Section: New Results
Computational Physiology
Tumor Growth Modeling
Participants : Erin Stretton [Correspondant] , Emmanuel Mandonnet, Bjoern Menze, Hervé Delingette, Nicholas Ayache.
This work was funded by Care4me, EU program.
DTI, MRI, simulation, clinical, tumor, brain, glioma
We aim at developing image analysis methods and biophysical models in order to guide the planning of therapies (surgical removal and radiotherapy) for brain cancer (glioma) patients. Our work is focused on those objectives :
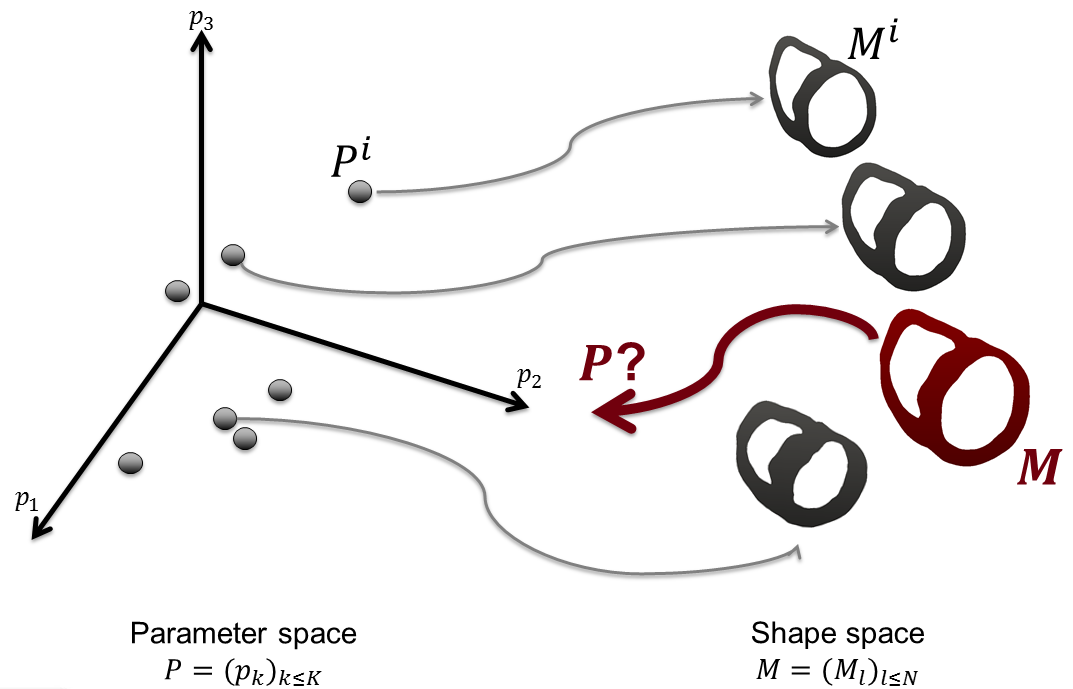
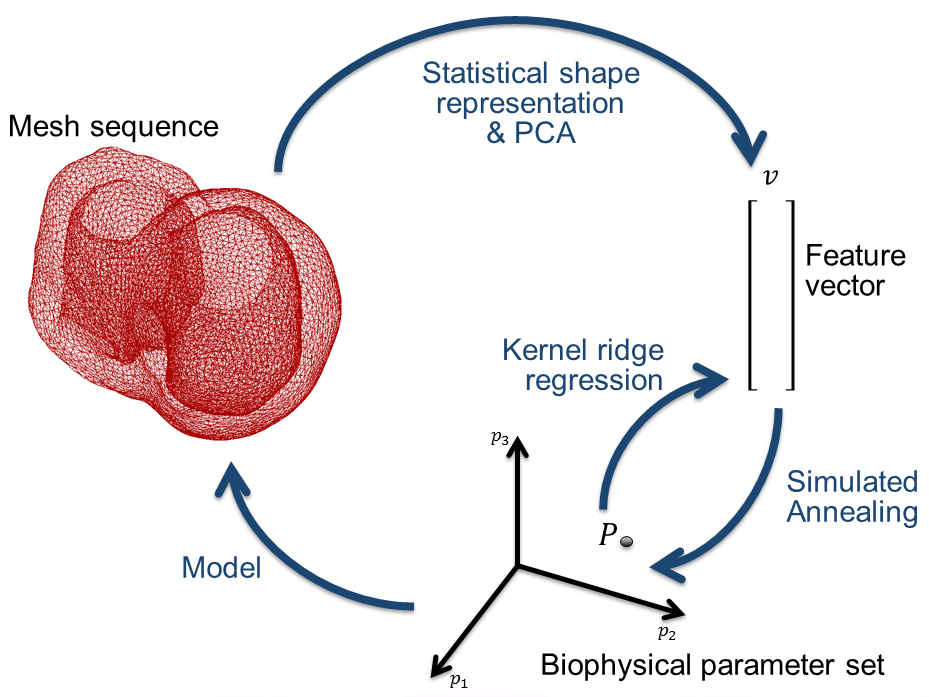
Predicting the location of glioma recurrence after a resection surgery [50] .
