are still challenging situations; They also seldom exploit time redundancy; Additionally, data driven strategies are yet to be fully investigated in the field. The MORPHEO team builds on the Kinovis platform for data acquisition and has addressed these issues with, in particular, contributions on time integration, in order to increase the resolution for both shapes and appearances, on representations, as well as on exploiting machine learning tools when modeling dynamic scenes. Our originality lies, for a large part, in the larger scale of the dynamic scenes we consider as well as in the time super resolution strategy we investigate. Another particularity of our research is a strong experimental foundation with the multiple camera Kinovis platforms.
3.2 Dynamic Shape Vision
Dynamic Shape Vision refers to research themes that consider the motion of dynamic shapes, with e.g. shapes in different poses, or the deformation between different shapes, with e.g. different human bodies. This includes for instance shape tracking and shape registration, which are themes covered by MORPHEO. While progress has been made over the last decade in this domain, challenges remain, in particular due to the required essential task of shape correspondence that is still difficult to perform robustly. Strategies in this domain can be roughly classified into two categories: (i) data driven approaches that learn shape spaces and estimate shapes and their variations through space parameterizations; (ii) model based approaches that use more or less constrained prior models on shape evolutions, e.g. locally rigid structures, to recover correspondences. The MORPHEO team is substantially involved in both categories. The second one leaves more flexibility for shapes that can be modeled, an important feature with the Kinovis platform, while the first one is interesting for modeling classes of shapes that are more likely to evolve in spaces with reasonable dimensions, such as faces and bodies under clothing. The originality of MORPHEO in this axis is to go beyond static shape poses and to consider also the dynamics of shape over several frames when modeling moving shapes, and in particular with shape tracking and animation.
3.3 Inside Shape Vision
Another research axis is concerned with the ability to perceive inside moving shapes. This is a more recent research theme in the MORPHEO team that has gained importance. It was originally the research associated to the Kinovis platform installed in the Grenoble Hospitals. This platform is equipped with two X-ray cameras and ten color cameras, enabling therefore simultaneous vision of inside and outside shapes. We believe this opens a new domain of investigation at the interface between computer vision and medical imaging. Interesting issues in this domain include the links between the outside surface of a shape and its inner parts, especially with the human body. These links are likely to help understanding and modeling human motions. Until now, numerous dynamic shape models, especially in the computer graphics domain, consist of a surface, typically a mesh, rigged to a skeletal structure that is never observed in practice but allows to parameterize human motion. Learning more accurate relationships using observations can therefore significantly impact the domain.
3.4 Shape Animation
3D animation is a crucial part of digital media production with numerous applications, in particular in the game and motion picture industry. Recent evolutions in computer animation consider real videos for both the creation and the animation of characters. The advantage of this strategy is twofold: it reduces the creation cost and increases realism by considering only real data. Furthermore, it allows to create new motions, for real characters, by recombining recorded elementary movements. In addition to enable new media contents to be produced, it also allows to automatically extend moving shape datasets with fully controllable new motions. This ability appears to be of great importance with deep learning techniques and the associated need for large learning datasets. In this research direction, we investigate how to create new dynamic scenes using recorded events. More recently, this also includes applying machine learning to datasets of recorded human motions to learn motion spaces that allow to synthesize novel realistic animations.
4 Application domains
4.1 4D modeling
Modeling shapes that evolve over time, analyzing and interpreting their motion has been a subject of increasing interest of many research communities including the computer vision, the computer graphics and the medical imaging communities. Recent evolutions in acquisition technologies including 3D depth cameras (Time-of-Flight and Kinect), multi-camera systems, marker based motion capture systems, ultrasound and CT scanners have made those communities consider capturing the real scene and their dynamics, create 4D spatio-temporal models, analyze and interpret them. A number of applications including dense motion capture, dynamic shape modeling and animation, temporally consistent 3D reconstruction, motion analysis and interpretation have therefore emerged.
4.2 Shape Analysis
Most existing shape analysis tools are local, in the sense that they give local insight about an object's geometry or purpose. The use of both geometry and motion cues makes it possible to recover more global information, in order to get extensive knowledge about a shape. For instance, motion can help to decompose a 3D model of a character into semantically significant parts, such as legs, arms, torso and head. Possible applications of such high-level shape understanding include accurate feature computation, comparison between models to detect defects or medical pathologies, and the design of new biometric models.
4.3 Human Motion Analysis
The recovery of dense motion information enables the combined analysis of shapes and their motions. This allows to classify based on motion cues, which can help in the identification of pathologies or the design of new prostheses. Analyzing human motion can also be used to learn generative models of realistic human motion, which is a recent research topic within Morpheo.
5 Social and environmental responsibility
5.1 Footprint of research activities
The footprint of our research activity is dominated by dissemination and data costs.
Dissemination strategy: Traditionally, Morpheo's dissemination strategy has been to publish in the top conferences of the field (CVPR, ECCV, ICCV, MICCAI), as well as to give invited talks, leading to some long disance work trips. We currently try to increase submissions to top journals in the field directly, hence reducing travels, while still attending some in-person conferences or seminars to allow for networking.
Data management: The data produced by the Morpheo team occupies large memory volumes. The Kinovis platform typically produces around 1.5GB per second when capturing one actor at 50fps. The platform that also captures X-ray images at CHU produces around 1.3GB of data per second at 30fps for video and X-rays. For practical reasons, we reduce the data as much as possible with respect to the targeted application by only keeping e.g. 3D reconstructions or down-sampled spatial or temporal camera images. Yet, acquiring, processing and storing these data is costly in terms of resources. In addition, the data captured by these platforms are personal data with highly constrained regulations. Our data management therefore needs to consider multiple aspects: data encryption to protect personal information, data backup to allow for reproducibility, and the environmental impact of data storage and processing. For all these aspects, we are constantly checking for new and more satisfactory solutions.
For data processing, we rely heavily on cluster uses, in particular with neural networks which are known to have a heavy carbon footprint. Yet, in our research field, these types of processing methods have been shown to lead to significant performance gains. For this reason, we continue to use neural networks as tools, while attempting to use architectures that allow for faster and more energy efficient training, and simple test cases that can often be trained on local machines (with GPU) to allow for less costly network design. A typical example of this type of research is the work of Briac Toussaint, whose first thesis contribution exhibits a high quality reconstruction algorithm for the Kinovis platform, with under a minute of GPU computation time per frame, an order of magnitude faster than our previous research solution, yet it achieves improved precision.
5.2 Impact of research results
Morpheo's main research topic is not related to sustainability. Yet, some of our research directions could be applied to solve problems relevant to sustainability.
Realistic digital human modeling holds the potential to allow for realistic interactions through large geographic distances, leading to more realistic and satisfactory teleconferencing systems. Morpheo captures and analyzes humans in 4D, thereby allowing to capture both their shapes and movement patterns, and actively works on human body modeling. In this line of work, Morpheo works on the project 3DMOVE to model realistic human motion, and participates in the Nemo.AI joint lab with Interdigital on advancing this topic.
Of course, as with any research direction, ours can also be used to generate technologies that are resource-hungry and whose necessity may be questionable, such as inserting animated 3D avatars in scenarios where simple voice communication would suffice, for instance.
6 Highlights of the year
6.1 Awards
- Prix Innovation INRIA: Kinowild
- Marilin Keller (co-supervised by Michael Black and Sergi Pujades) wins Honorable Mention for the 2024 MPI-IS Outstanding Female doctoral Student Prize.
- Best Paper (Nagao Award) at MIRU 2024 - Meeting on Image Recognition and Understanding (Award page), biggest Japanese Symposium on image recognition and understanding. 3D Shape Modeling with Adaptive Centroidal Voronoi Tesselation on Signed Distance Field 13.
