Section:
Research Program
Grammatical inference and highly expressive structures
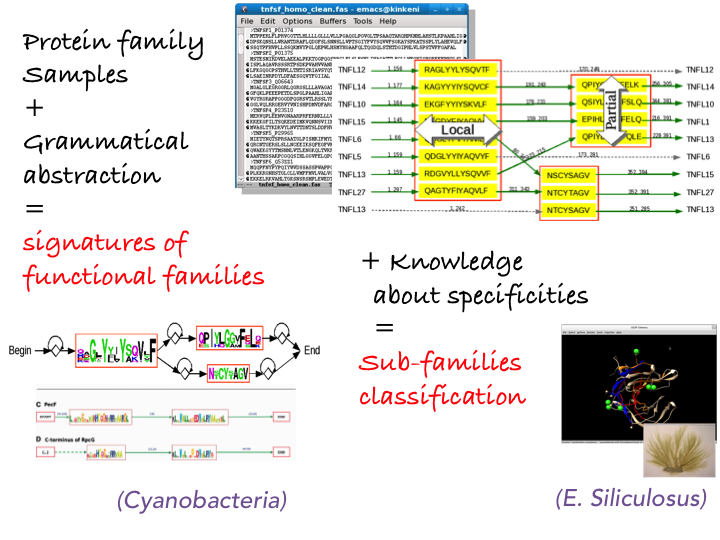
Our main field of expertise in machine learning concerns grammatical models with a strong expertise in finite state automata learning. By introducing a similar fragment merging heuristic approach, we have proposed an algorithm that learns successfully automata modeling families of (non homologous) functional families of proteins [3] , leading to a tool named Protomata-learner. As an example, this tools allows us to properly model the function of the protein family TNF, which is impossible with other existing probabilistic-based approach (see Fig. 3 ). It was also applied to model families of proteins in cyanobacteria [2] . Our future goal is to further demonstrate the relevance of formal language modelling by addressing the question of enzyme prediction, from their genomic or protein sequences, aiming at better sensitivity and specificity. As enzyme-substrate interactions are very specific central relations for integrated genome/metabolome studies and are characterized by faint signatures, we shall rely on models for active
sites involved in cellular regulation or catalysis mechanisms. This requires to build models gathering both structural and sequence information is order to describe (potentially nested or crossing) long-term dependencies such as contacts of amino-acids that are far in the sequence but close in the 3D protein folding. We wish to extend our expertise towards inferring Context-Free Grammars including the topological information coming from the structural characterization of active sites.
Figure
3. Protomata Learner workflow. Starting from a set of protein sequences, a partial local alignment is computed and an automaton is inferred, which can be considered as a signature of the family of proteins.
This allows searching for new members of the family [2] . Adding further information about the specific properties of proteins within the family allows to exhibit a refined classification.
|
|
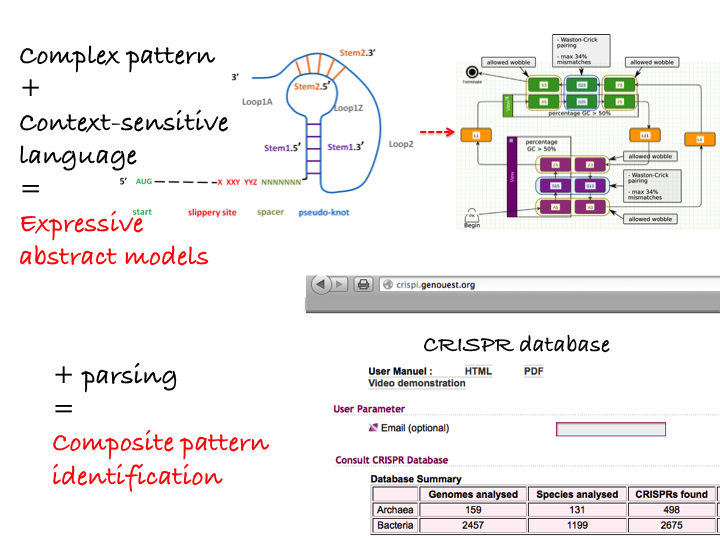
Using context-free grammars instead of regular patterns increases the complexity of parsing issues. Indeed, efficient parsing tools have been developed to identify patterns within genomes but most of them are restricted to simple regular patterns. Definite Clause Grammars (DCG), a particular form of logical context-free grammars have been used in various works to model DNA sequence features [54] . An extended formalism, String Variable Grammars (SVGs), introduces variables that can be associated to a string during a pattern search (see Fig. 4 ) [59] , [58] . This increases the expressivity of the formalism towards midly context sensitive grammars. Thus, those grammars model not only DNA/RNA sequence features but also structural features such as repeats, palindromes, stem/loop or pseudo-knots. We have designed a tool, STAN (suffix-tree analyser) which makes it possible to search for a subset of SVG patterns in full chromosome sequences [7] . This tool was used for the recognition of transposable elements in Arabidopsis thaliana [60] or for the design of a CRISPR database [9] . See Figure 4 for illustration.
Our goal is to extend the framework of STAN. Generally, a suitable language for the search of particular components in languages has to meet several needs : expressing existing structures in a compact way, using existing databases of motifs, helping the description of interacting components. In other words, the difficulty is to find a good tradeoff between expressivity and complexity to allow the specification of realistic models at genome scale. In this direction, we are working on Logol, a language and framework based on a systematic introduction of constraints on string variables.
Figure
4. Graphical modeling of a pseudo-knot (RNA structure) based on the expressivity of String Variable Grammars used in the Logol framework. Combined with
parsers, this leads to composite pattern identification such as CRISPR [56] .
|
|