Determining the best description of tumor cell diffusion tensor in white matter (patient-based, atlas-based or isotropic) which leads to the most accurate results for predicting future tumor growth.
Comparing tumor growth speeds on 3 patient cases. This is a work in progress and the objective is to reach 30 patients when the work is complete.
Generation of Synthetic but Visually Realistic Time Series of Cardiac Images Combining a Biophysical Model and Clinical Images
Participants : Adityo Prakosa [Correspondant] , Maxime Sermesant, Hervé Delingette, Stéphanie Marchesseau, Eric Saloux [CHU Caen] , Pascal Allain [Philips Healthcare] , Nicolas Villain [Philips Healthcare] , Nicholas Ayache.
This work was done in collaboration with Medisys, Philips Healthcare Suresnes, France, and the Cardiology Department of CHU Caen, France. This work was partially supported by the European Research Council through the ERC Advanced Grant MedYMA on Biophysical Modelling and Analysis of Dynamic Medical Images and the European project euHeart.
synthetic 4D cardiac sequences, cardiac electromechanical model, non-rigid registration
A pipeline to create visually realistic synthetic 4D cardiac sequence using the cardiac motion simulated by an electromechanical model is developed. This pipeline combines the simulated myocardium displacement field with the estimated myocardium displacement field from a registration method. This combined displacement field is then used to warp the original images in order to create the synthetic cardiac sequence.
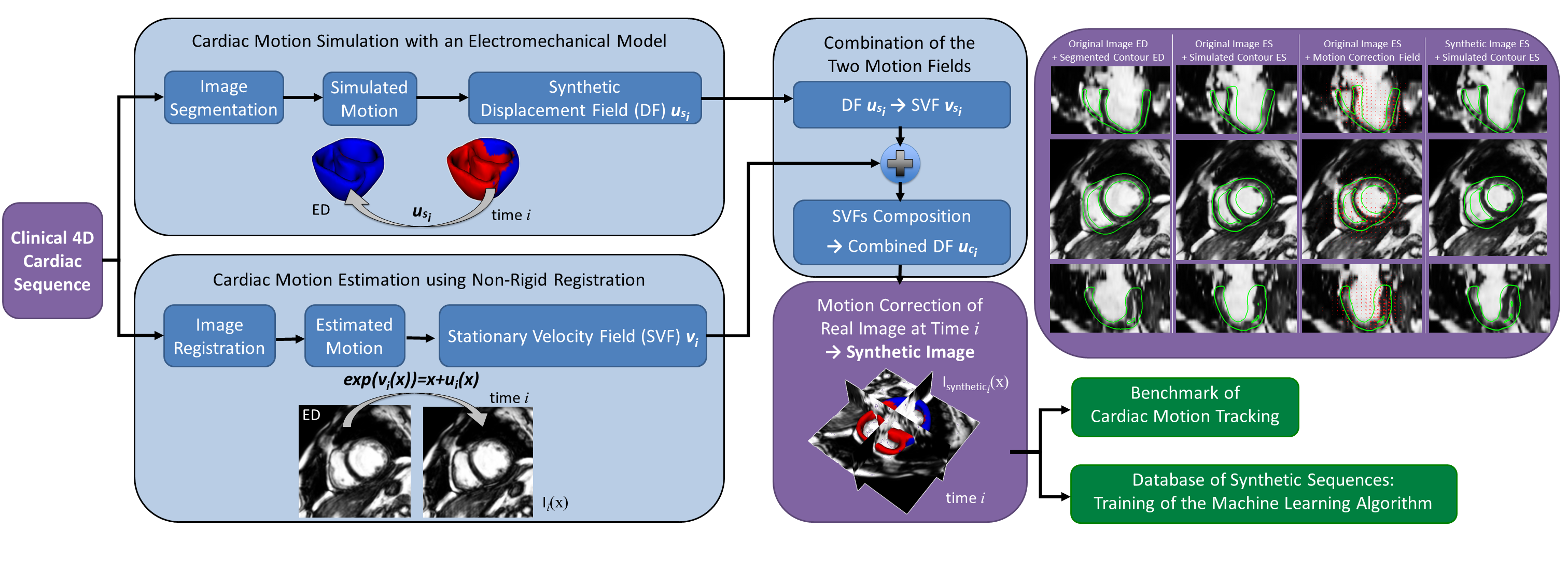
In [20] ,we proposed a new approach based on Stationary Velocity Fields to combine the two motions (see Figure 8 ). We also proposed a new method that diffuses displacement fields in order to maintain the continuity between the simulation and the real image with minimal texture distortion. Thanks to the detailed interplay between image processing and biophysical modeling, we can fully use a complete sequence in order to generate several new ones. This method also gives better realism compared to traditional methods based on the deformation of an end-diastolic image, since the generated synthetic sequence will also contain the motion of surrounding tissues such as the motion of the mitral valve.