6.2 Team Hiring
Jean-Sébastien Franco, Morpheo group leader and previously associate professor, was recruited as full time Senior Inria Researcher on October 1st, 2024.
6.3 CHU collaboration strengthened
- Installation of the Kinovis platform at the CHU
In April 2024, the mobile Kinovis platform was installed in the facilities of the LAGAME laboratory at the Pediatric Hospital in Grenoble (CHUGA), led by Aurélien Courvoisier. The deployment of the platform has allowed to build a first Proof of Concept in which patients with scoliosis are acquired during their regular visits at the hospital. This collaboration aims at better understanding the underlying factors of Adolescent Idiopathic Scoliosis through shape and motion analysis.
6.4 Organization of the IABM conference
The second edition of the Colloque Français d'Intelligence Artificielle en Imagerie Biomédicale (IABM 2024) was held in Grenoble on the 25, 26 and 27 March 2024. The objective of this Event is to gather the main actors in France from the academy, industry and the clinics working on Artificial Intelligence applied to medical images. The event is co-organized by the 3IA institures of Grenoble (MIAI), Nice (3IA Côte d’Azur) and Paris (PRAIRIE). The event was a success gathering 240 participants. Sergi Pujades was in the organizing comittee.
7 New software, platforms, open data
7.1 New software
7.1.1 Millimetric humans
-
Name:
Millimetric Human Surface Capture in Minutes
-
Keywords:
Multi-View reconstruction, Differentiable Rendering, Recognition of human movement
-
Functional Description:
The code allows to reconstruct geometry from human multi-view capture datasets.
- URL:
- Publication:
-
Contact:
Jean Franco
7.1.2 HIT
-
Name:
Human Implicit Tissues
-
Keywords:
Implicit surface, Geometry
-
Functional Description:
This software takes as input the shape of the surface of a person, in the SMPL parametrization, and predicts the volumetric tissues (lean tissue, adipose tissue and bone tissue) inside it.
- URL:
-
Contact:
Sergi Pujades
-
Partner:
Max Planck Institute for Intelligent Systems
7.1.3 SKELJ
-
Name:
On predicting 3D bone locations inside the human body
-
Keywords:
Human Body Surface, Shape approximation, 3D, 3D modeling
-
Functional Description:
This software takes as input the shape of the surface of a person, in the SMPL parametrization, and outputs the 3D location of the bones, parametrized as a the SKEL mesh.
- URL:
-
Contact:
Sergi Pujades
-
Partner:
Max Planck Institute for Intelligent Systems
7.1.4 Osatta
-
Name:
One-Shot Automatic Test Time Augmentation for Domain Adaptation
-
Keywords:
Image analysis, Image processing, Machine learning
-
Functional Description:
This software computes a family of image transformations that allow to improve the accuracy of a fundamental model on biased cohorts with few annotations.
- URL:
-
Contact:
Sergi Pujades
7.1.5 MotionRetargeter
-
Name:
Correspondence-free online human motion retargeting
-
Keywords:
3D animation, 3D modeling
-
Functional Description:
Allows to retarget the motion of one human body to another body shape, without needing any correspondence information. Code allows to reproduce results in: Mathieu Marsot, Rim Rekik, Stefanie Wuhrer, Jean-Sébastien Franco, Anne-Hélène Olivier. Correspondence-free online human motion retargeting. arXiv 2302.00556, 2023.
- URL:
-
Contact:
Rim Rekik Dit Nekhili
7.2 New platforms
Kinovis platform deployment.
The Kinovis platform has been partly moved to the Grenoble Hospital (CHU), where is is used on patients by clinicians. This successful Proof-of-Concept demonstrates:
- the platform mobility at a technical level
- the ability to capture in varied environments at a scientific level
- the tremendous potential in real-life applications
Participants:Julien Pansiot , Sergi Pujades
7.3 Open data
HIT and SKEL-J Dataset.
In collaboration with the Max Planck Institute for Perceiving Systems, two datasets where created and shared in an Open Data policy. The data is described in the publications HIT10 and SKEL-J 9, and is available on the corresponding web pages https://hit.is.tue.mpg.de/ and https://3dbones.is.tue.mpg.de.
ShowMe Dataset.
Morpheo and Naver Labs Europe have released a dataset of hand-object 3D reconstruction from monocular video, which provides a set of diverse hand-object videos where objects in the dataset are presented to the the camera. This dataset, which is freely available at https://europe.naverlabs.com/research/showme/, was the evaluation basis for the adjoining CVIU journal article 7, which describes the protocol and dataset in detail.
MVMannequin Dataset.
For the purpose of evaluating high quality dressed human reconstruction from multiple videos, we created the MVMannequin dataset, a dataset of 14 dressed mannquins captured with the Inria Kinovis multi-camera capture system in Montbonnot, and their submillimetric accurate static 3D scans as reference. It is freely available on the webpage https://kinovis.inria.fr/mvmannequin/. The dataset and its purpose are described in more detail in the Siggraph Asia publication Millimetric Human Surface Capture in Minutes 14.
8 New results
8.1 SHOWMe: Robust object-agnostic hand-object 3D reconstruction from RGB video
Participants: Anilkumar Swamy, Vincent Leroy, Philippe Weinzaepfel, Fabien Baradel, Salma Galaaoui, Romain Brégier, Matthieu Armando, Jean-Sébastien Franco, Grégory Rogez.
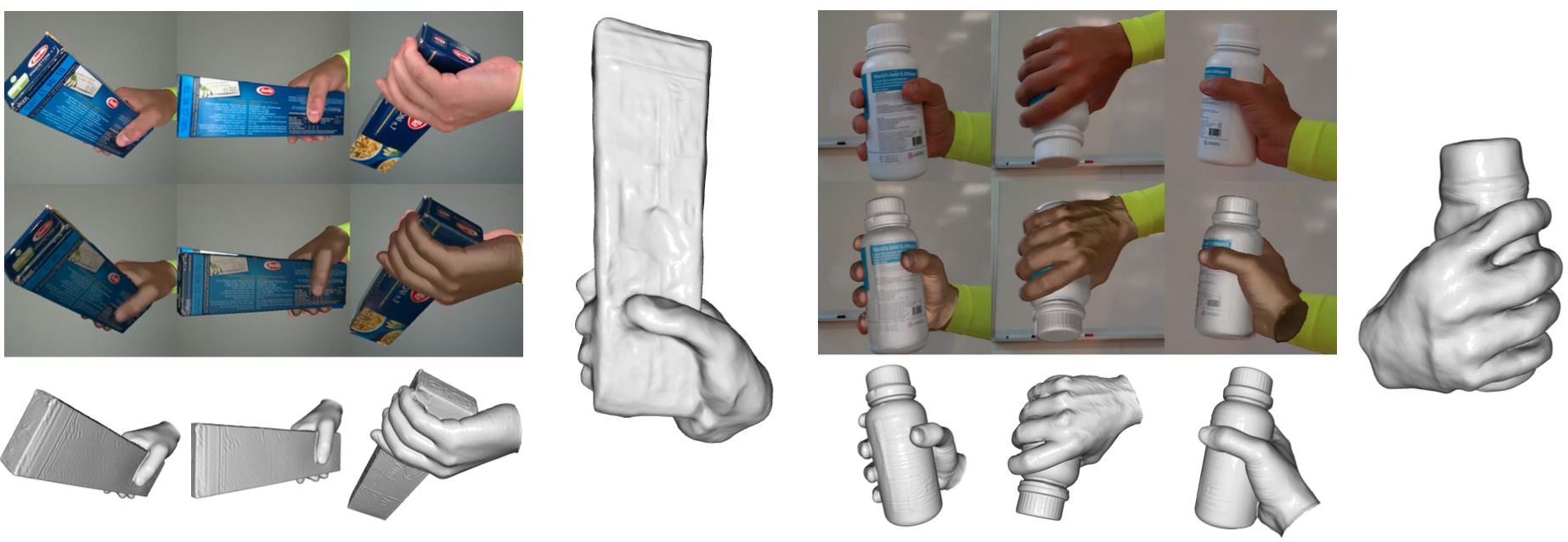
Exerpts of the SHOWME dataset.
