Section: New Results
Computational Anatomy
Statistical Analysis of Transformations on Lie groups and longitudinal studies
Participants : Xavier Pennec [Correspondant] , Marco Lorenzi, Nicolas Duchateau [Hospital Clinic, Univ. Barcelona] .
Lie groups, transformations, mean value, non-linear registration
In order to perform statistics on transformations for computational anatomy purposes, we investigate alternative theoretical structures to the right- (or left-) invariant Riemannian setting usually used.
In order to define a notion of a mean which is consistent with Lie group operations we propose in [58] to replace the Riemannian metric by an affine connection structure on the group. We show that the canonical Cartan connections of a connected Lie group provides group geodesics including one-parameter subgroups which are completely consistent with the composition and inversion. To extend statistical operations to such a non-metric structure, we propose an implicit definition as an exponential barycenter (there is no Fréchet mean like in Riemannian Manifolds) and a linearly convergent iterative fixed point algorithm to reach it. This results into naturally bi-invariant means which are unique when the dispersion of the data is small enough. In some cases including rotations and rigid-body transformations, there is even a global existence and uniqueness theorem which is similar to the Riemannian case.
In [15] , we investigate the canonical Cartan connections and their associated parallel transport for diffeomorphisms, which justifies the use of one-parameter subgroups (the flow of stationary velocity fields or SVF) for diffeomorphic image registration. In particular, we derive closed-forms for different parallel transports and we compare SVF and LDDMM approaches with experiments on longitudinal and inter-subject registration.
In [34] , we analyses with practical expriments what kind of parallel transport is needed to reorient the deformation characteristics along the time sequences of the cardiac motion. Contrarily to the case of the brain, inter-subject transformations to normalize the heart between different subjects are of the same order than deformations along the sequence.
Statistical Analysis of Longitudinal Transformations in the LDDMM framework
Participants : Stanley Durrleman [Correspondant] , Xavier Pennec, Alain Trouvé [CMLA, ENS Cachan] , Nicholas Ayache, José Braga [UMR 5288 CNRS-Université Toulouse Paul Sabatier] .
Lie groups, transformations, mean value, non-linear registration
The work initiated the previous years with the PhD of S. Durrleman on the spatio-temporal modeling of shapes was applied with J. Braga to quantify ontogenetic differences between bonobo (Pan paniscus) and chimpanzee (Pan troglodytes) endocrania, using dental development as a timeline. We perform a temporal surface regression that estimates typical endocranial ontogenetic trajectories separately for bonobos and chimpanzees which highlights non-linear patterns of endocranial ontogenetic change and significant differences between species at local anatomical levels rather than considering the endocranium as a uniform entity. The decomposition of the spatio-temporal inter-species difference into a morphological deformation (accounting for size and shape differences independently of age) and a time warp (accounting for changes in the dynamics of development) indicates that juvenile bonobos develop much slower than juvenile chimpanzees, suggesting that inter-specific ontogenetic shifts do not only concern endocranial volume increase, but also the rate of shape changes over time. Our method provides, for the first time, a quantitative estimation of inter-specific ontogenetic shifts that appear to differentiate non-linearly. This work was pusblished in the journal of human evolution [10] .
The Kernel Bundle Framework for Diffeomorphic Image Registration
Participants : Xavier Pennec [Correspondant] , Stefan Sommer [Computer Science Dpt, University of Copenhagen, DK] , François Lauze [Computer Science Dpt, University of Copenhagen, DK] , Mads Nielsen [Computer Science Dpt, University of Copenhagen, DK] .
This work in collaboration with the Computer Science Department of the University of Copenhagen (DK) was initiated during the 6 month visit of S. Sommer at Asclepios in 2010-2011 and was continued remotely since then.
non-rigid registration algorithm, statistics, deformations, shapes, locally affine deformations, sparsity
In order to detect small-scale deformations during longitudinal registration while allowing large-scale deformation needed for inter-subject normalization, we wish to model deformation at multiple scales and represent the deformation at the relevant scales only. We combined in [49] , [27] a sparsity prior with the multi-scale Kernel Bundle framework, resulting in an algorithm allowing compact representation of deformation across scales.