The new synthetic images are similar to the original ones except for the motion of the heart which is modified to follow the motion provided by a biophysical model. The parameters of the biophysical model can be modified to create variations around this motion. This pipeline has been applied to generate different synthetic sequences from different imaging modalities. It is generic and can be used with a different biophysical model or a different image registration algorithm, and it can be extended to other organs.
As these synthetic 4D cardiac sequences have kinematic ground truth information, those sequences represent in themselves a valuable resource to benchmark motion tracking methods or to train machine-learning algorithm.
|
Real-Time Cardiac Electrophysiology Computing for Training Simulator
Participants : Hugo Talbot [Correspondant] , Hervé Delingette, Stephane Cotin, Maxime Sermesant, Christian Duriez.
This work was funded by the ADT Sofa and is conducted in collaboration with project teams Shacra and Evasion.
Cardiac electrophysiology simulation, real-time, GPU computing, patient-specific study
Cardiac arrhythmia is a very frequent pathology related to an abnormal electrical activity in the myocardium. This work aims at developing a training simulator for interventional radiology and thermo-ablation of these arrhythmias.
The latest improvements lead on electrophysiology simulation (see Figure 9 ) using GPU computing allowed us to reach real-time performance[52] . The issue of fast electrophysiology was a major bottleneck in the development of our simulator.
Coupling between the cardiac electrophysiology model with cardiac mechanical models has been achieved, thus leading to an interactive framework. Moreover, the electrophysiology simulation has been also coupled with a navigation simulation.
In the context of his work on cardiac electrophysiology, we initiated two different collaborations. Joint work has been performed with the team CARMEN from Inria Bordeaux on bidomain modeling for cardiac electrophysiology. This exchange targeted the implementation in SOFA of these models. Secondly, a collaboration with the MACS team in Saclay has been initiated to personalize the cardiac electrophysiology model based on the Verdandi library.
Personalized model of the heart for cardiac therapy planning
Participants : Stéphanie Marchesseau [Correspondant] , Hervé Delingette, Nicholas Ayache, Maxime Sermesant.