In this paper, we tackle the problem of detailed hand-object 3D reconstruction from monocular video with unknown objects, for applications where the required accuracy and level of detail is important, e.g. object hand-over in human-robot collaboration, or manipulation and contact point analysis. While the recent literature on this topic is promising, the accuracy and generalization abilities of existing methods are still lacking. This is due to several limitations, such as the assumption of known object class or model for a small number of instances, or over-reliance on off-the-shelf keypoint and structure-from-motion methods for object-relative viewpoint estimation, prone to complete failure with previously unobserved, poorly textured objects or hand-object occlusions. To address previous method shortcomings, this work presents a 2-stage pipeline superseding state-of-the-art performance on several metrics.
First, the method robustly retrieves viewpoints relying on a learned pairwise camera pose estimator trainable with a low data regime, followed by a globalized Shonan pose averaging. Second, it simultaneously estimates detailed 3D hand-object shapes and refines camera poses using a differential renderer-based optimizer. To better assess the out-of-distribution abilities of existing methods, and to showcase our methodological contributions, this work introduces the new SHOWMe benchmark dataset with 96 sequences annotated with poses, millimetric textured 3D shape scans, and parametric hand models, introducing new object and hand diversity. The work shows that our method is able to reconstruct 100% of these sequences as opposed to SotA Structure-from-Motion (SfM) or hand-keypoint-based pipelines, and obtains reconstructions of equivalent or better precision when existing methods do succeed in providing a result. We hope these contributions lead to further research under harder input assumptions.
This work was published as an article in the Computer Vision and Image Understanding journal 7. The dataset can be downloaded at SHOWME webpage.
8.2 Millimetric Human Surface Capture in Minutes
Participants: Briac Toussaint, Laurence Boissieux, Diego Thomas, Edmond Boyer, Jean-Sébastien Franco.

We present an implicit surface reconstruction algorithm achieving millimetric precision in 3 minutes of training time for fast and accurate multi-view human acquisition.
Detailed human surface capture from multiple images is an essential component for many 3D production, analysis and transmission tasks. Yet producing millimetric precision 3D models in practical time, and actually verifying their 3D accuracy in a real-world capture context, remain key challenges due to the lack of specific methods and data for these goals. This work proposes two complementary contributions to this end. The first one is a highly scalable neural surface radiance field approach able to achieve millimetric precision by construction, while demonstrating high compute and memory efficiency. The second one is a novel dataset MVMannequin of clothed mannequin geometry captured with a high resolution hand-held 3D scanner paired with calibrated multi-view images, that allow to verify the millimetric accuracy claim.
Although the proposed approach can produce such a highly dense and precise geometry, the work shows how aggressive sparsification and optimizations of the neural surface pipeline lead to estimations requiring only minutes of computation time and a few GB of VRAM memory on GPU, while allowing for real-time millisecond neural rendering. On the basis of the presented framework and dataset, we provide a thorough experimental analysis of how such accuracies and efficiencies are achieved in the context of multi-camera human acquisition.
The paper was published at the Sigraph Asia 2024 conference program 14, and more resources are provided on the MillimetricHumans project webpage.
8.3 On predicting 3D bone locations inside the human body
Participants: Abdelmoutaleb Dakri, Vaibhav Arora, Léo Challier, Marilyn Keller, Michael Black, Sergi Pujades.
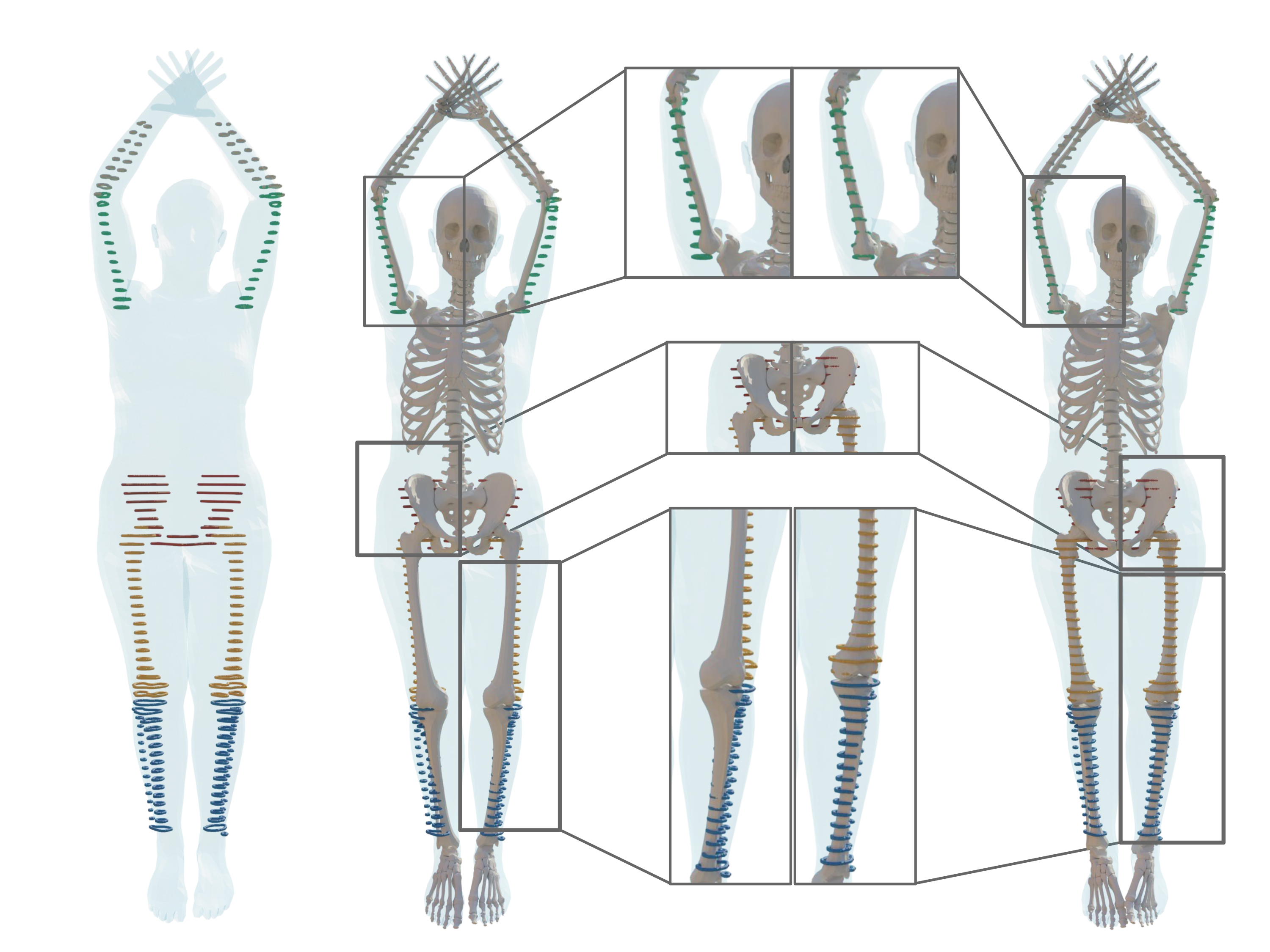
In this work we tackle the problem of predicting the location of the bones inside the human body.