In [28] , we further extend the framework by introducing higher-order momentum distributions in the LDDMM registration framework. While the zeroth order moments previously used in LDDMM only describe local displacement, the first-order momenta that are proposed here represent a basis that allows local description of affine transformations. Beyond the careful mathematical construction, we show the implications for sparse image registration and we provide examples of how the parametrization enables registration with a very low number of parameters.
Spectral Correspondances in Non-linear Image Registration
Participants : Xavier Pennec [Correspondant] , Hervé Lombaert, Nicholas Ayache, Leo Grady [SCR, Princeton, US] , Farida Cheriet [Saint-Justine Hospital, Montreal, CA] .
This work was performed in collaboration with Saint-Justine Hospital in Montreal (CA) and Siemens Corporate Research in Princeton (US).
non-rigid registration algorithm
The demons algorithm was enhanced to include spectral feature correspondences between the images [38] . This feature proves to drastically enhance the robustness of the registration algorithm, which turns out to have a major impact on the construction of atlases. This work was awarded the best paper award at the Medical Computer Vision Workshop [39] and was protected by a patent filling in the US [63]
Longitudinal Analysis of Brain Atrophy in Alzheimer's Disease
Participants : Marco Lorenzi [Correspondant] , Xavier Pennec, Nicholas Ayache, Giovanni B. Frisoni [IRCCS San Giovanni di Dio Fatebenefratelli, Brescia, Italy] .
This work is done in collaboration with LENITEM, IRCCS San Giovanni di Dio Fatebenefratelli, Brescia, Italy.
Alzheimer's Disease, non-rigid registration algorithm, longitudinal analysis.
The accurate analysis of the longitudinal structural changes in the brain plays a central role in the study of Alzheimer's disease (AD), for diagnostic purposes and for the assessment of the drugs efficacy in clinical trials. The goal of this project is to provide robust and effective instruments based on non-rigid registration of serial MR images for the modeling and the quantification of the brain atrophy evolution in AD. In 2012, our main scientific developments were the following:
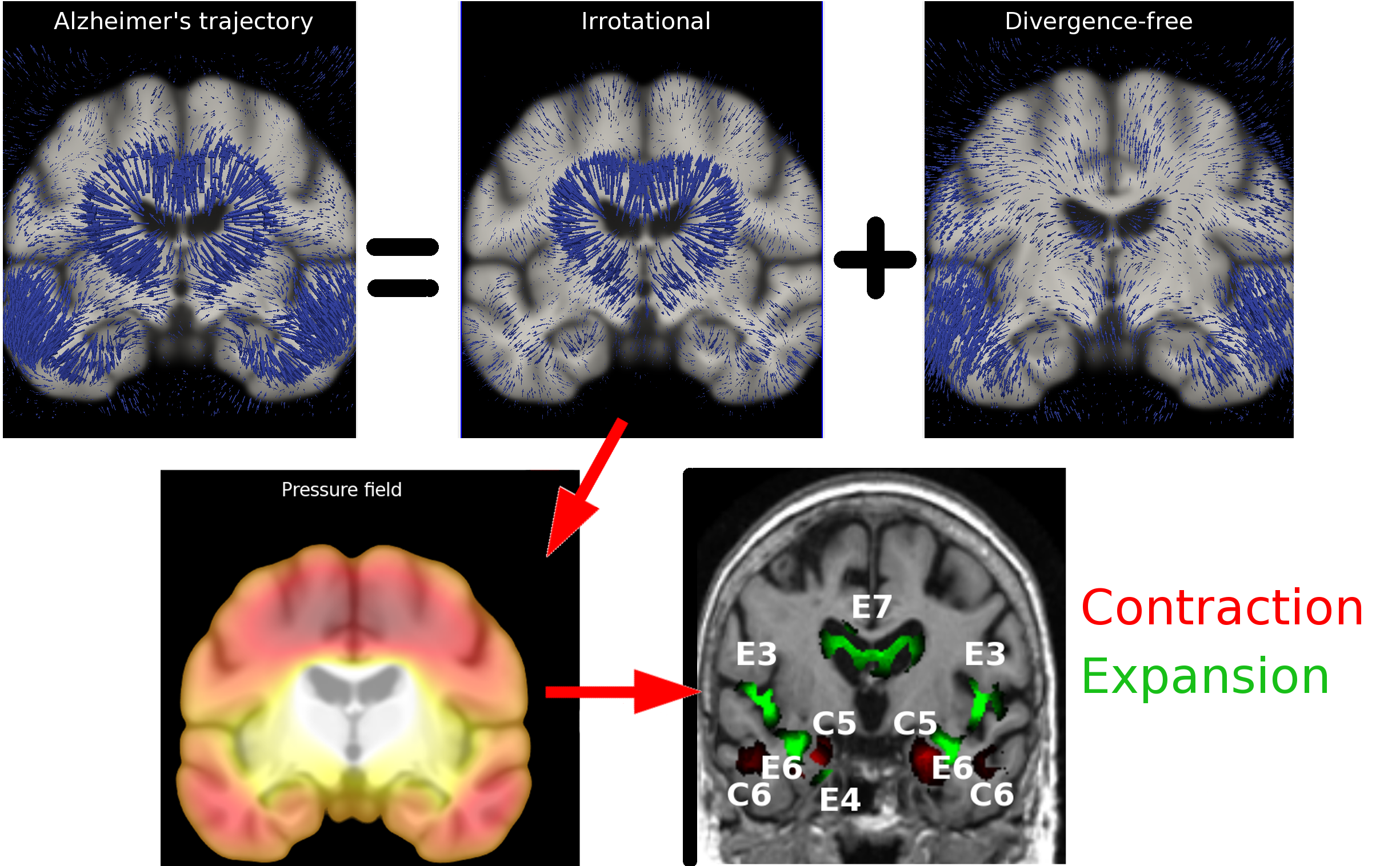