An award has been won for this work at the MICCAI 2012 Conference. It was partially funded by the European Community's euHeart project under grant agreement 224495 and by the ERC advanced Grant MedYMA 291080.
Cardiac simulation, sensitivity analysis, calibration algorithm, specificity study
We implemented the full Bestel-Clement-Sorine electromechanical model of the heart in SOFA [55] , [52] .
We ran a complete sensitivity analysis to check its behaviour for healthy and pathological cases [16] .
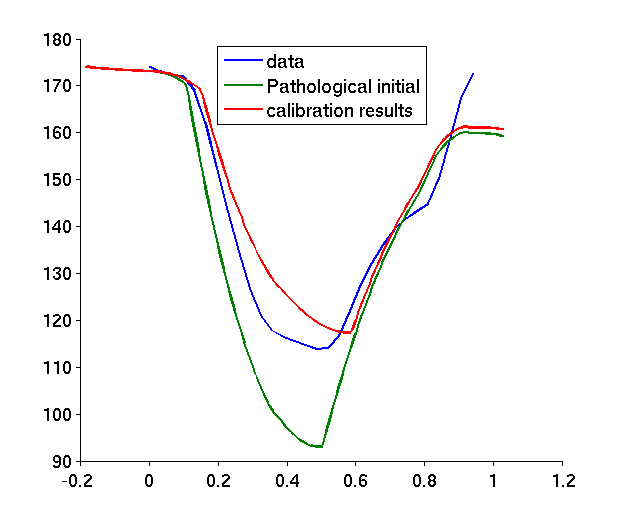
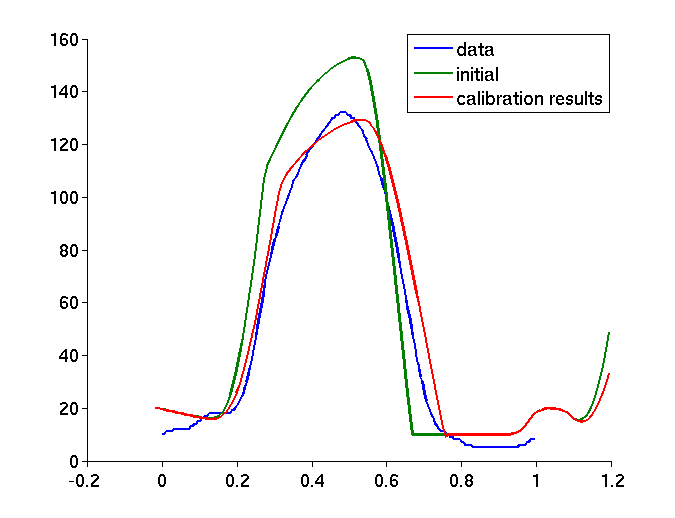
A new calibration algorithm was proposed [16] in order to initialize global mechanical parameters from the volume and pressure curves, before further personalization (see Figure 10 ).
The application of this new method on 6 healthy and 2 pathological cases allowed to draw preliminary conclusions on specific parameters to a given pathology [43] , [17] .
The model has also been used to create synthetic images in [20] and for the data of the STACOM 2012 challenge [33] .
Image-based glioma modeling for radiotherapy planning
Participants : Bjoern Menze [Correspondant] , Ender Konukoglu [MSR Cambridge] , Jan Unkelbach [Harvard MGH] .
Implemented the generative tumor segmentation model together with the E. Konukoglu's tumor infiltration model for evaluation at the MGH Department of Radiation Oncology.
Integrated tumor infiltration model with radiation therapy model.
Cardiac Arrhythmia Radio-frequency Ablation Planning
Participants : Rocio Cabrera Lozoya [Correspondant] , Maxime Sermesant, Hervé Delingette, Nicholas Ayache.
This work is performed in the context of the the PhD of Rocio Cabrera Lozoya in collaboration with the IHU LIRYC Bordeaux and is funded by ERC MedYMA.
Biophysical model development for the prediction of radio frequency ablation sites for ventricular tachycardias.