Knowing the precise location of the bones inside the human body is key in several medical tasks, such as patient placement inside an imaging device or surgical navigation inside a patient. Our goal is to predict the bone locations using only an external 3D body surface obser- vation. Existing approaches either validate their predictions on 2D data (X-rays) or with pseudo-ground truth computed from motion capture using biomechanical models. Thus, methods either suffer from a 3D-2D projection ambiguity or directly lack validation on clinical imaging data. In this work 9, we start with a dataset of segmented skin and long bones obtained from 3D full body MRI images that we refine into individual bone segmentations. To learn the skin to bones correlations, one needs to register the paired data. Few anatomical models allow to register a skeleton and the skin simultaneously. One such method, SKEL, has a skin and skeleton that is jointly rigged with the same pose parameters. However, it lacks the flexibility to adjust the bone locations inside its skin. To address this, we extend SKEL into SKEL-J to allow its bones to fit the segmented bones while its skin fits the segmented skin. These precise fits allow us to train SKEL-J to more accurately infer the anatomical joint locations from the skin surface. Our qualitative and quantitative results show how our bone location predictions are more accurate than all existing approaches. o foster future research, we make available for research purposes the individual bone segmentations, the fitted SKEL-J models as well as the new inference methods on the project webpage.
8.4 Pose-independent 3D Anthropometry from Sparse Data
Participants: David Bojanić, Stefanie Wuhrer, Tomislav Petković, Tomislav Pribanić.
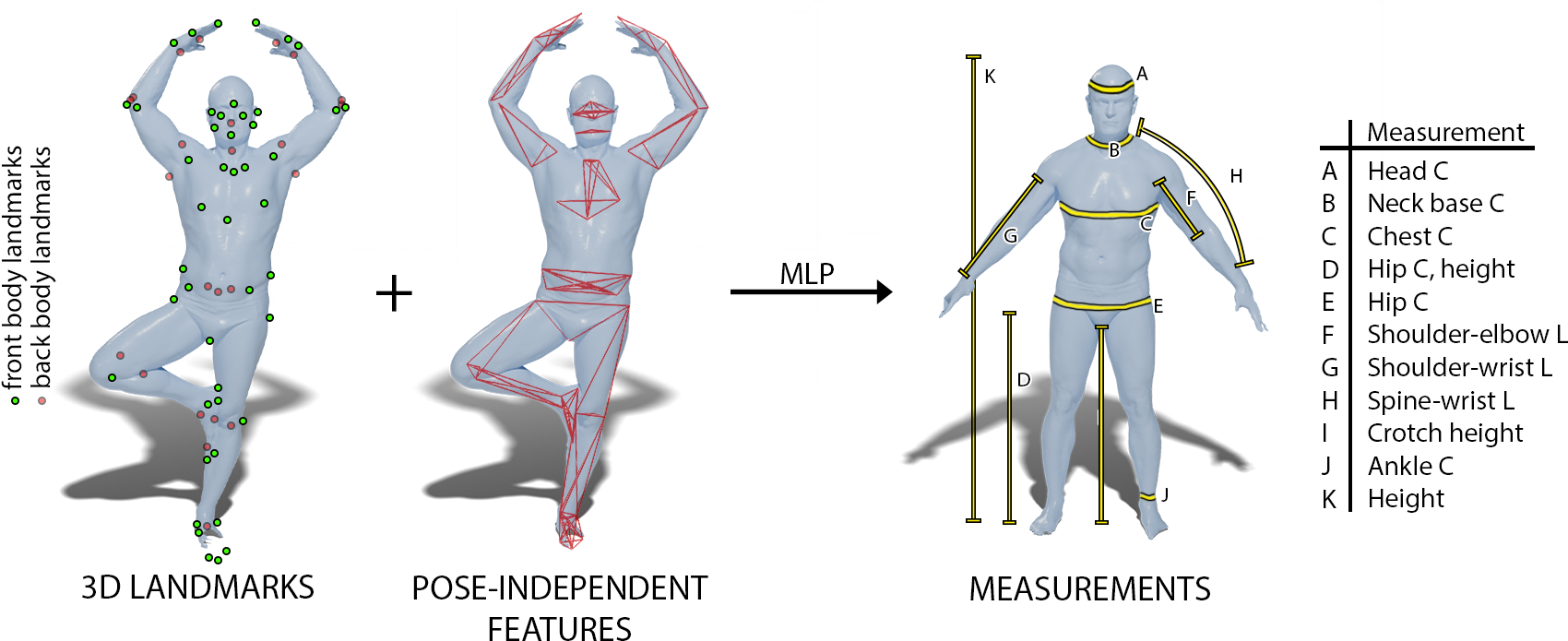
We propose an approach to estimate body measurements from 3D landmark locations on a body in any given pose.
3D digital anthropometry is the study of estimating human body measurements from 3D scans. Precise body measurements are important health indicators in the medical industry, and guiding factors in the fashion, ergonomic and entertainment industries. The measuring protocol consists of scanning the whole subject in the static A-pose, which is maintained without breathing or movement during the scanning process. However, the A-pose is not easy to maintain during the whole scanning process, which can last even up to a couple of minutes. This constraint affects the final quality of the scan, which in turn affects the accuracy of the estimated body measurements obtained from methods that rely on dense geometric data. Additionally, this constraint makes it impossible to develop a digital anthropometry method for subjects unable to assume the A-pose, such as those with injuries or disabilities. We propose a method that can obtain body measurements from sparse land marks acquired in any pose. We make use of the sparse landmarks of the posed subject to create pose-independent features, and train a network to predict the body measurements as taken from the standard A-pose. We show that our method achieves comparable results to competing methods that use dense geometry in the standard A-pose, but has the capability of estimating the body measurements from any pose using sparse landmarks only.
8.5 A Survey on Realistic Virtual Human Animations: Definitions, Features and Evaluations
Participants: Rim Rekik, Stefanie Wuhrer, Ludovic Hoyet, Katja Zibrek, Anne-Hélène Olivier.
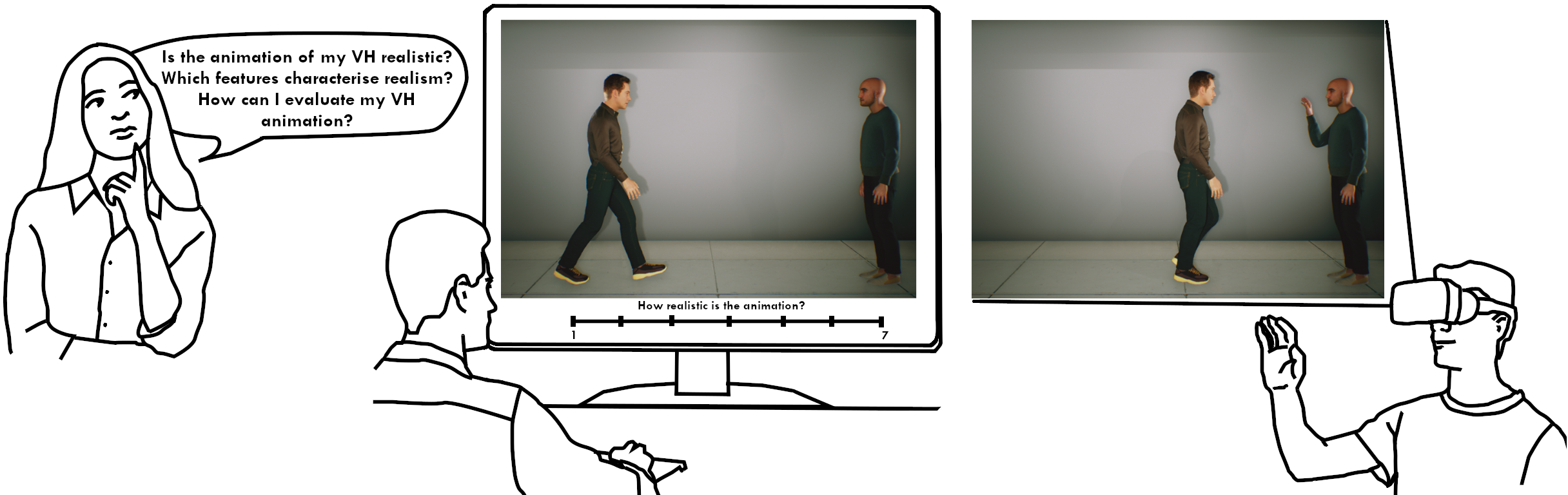
The central goal of this survey is to explore the aspects of animation realism of virtual humans.