We developed a framework for the consistent definition of anatomical regions of longitudinal brain atrophy, and for the robust quantification of longitudinal regional percentage volume loss. The framework is based on the analysis of the flux associated to longitudinal deformations (see Figure 4 ), and was successfully applied to large public dataset of brain images (ADNI - http://adni.loni.ucla.edu/ ). The work was accepted for oral podium presentation at the MICCAI conference 2012 [41] .
We applied the flux analysis for the quantification of the longitudinal hippocampal and ventricular atrophy in AD. The proposed framework was presented at the NIBAD MICCAI Challenge 2012 [42] , and compared favorably with state-of-art methods in terms of accuracy and stability when applied on the challenge dataset.
We proposed in [40] a model of the morphological changes in Alzheimer's based on the disentangling of the normal aging component from the pathological atrophy. The model was promoted and presented to the neuroscience community during international scientific conferences [72] , [71] .
These scientific advances were also included along with the previous ones in the PhD manuscript [1] .
|
Statistical Modelling of Cardiac Growth, Deformation and Blood Flow from Medical Images
Participants : Kristin McLeod [Correspondant] , Adityo Prakosa, Christof Seiler, Maxime Sermesant, Xavier Pennec.
This work was partially funded by the EU project Care4me ITEA2.
Image registration, Demons algorithm, LDDMM, reduced models, CFD, polyaffine, cardiac motion tracking
This work involves developing reduced models of cardiac motion, blood flow and growth.
Extending the 2011 motion tracking challenge [44] , the iLogDemons registration algorithm was applied this year to a data-set of synthetic echocardiography sequences with a training set (provided with ground truth) and testing set to quantitavely compare this algorithms with other cardiac motion tracking algorithms [46] .
A reduced order model of cardiac motion based on a polyaffine log-demons registration was developed to represent the motion along the cardiac cycle with a smaller number of parameters compared to previously proposed methods. The method was applied to a data-set of 10 volunteers and the results were presented at the 2012 STACOM workshop at the MICCAI conference [45] .
The analysis of a statistical model for reduced blood flow simulations in the pulmonary artery proposed in the 2010 STACOM workshop is currently being extended to a journal version with an improved method and a larger data-set.