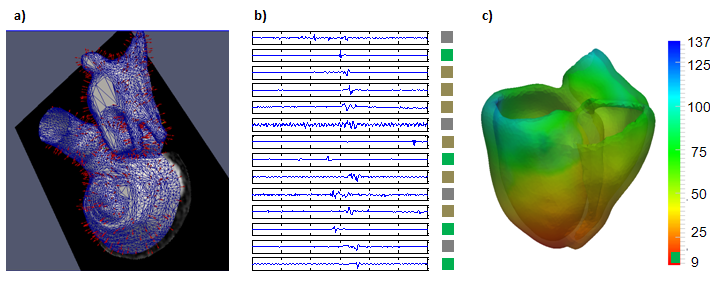
Structural and functional characterization of target sites using 3D imaging and EP measurements through machine learning algorithms (see Figure 11 ).
Computational modeling of radiofrequency ablation for the planning and guidance of abdominal tumor treatment
Participants : Chloe Audigier [Correspondant] , Herve Delingette, Tommaso Mansi [Siemens Corporate Research] , Nicholas Ayache.
This PhD is carried out between Asclepios research group, Inria Sophia Antipolis, France and the Image Analytics and Informatics global field, Siemens Corporate Research, Princeton, USA.
Therapy planning, radio-frequecy ablation, Liver
The objective of this work is to develop a computational framework for patient-specific planning of radiofrequency ablation:
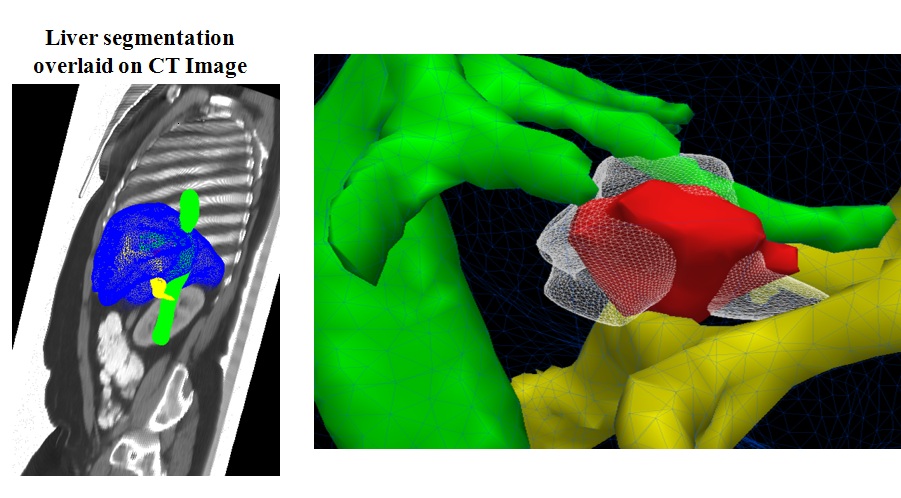
A patient-specific detailed anatomical model of the liver is estimated from standard CT image and meshed to generate a tetrahedral volume mesh.
A porous media model is used to compute the patient-specific blood flow in the hepatic circulatory system.
Bio-heat equations have been implement in SOFA to model the heat propagation in biological tissues.
A cell death model is included to account for the cellular necrosis.
|
Tumor Growth Simulation for the creation of a database of virtual patients
Participants : Nicolas Cordier [Correspondant] , Nicholas Ayache, Hervé Delingette, Bjoern Menze, Ezequiel Geremia.
This work was funded by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain MRI, Tumor simulation.
Synthesizing multi-channel MR images with healthy and glial tumors.
Creating a database of synthetic images for training and validating of brain tumor segmentation algorithms.
Learning approach for the Mechanical personalization of cardiac models
Participants : Loic Le Folgoc [Correspondant] , Hervé Delingette, Antonio Criminisi, Nicholas Ayache.
This work was partly funded by Microsoft Research through its PhD Scholarship Programme and by the ERC Advanced Grant MedYMA.