Generating realistic animated virtual humans is a problem that has been extensively studied with many applications in different types of virtual environments. However, the creation process of such realistic animations is challenging, especially because of the number and variety of influencing factors, that should then be identified and evaluated. In this paper, we attempt to provide a clearer understanding of how the multiple factors that have been studied in the literature impact the level of realism of animated virtual humans, by providing a survey of studies assessing their realism. This includes a review of features that have been manipulated to increase the realism of virtual humans, as well as evaluation approaches that have been developed. As the challenges of evaluating animated virtual humans in a way that agrees with human perception are still active research problems, this survey further identifies important open problems and directions for future research.
8.6 Correspondence-free online human motion retargeting
Participants: Rim Rekik, Mathieu Marsot, Anne-Hélène Olivier, Jean-Sébastien Franco, Stefanie Wuhrer.
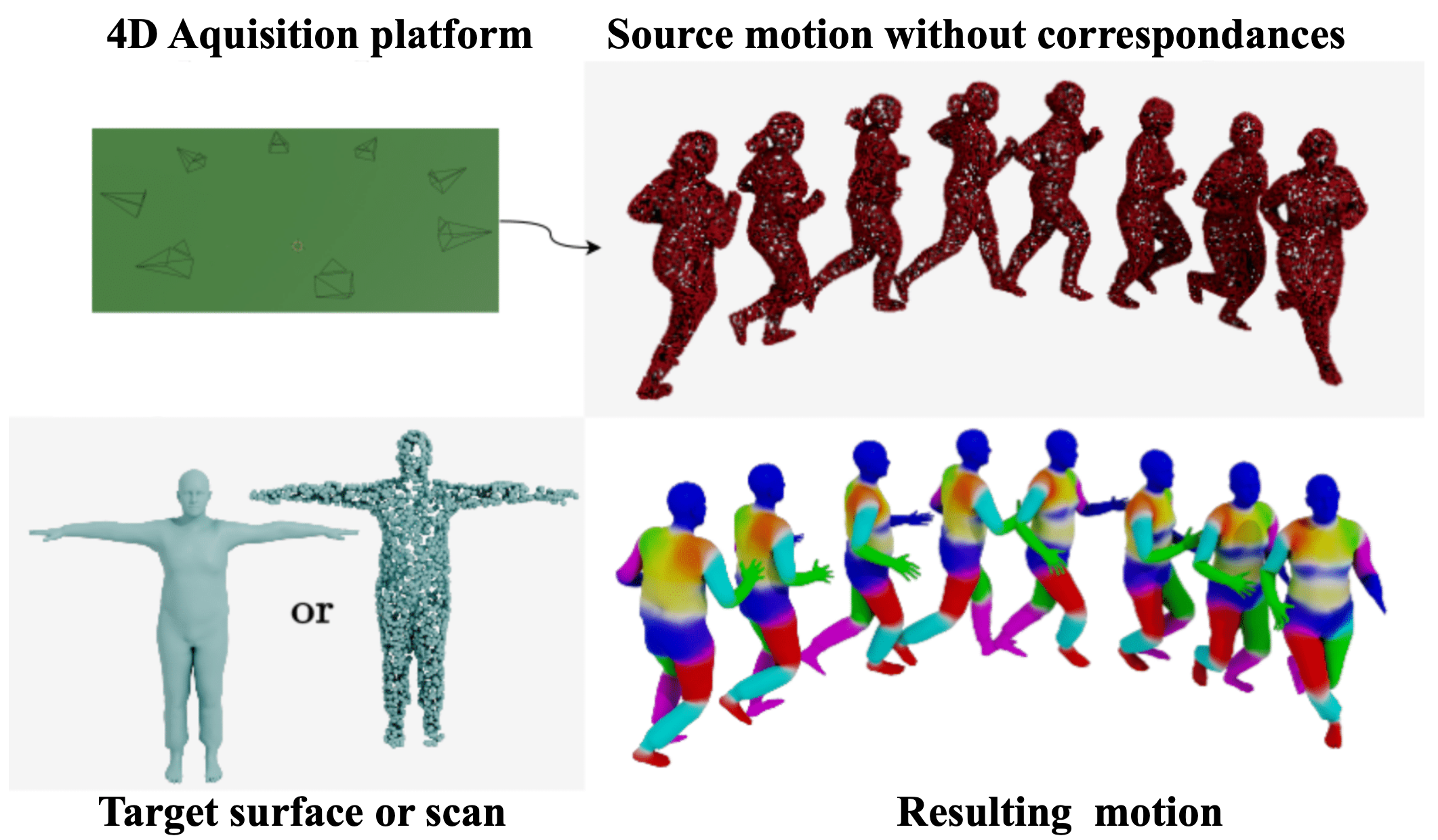
This work achieves high-quality motion retargeting between unregistered models, and can be used on the output of the Kinovis platform directly.
We present a data-driven framework for unsupervised human motion retargeting that animates a target subject with the motion of a source subject. Our method is correspondence-free, requiring neither spatial correspondences between the source and target shapes nor temporal correspondences between different frames of the source motion. This allows to animate a target shape with arbitrary sequences of humans in motion, possibly captured using 4D acquisition platforms or consumer devices. Our method unifies the advantages of two existing lines of work, namely skeletal motion retargeting, which leverages long-term temporal context, and surface-based retargeting, which preserves surface details, by combining a geometry-aware deformation model with a skeleton-aware motion transfer approach. This allows to take into account long-term temporal context while accounting for surface details. During inference, our method runs online, i.e. input can be processed in a serial way, and retargeting is performed in a single forward pass per frame. Experiments show that including long-term temporal context during training improves the method’s accuracy for skeletal motion and detail preservation. Furthermore, our method generalizes to unobserved motions and body shapes. We demonstrate that our method achieves state-of-the-art results on two test datasets and that it can be used to animate human models with the output of a multi-view acquisition platform.
8.7 3D Shape Modeling with Adaptive Centroidal Voronoi Tesselation on Signed Distance Field
Participants: Diego Thomas, Briac Toussaint, Jean-Sébastien Franco, Edmond Boyer.

Sampling is shown, left figure shows the reconstructed geometry, right figure a color-coded 3D model
Volumetric shape representations have become ubiquitous in multi-view reconstruction tasks. They often build on regular voxel grids as discrete representations of 3D shape functions, such as SDF or radiance fields, either as the full shape model or as sampled instantiations of continuous representations, as with neural networks. Despite their proven efficiency, voxel representations come with the precision versus complexity trade-off. This inherent limitation can significantly impact performance when moving away from simple and uncluttered scenes.