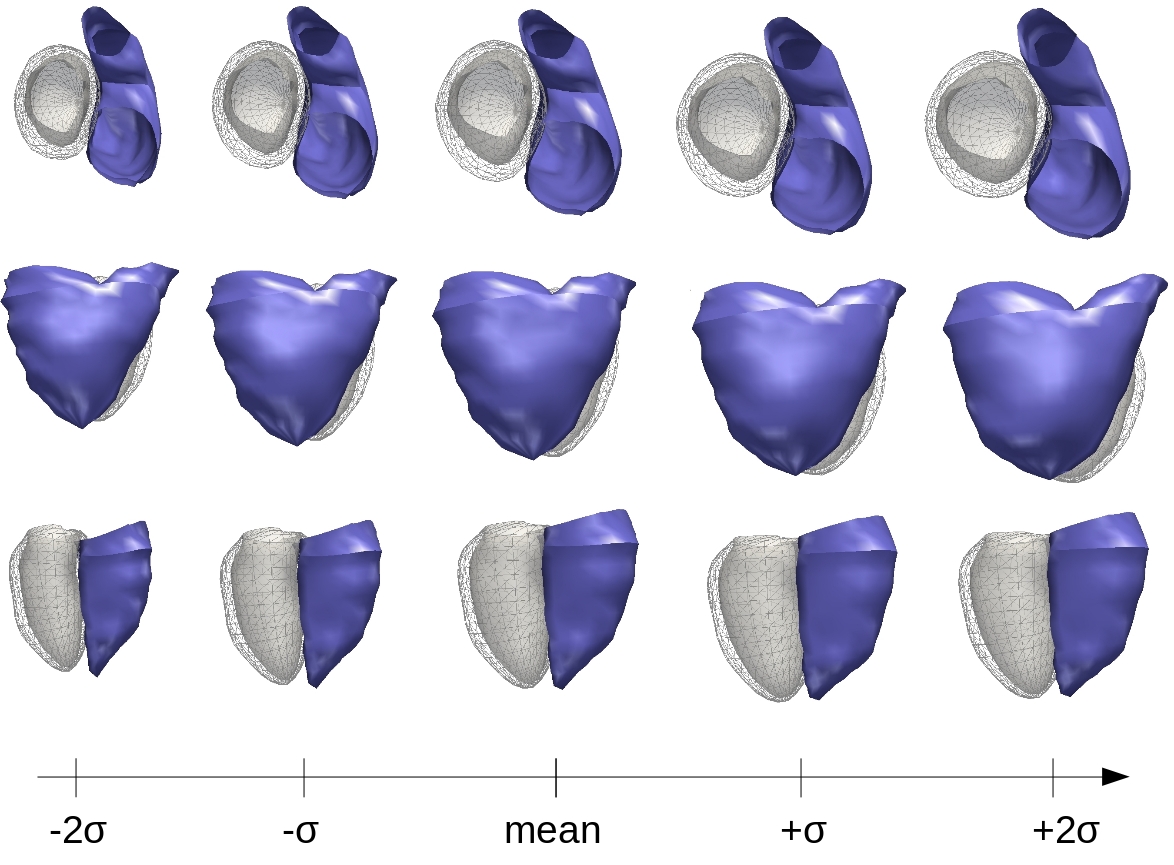
The statistical modeling of the right ventricle growth in a population of Tetralogy of Fallot patients was extended to a full bi-ventricular growth model on different data [56] . Results confirm the previous findings which were shown to be useful in providing insights for patient treatment [13] (see Figure 5 ).
|
Trees on Geometrical Deformations to Model the Statistical Variability of Organs in Medical Images
Participants : Christof Seiler [Correspondant] , Xavier Pennec, Mauricio Reyes [Institute for Surgical Technology and Biomechanics, University of Bern, Switzerland] .
This work is performed in the context of the joint PhD of Christof Seiler at the Institute for Surgical Technology and Biomechanics, University of Bern, Switzerland and Asclepios Inria [3] .