Inverse problem, machine learning, patient-specific, current, kinematics
A machine-learning framework for the mechanical personalization of the Bestel-Clement-Sorine model of the heart from patient-specific kinematics
The computational burden is moved to an offline stage, where the inter-subject variability in motion is captured via the statistical analysis of training samples
Towards a probabilistic framework for the personalization and therapy planning problems, to better account for significant and diverse uncertainty sources
Published at the MICCAI 2012 Workshop on Medical Computer Vision[37]
Brain Tumor Growth Modeling
Participants : Matthieu Lê [Correspondant] , Nicholas Ayache, Hervé Delingette.
Gliomas simulations, reaction-diffusion, brain tumors
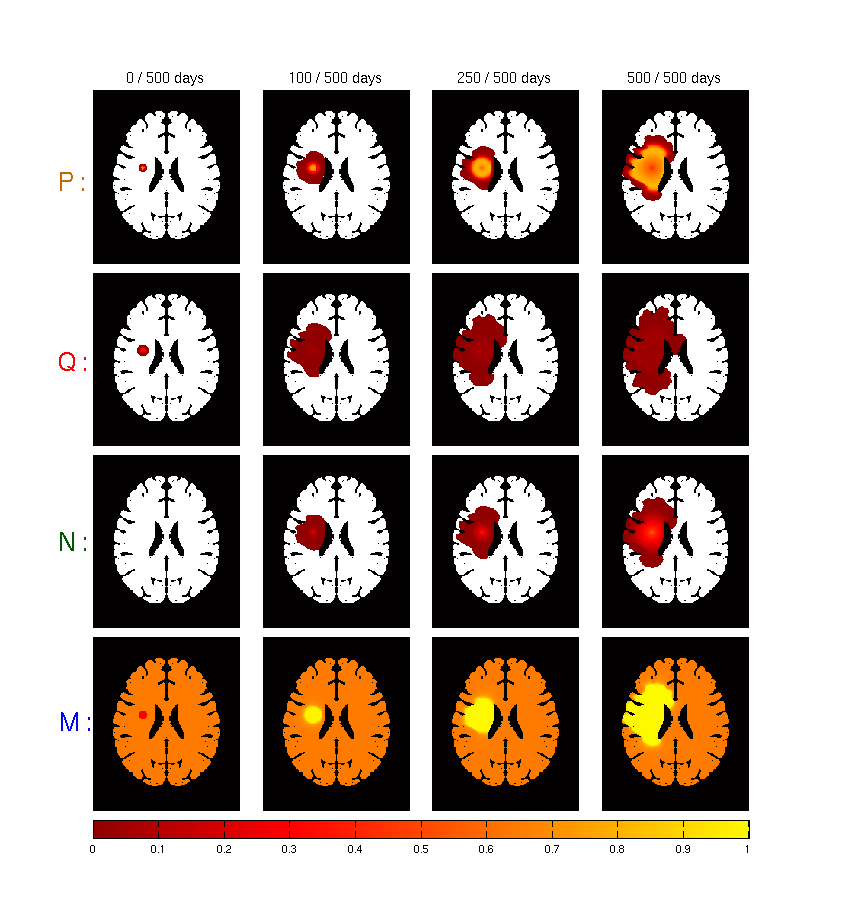
In collaboration with the MC2 research team in Bordeaux, we developed a tumor growth model based on different types of cell : necrotic, proliferative and quiescent cells (see Figure 14 ). It is also based on the underlying vascularization of the brain.
We studied the impact of the vascularization angiogenesis factor and degradation factor.
|
Modeling of atrophy of the brain in Alzheimer's Disease
Participants : Bishesh Khanal [Correspondant] , Xavier Pennec, Nicholas Ayache.
Alzheimer's Disease (AD), modeling atrophy, biomechanical model
The idea is to have a model which produces deformation of the brain when a known distribution of local volume change (atrophy) is prescribed to the model. The study is to understand how brain deformation evolve in time with respect to temporal and spatial variation of atrophy.
During the masters internship period a simple model was tested in 2D square and 3D cube where high atrophy regions acted as sinks for the displacement field [70] (see Figure 15 ).