This work investigates an alternative discretization strategy with the Centroidal Voronoi Tessellation (CVT). CVTs allow to better partition the observation space with respect to shape occupancy and to focus the discretization around shape surfaces. To leverage this discretization strategy for multi-view reconstruction, this work introduced a volumetric optimization framework that combines explicit SDF fields with a shallow color network, in order to estimate 3D shape properties over tetrahedral grids. Experimental results with Chamfer statistics validate this approach with unprecedented reconstruction quality on various scenarios such as objects, open scenes or human.
This work has lead to several publicatons, a research report 16, and a MIRU 2024 Japanese computer vision conference, where is obtained a best paper award 13. It is also the subject of an accepted submission to the WACV 2025 computer vision conference.
8.8 HIT: Human Implicit Tissues
Participants: Marilyn Keller, Vaibhav Arora, Abdelmoutaleb Dakri, Shivam Chandhok, Jürgen Machann, Andreas Fritsche, Michael J. Black, Sergi Pujades.
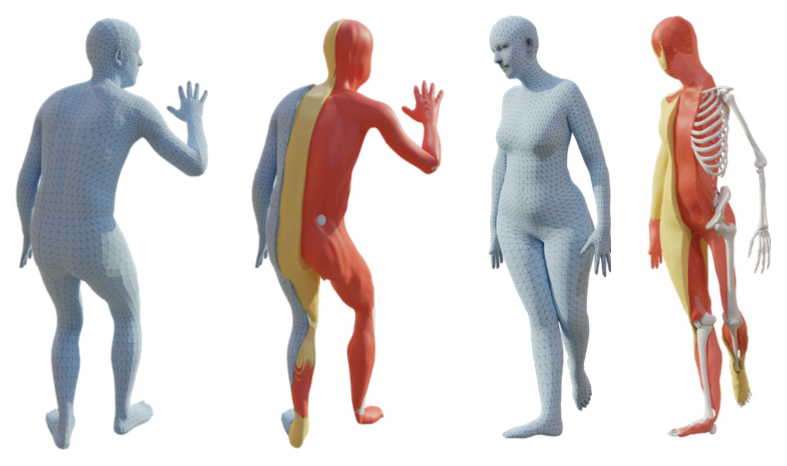
In this work we learn how to predict the volumetric internal tissues from a person from the shape of its body surface.
The creation of personalized anatomical digital twins is important in the fields of medicine, computer graphics, sports science, and biomechanics. To observe a subject's anatomy, expensive medical devices (MRI or CT) are required and the creation of the digital model is often time-consuming and involves manual effort. Instead, we leverage the fact that the shape of the body surface is correlated with the internal anatomy; for example, from surface observations alone, one can predict body composition and skeletal structure. In this work 10, we go further and learn to infer the 3D location of three important anatomic tissues: subcutaneous adipose tissue (fat), lean tissue (muscles and organs), and long bones. To learn to infer these tissues, we tackle several key challenges. We first create a dataset of human tissues by segmenting full-body MRI scans and registering the SMPL body mesh to the body surface. With this dataset, we train HIT (Human Implicit Tissues), an implicit function that, given a point inside a body, predicts its tissue class. HIT leverages the SMPL body model shape and pose parameters to canonicalize the medical data. Unlike SMPL, which is trained from upright 3D scans, the MRI scans are taken of subjects lying on a table, resulting in significant soft-tissue deformation. Consequently, HIT uses a learned volumetric deformation field that undoes these deformations. Since HIT is parameterized by SMPL, we can repose bodies or change the shape of subjects and the internal structures deform appropriately. We perform extensive experiments to validate HIT's ability to predict plausible internal structure for novel subjects. The dataset and HIT model are available on the HIT project webpage to foster future research in this direction.
8.9 OSATTA
Participants: Felix Kuper, Sergi Pujades.
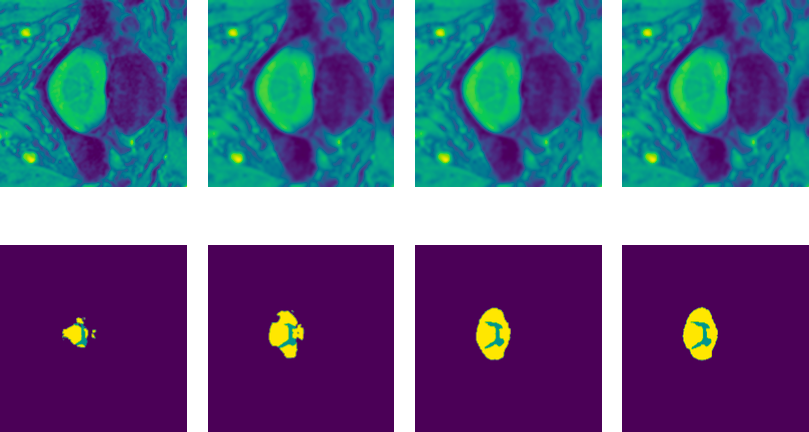
In this work we learn how to find image transformation families which can correct for the individual bias present in a small cohort of data. These transformations align the source data to the target Fundamental Model distribution to improve its performance.
Fundamental models (FM) are reshaping the research paradigm by providing ready-to-use solutions to many challenging tasks, such as image classification, registration, or segmentation. Yet, their performance on new dataset cohorts significantly drops, particularly due to domain gaps between the training (source) and testing (target) data. Recently, test time augmentation strategies aim at finding target-to-source- mappings (t2sm), which improve the performance of the FM on the target dataset, by inspecting the the FM weights, thus assuming access to them. While this assumption holds for open research models, it does not for commercial ones (e.g., Chat-GPT). These are provided as black boxes, and the training data and the model weights are not available. In our work11, we propose a new generic few-shot method that enables the computation of a target-to-source mapping by only using the black-box model’s outputs. We start by defining a parametric family of functions for the t2sm, and with a simple loss function, we optimize the t2sm parameters based on a single labeled image volume. This effectively provides a mapping between the source domain and the target domain. In our experiments, we investigate how to improve the segmentation performance of a given FM (a UNet), and we outperform state-of-the-art accuracy in the 1-shot setting, with further improvement in a few-shot setting. Our approach is invariant to the model architecture as the FM is treated as a black box, which significantly increases our method’s practical utility in real-world scenarios. We make the code available for reproducibility purposes on the OSATTA project webpage.
9 Bilateral contracts and grants with industry
9.1 Bilateral contracts with industry
Participants: Antoine Dumoulin, Youssef Ben Cheikh, Stefanie Wuhrer, Jean-Sébastien Franco.
- The Morpheo INRIA team has a collaboration with Interdigital in Rennes through the Nemo.AI joint lab. The kickoff to this collaboration has happened in November 2022. The collaboration involves two PhD co-supervisions, Antoine Dumoulin and Youssef Ben Cheikh, at the INRIA de l'Université Grenoble Alpes. The subject of the collaboration revolves around digital humans, one the one hand to estimate the clothing of humans from images, and on the other to estimate hair models from one or several videos.
9.2 Bilateral grants with industry
Participants: Jean-Sébastien Franco.
- The Morpheo team is also involved in a CIFRE PhD co-supervision (Anilkumar Swamy) with Naver Labs since July 2021, with Gregory Rogez, Mathieu Armando and Vincent Leroy. The work revolves around monocular hand-object reconstruction by showing the object to the camera.
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Participation in other International Programs
Shape Up! Keiki
Participants: Sergi Pujades, Abdelmoutaleb Dakri, Vaibhav Arora.