Parametrization of diffeomorphisms, Shape statistics, Multiscale and hierarchical trees, Log-Euclidean polyaffine transformations, Polyaffine registration, Log-Demons registration, Generative statistical model, Bayesian registration, Mandibles, Femurs
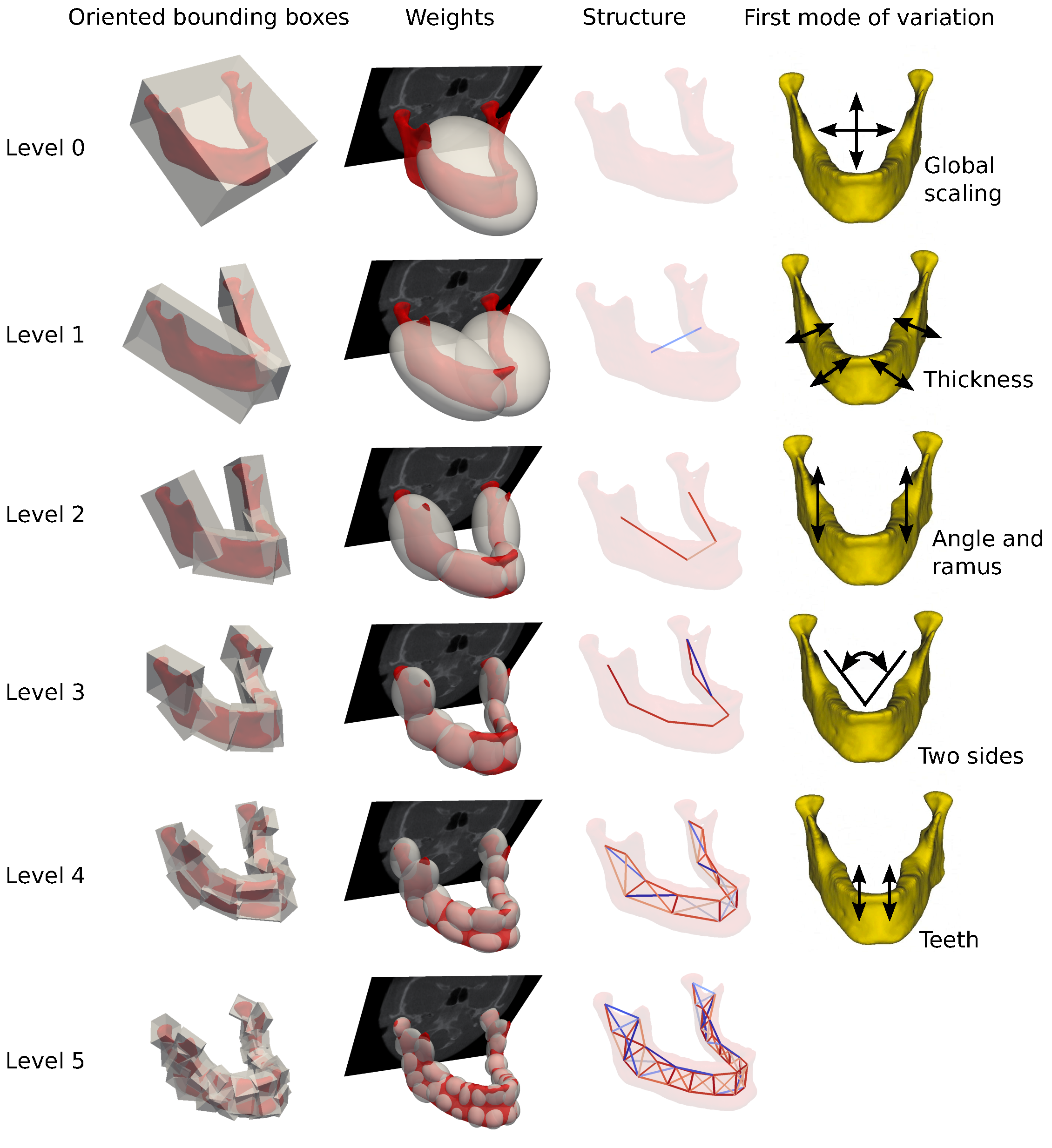
Intersubject anatomical deformations between patients can be found on coarse and fine scales. Each level of granularity has specific regions of interest in clinical applications. The challenge is to connect geometrical deformations to clinical regions across scales.
We presented this connection by introducing structured diffeomorphic registration [25] . At the core of our method is the parametrization of geometrical deformations with trees of locally affine transformations describing intersubject variability across scales (see Figure 6 ).
The methodology of [25] was successfully applied to mandible implant design [32] and in a clinical journal paper on allograft selection [22] .
We statistically modeled the deformation parameters in a population by formulating a generative statistical model [48] . This model allowed us to incorporate deformation statistics as a prior in a Bayesian setting and it enabled us to extend the classical sequential coarse to fine registration to a simultaneous optimization of all scales.
We explored cell shape statistics to classify stem cells [24] .