- Shape Up! Keiki
-
Partner Institution(s):
- University of Hawai'i, Cancer Center, USA
- Pennington Biomedical Research Center, USA
- University of California San Francisco, USA
- University of Washington, USA
- Inria Grenoble - Rhone Alpes, France
-
Date/Duration:
5 years, 2022-2026
-
Additionnal info/keywords:
The long term goal of the Shape Up! Keiki is 1) to provide pediatric phenotype descriptors of health using body shape, and 2) to provide the tools to visualize and quantify body shape in research and clinical practice. Our overall approach is to first derive predictive models of how body shape relates to regional and total body composition (trunk fat, muscle mass, lean mass, and percent fat) with consideration to the rapidly changing hydration over the early years of life (birth to 5 years). We will then identify covariates that impact the precision and accuracy of our optical models. We will then show how our optical body composition estimates are associated with important developmental and dietary factors. Our central hypothesis is that optical estimates of body composition suitably represent a 5-compartment (5C) body composition model for studies of adiposity and health in young children and are superior to that of simple anthropometry and demographics. The rationale for this study is that early life access to accurate body composition data will enable identification of factors that increase obesity, metabolic disease, and cancer risk, and provide a means to target interventions to those that would benefit. The expected outcome is that our findings would be immediately applicable to accessible gaming and imaging sensors found on modern computers.
10.2 International research visitors
10.2.1 Visits of international scientists
Other international visits to the team
- David Bojanić visited Morpheo from February until July 2024 to work on estimating measurements of 3D humans in a pose-invariant way.
10.3 National initiatives
10.3.1 ANR
ANR JCJC SEMBA – Shape, Motion and Body composition to Anatomy.
Existing medical imaging techniques, such as Computed Tomography (CT), Dual Energy X-Ray Absorption (DEXA) and Magnetic Resonance Imaging (MRI), allow to observe internal tissues (such as adipose, muscle, and bone tissues) of in-vivo patients. However, these imaging modalities involve heavy and expensive equipment as well as time consuming procedures. External dynamic measurements can be acquired with optical scanning equipment, e.g. cameras or depth sensors. These allow high spatial and temporal resolution acquisitions of the surface of living moving bodies. The main research question of SEMBA is: “can the internal observations be inferred from the dynamic external ones only?”. SEMBA’s first hypothesis is that the quantity and distribution of adipose, muscle and bone tissues determine the shape of the surface of a person. However, two subjects with a similar shape may have different quantities and distributions of these tissues. Quantifying adipose, bone and muscle tissue from only a static observation of the surface of the human might be ambiguous. SEMBA's second hypothesis is that the shape deformations observed while the body performs highly dynamic motions will help disambiguating the amount and distribution of the different tissues. The dynamics contain key information that is not present in the static shape. SEMBA’s first objective is to learn statistical anatomic models with accurate distributions of adipose, muscle, and bone tissue. These models are going to be learned by leveraging medical dataset containing MRI and DEXA images. SEMBA's second objective will be to develop computational models to obtain a subject-specific anatomic model with an accurate distribution of adipose, muscle, and bone tissue from external dynamic measurements only.
ANR JCJC 3DMOVE - Learning to synthesize 3D dynamic human motion.
It is now possible to capture time-varying 3D point clouds at high spatial and temporal resolution. This allows for high-quality acquisitions of human bodies and faces in motion. However, tools to process and analyze these data robustly and automatically are missing. Such tools are critical to learning generative models of human motion, which can be leveraged to create plausible synthetic human motion sequences. This has the potential to influence virtual reality applications such as virtual change rooms or crowd simulations. Developing such tools is challenging due to the high variability in human shape and motion and due to significant geometric and topological acquisition noise present in state-of-the-art acquisitions. The main objective of 3DMOVE is to automatically compute high-quality generative models from a database of raw dense 3D motion sequences for human bodies and faces. To achieve this objective, 3DMOVE will leverage recently developed deep learning techniques. The project also involves developing tools to assess the quality of the generated motions using perceptual studies. This project involves two Ph.D. students: Mathieu Marsot who graduated in May 2023 and Rim Rekik hired November 2021.
ANR Human4D - Acquisition, Analysis and Synthesis of Human Body Shape in Motion
Human4D is concerned with the problem of 4D human shape modeling. Reconstructing, characterizing, and understanding the shape and motion of individuals or groups of people have many important applications, such as ergonomic design of products, rapid reconstruction of realistic human models for virtual worlds, and an early detection of abnormality in predictive clinical analysis. Human4D aims at contributing to this evolution with objectives that can profoundly improve the reconstruction, transmission, and reuse of digital human data, by unleashing the power of recent deep learning techniques and extending it to 4D human shape modeling. It involves 4 academic partners: The laboratory ICube at Strasbourg which is leading the project, the laboratory Cristal at Lille, the laboratory LIRIS at Lyon and the INRIA Morpheo team. This project involves Ph.D. student Aymen Merrouche hired in November 2021.
ANR Equipex+ CONTINUUM - Collaborative continuum from digital to human
The CONTINUUM project will create a collaborative research infrastructure of 30 platforms located throughout France, including Inria Grenoble's Kinovis, to advance interdisciplinary research based on interaction between computer science and the human and social sciences. Thanks to CONTINUUM, 37 research teams will develop cutting-edge research programs focusing on visualization, immersion, interaction and collaboration, as well as on human perception, cognition and behaviour in virtual/augmented reality, with potential impact on societal issues. CONTINUUM enables a paradigm shift in the way we perceive, interact, and collaborate with complex digital data and digital worlds by putting humans at the center of the data processing workflows. The project will empower scientists, engineers and industry users with a highly interconnected network of high-performance visualization and immersive platforms to observe, manipulate, understand and share digital data, real-time multi-scale simulations, and virtual or augmented experiences. All platforms will feature facilities for remote collaboration with other platforms, as well as mobile equipment that can be lent to users to facilitate onboarding.
ANR PRC Inora
The INORA project aims at understanding the mechanisms of action of shoes and orthotic insoles on Rheumatoid arthritis (RA) patients through patient-specific computational biomechanical models. These models will help in uncovering the mechanical determinants to pain relief, which will enable the long-term well-being of patients. Motivated by the numerous studies highlighting erosion and joint space narrowing in RA patients, we postulate that a significant contributor to pain is the internal joint loading when the foot is inflamed. This hypothesis dictates the need of a high-fidelity volumetric segmentation for the construction of the patient-specific geometry. It also guides the variables of interests in the exploitation of a finite element (FE) model. The INORA project aims at providing numerical tools to the scientific, medical and industrial communities to better describe the mechanical loading on diseased distal foot joints of RA patients and propose a patient-specific methodology to design pain-relief insoles. The Mines de St Etienne is leading this project.
ANR 4DPlants - Learning Plant Growth Models
The overall goal of the 4DPlants project is to invent solutions that will make plant phenotyping in 3D and over time reliable and accessible to a majority of plant scientists. This is achieved in a consortium including biologists and computer scientists by acquiring growing plants in 3D, and by analyzing growth changes using data-driven methods. The project, which started in December 2024, includes partners from Université de Strasbourg, ENS Lyon, and Morpheo team at Inria. This projet involves Ph.D. student Samara Ghrer, hired in December 2024.
10.4 Regional initiatives
Persiyval Phd Grant
In collaboration with the IIHM team from the LIG, a co-supervision started on the topic of high precision real-time tracking of fingers with markerless vision systems.
Participants: Quentin Zoppis Sergi PUJADES Laurence Nigay François Berard
11 Dissemination
11.1 Promoting scientific activities
11.1.1 Scientific events: organisation
Member of the organizing committees
- Sergi Pujades was in the organizing committe of IABM 2024
11.1.2 Scientific events: selection
Reviewer
- Sergi Pujades reviewed for CVPR and MICCAI
- Jean-Sébastien Franco reviewed for CVPR, ECCV, 3DV
- Stefanie Wuhrer reviewed for CVPR, ECCV, 3DV
11.1.3 Journal
Member of the editorial boards
- Stefanie Wuhrer was associate editor for IEEE Transactions on Circuits and Systems for Video Technology
- Jean-Sébastien Franco was associate editor for the International Journal on Computer Vision.