We investigated the benefits of considering patient metadata and morphometric measures to enhance bone surface shape prediction [6] .
|
Evaluation of iLogDemons Algorithm for Cardiac Motion Tracking in Synthetic Ultrasound Sequence
Participants : Adityo Prakosa [Correspondant] , Kristin McLeod, Maxime Sermesant, Xavier Pennec.
This work was partially funded by the European Research Council (ERC) through the support of the MedYMA advanced grant 291080 and the European project euHeart.
synthetic echocardiography, iLogDemons, cardiac motion tracking
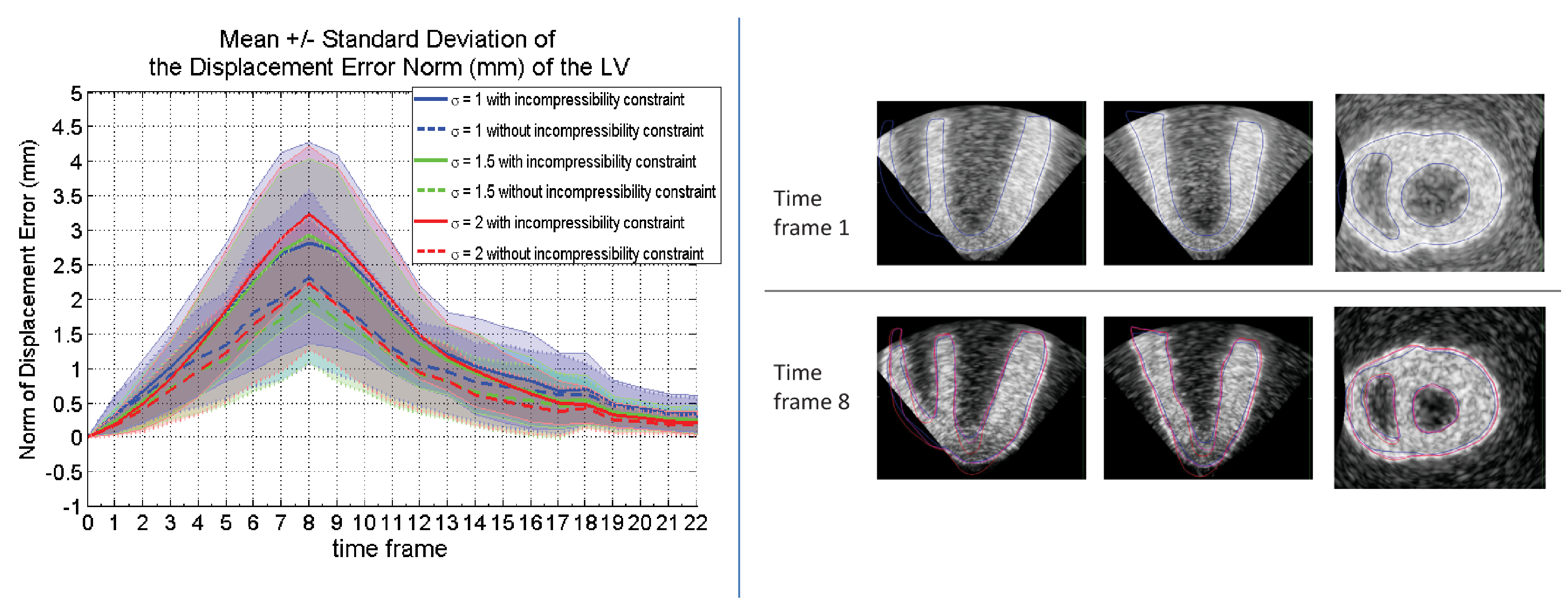
The LogDemons and iLogDemons non-linear registration algorithms were evaluated on a dataset of synthetic cardiac ultrasound sequences [33] , [46] . With these synthetic sequences, it is possible to quantify the performance of these registration algorithms since the ground truth motion was given. Therefore the LogDemons/ iLogDemons can be evaluated objectively (see Figure 7 ).
|
Simulation of Atrophy in Alzheimer's disease
Participants : Arnaud Le Carvennec [Correspondant] , Sebastien Ourselin [UCL] , Nick Fox [UCL] , Xavier Pennec [Inria] , Nicholas Ayache [Inria] .
Thesis in collaboration between Asclepios team at Inria and Center for Medical Image Computing (CMIC)-Dementia Research Center (DRC) at University College London (UCL).
Simulation, Alzheimer's disease, registration