Reviewer - reviewing activities
- Sergi Pujades reviewed for MedIA
- Stefanie Wuhrer reviewed for CVIU
11.1.4 Invited talks
- Sergi Pujades did an invited talk at IABM 2024 "Externe - Interne : forme du corps et anatomie interne" on 26th March 2024.
- Sergi Pujades did an invited talk at TIMC GMCAO "Externe - Interne : forme du corps et anatomie interne" on 17th September 2024.
- Sergi Pujades did an invited talk at MPI Perceiving Systems on "How to predict the inside from the outside? Segment, register, model and infer!" on 28th November 2024.
- Jean-Sébastien Franco did an invited talk at Kyushu University on "Capture and Analysis of Shapes in Motion", on 29th of November, 2024.
- Stefanie Wuhrer presented “Learning representations of 4D human motion”. Online expert lecture at 3D Vision Summer School, Bangalore, India (remote attendance), May 2024.
11.1.5 Scientific expertise
- Sergi Pujades was on the scientific committee for the study of the "Delegations INRIA" applications
- Jean-Sébastien Franco was on the recruiting committee (COS) for two assistant professor positions at University of Nancy / Loria.
- Stefanie Wuhrer was member of jury d’admission CRCN Inria 2024
11.1.6 Research administration
- Stefanie Wuhrer is référente données for the Inria centre of the Grenoble Alpes University
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
- Master: Sergi Pujades , Computer Vision, 54h EqTD, M2R Mosig GVR, Grenoble INP.
- Master: Sergi Pujades , Introduction to Visual Computing, 42h, M1R Mosig GVR, Université Grenoble Alpes.
- Master: Julien Pansiot , Introduction to Visual Computing, 15h EqTD, M1 MoSig, Université Grenoble Alpes.
- Master: Sergi Pujades , Internship supervision, 1h, Ensimag 3rd year, Grenoble INP.
- Master: Jean-Sébastien Franco , Introduction to Computer Graphics, 33h, Ensimag 2nd year, Grenoble INP.
- Master: Jean-Sébastien Franco , Introduction to Computer Vision, 45h, Ensimag 3rd year, Grenoble INP.
- Master: Jean-Sébastien Franco , Internship and project supervisions, 12h, Ensimag 2nd and 3rd year, Grenoble INP.
- Master: J.S. Franco, Leadership of the image pedagogic workgroup at Ensimag, 12, Ensimag 2nd and 3rd year, Grenoble INP.
11.2.2 Supervision
- Ph.D. defended: Marilyn Keller . Beyond the Surface: Statistical Approaches to Internal Anatomy Prediction, defended on 29th November 2024. Supervised by Sergi PUJADES and Michael Black .
- Ph.D. defended: Anilkumar Swamy . 3D hand-object reconstruction from monocular video. Defended 6th of November, 2024. Supervised by Jean-Sébastien Franco, Gregory Rogez (CIFRE Naverlabs Europe).
- Ph.D. ongoing: Vicente Estopier Castillo. INTENSIVE : Regional Lung Function Inference from Upper Body Surface Motion for Personalized Ventilation Protocols in Acute Respiratory Failure Patients, since 01.10.2023. Supervised by Jean Franco , Sergi Pujades and Sam Bayat .
- Ph.D. ongoing: Gabriel Ravelomanana . Automated medical image segmentation for the foot of patients with rheumatoid arthritis, since 01.11.2024. Supervised by Julien Pansiot and Sergi PUJADES .
- Ph.D. ongoing: Quentin Zoppis , since 01.10.2024. Supervised by François Berard , Laurence Nigay and Sergi Pujades .
- Ph.D. ongoing: Briac Toussaint , High precision alignment of non-rigid surfaces for 3D performance capture, since 01.10.2021, supervised by Jean-Sébastien Franco .
- Ph.D. ongoing: Rim Rekik . Human motion generation and evaluation, since 1.11.2021. Supervised by Anne-Hélène Olivier and Stefanie Wuhrer .
- Ph.D. ongoing: Aymen Merrouche . 4D correspondence computation, since 1.10.2021. Supervised by Edmond Boyer and Stefanie Wuhrer .
- Ph.D. ongoing: Antoine Dumoulin . Video-based dynamic garment representation and synthesis, since 1.11.2023. Supervised by Pierre Hellier , Adnane Boukhayma , and Stefanie Wuhrer .
- Ph.D. ongoing: Laura Neschen . Joint 4D reconstruction and correspondence computation, since 1.10.2024. Supervised by Jean-Sébastien Franco and Stefanie Wuhrer .
- Ph.D. ongoing: Samara Ghrer . Analysis of 3D plant growth, since 1.11.2024. Supervised by Christophe Godin , Franck Hétroy-Wheeler , and Stefanie Wuhrer .
11.2.3 Juries
- Sergi PUJADES was Ph.D comittee member of Dmitrii Zhemchuzhnikov, Université Grenoble Alpes
- Jean-Sébastien Franco reviewed for the Ph.D thesis of Boyang Yu, Université de Strasbourg.
- Jean-Sébastien Franco reviewed for the Ph.D thesis of Glenn Kerbiriou, INSA Rennes.
- Stefanie Wuhrer reviewed Ph.D. thesis of Souhaib Attaiki, Institut Polytechnique de Paris, Oct. 2024.
- Stefanie Wuhrer was Ph.D. committee member of Mehran Hatamzadeh, Universté Côte d’Azur, Nov. 2024.
- Stefanie Wuhrer reviewed Ph.D. thesis of Guénolé Fiche, CentraleSupélec Rennes, Nov. 2024.
- Stefanie Wuhrer reviewed Ph.D. thesis of Pierre Galmiche, Université de Strasbourg, Nov. 2024.
11.3 Popularization
11.3.1 Productions (articles, videos, podcasts, serious games, ...)
- The 4DHumanOutfit dataset was featured in an article on the Inria public relations website.
11.3.2 Others science outreach relevant activities
- Julien Pansiot was invited at "The Relationship between Research and Animation" round table at the MIFA - Annecy Animation Film Festival
- Julien Pansiot presented the Kinovis platform at the Tech &Fest event, Grenoble.
12 Scientific production
12.1 Major publications
- 1 inproceedingsOn predicting 3D bone locations inside the human body.MICCAI 2024 - 27th International Conference on Medical Image Computing and Computer Assisted InterventionMarrakech, Morocco2024, 1-11HAL
- 2 inproceedingsHIT: Estimating Internal Human Implicit Tissues from the Body Surface.CVPR 2024 - IEEE/CVF Conference on Computer Vision and Pattern RecognitionLecture Notes in Computer ScienceSeattle, United StatesIEEE2024, 3480-3490HALDOI
- 3 articleA Survey on Realistic Virtual Human Animations: Definitions, Features and Evaluations.Computer Graphics Forum432May 2024, e15064:1-21HALDOI
- 4 articleSHOWMe: Robust object-agnostic hand-object 3D reconstruction from RGB video.Computer Vision and Image Understanding247October 2024, 104073HALDOI
- 5 inproceedingsMillimetric Human Surface Capture in Minutes.SIGGRAPH Asia 2024 - 17th ACM SIGGRAPH Conference and Exhibition on Computer Graphics and Interactive Techniques in AsiaTokyo, JapanACM2024, 1-12HALDOI
12.2 Publications of the year
International journals
International peer-reviewed conferences
Reports & preprints

