Section: New Results
Background estimation and vesicle segmentation in live cell imaging
Participants : Thierry Pécot, Patrick Bouthemy, Charles Kervrann.
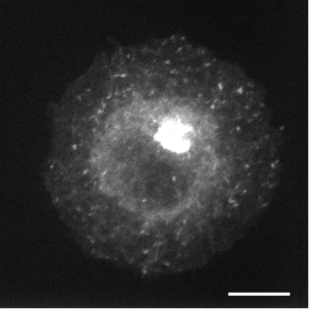
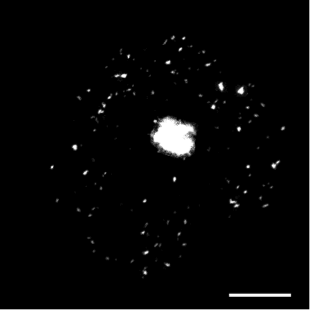
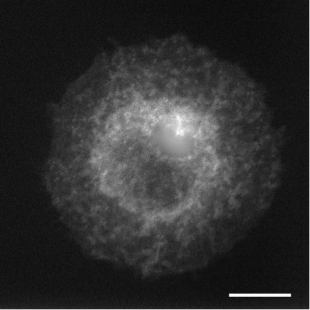
In live cell fluorescence microscopy images, the moving tagged structures of interest, such as vesicles, often appear as bright spots with intensity that varies along time over a time-varying and cluttered background. Localization and morphology assessment of these small objects over time is then crucial to provide valuable information for quantitative traffic analysis. In this study, we have focused on the Rab6 protein as a typical intracellular membrane-associated protein. Rab6 is known to promote vesicle trafficking from Golgi to Endoplasmic Reticulum or to plasma membrane. In our study, micro-fabricated patterns have been used to enforce cells to have circular or crossbow normalized shape. Micro-patterns impose constraints on the cytoskeleton and the location of organelles (e.g. Golgi apparatus) is thus better controlled. These micro-patterns also influence the spatial distribution of Rab6 transport carriers. However, the direct influence of the micro-patterns on the spatial dissemination of these trafficking vesicles has so far not been completely characterized. In this work, we have considered a statistical Bayesian approach in the framework of conditional random fields (CRF) for background estimation and vesicle segmentation [13] . Within this approach, we have designed a robust detection measure for fluorescence microscopy based on the distribution of neighbor patch similarity. We formulate the vesicle segmentation and background estimation as a global energy minimization problem. An iterative scheme to jointly segment vesicles and background is proposed for 2D-3D fluorescence image sequences. We have conducted a quantitative comparison with state-of-the-art methods on a large set of synthetic image sequences with a cluttered time-varying background and achieved a quantitative validation of the vesicle segmentation method on 2D and 3D micro-patterned cells expressing GFP-Rab6.
Reference: [13]
Collaborators: Jean Salamero (UMR 144 CNRS-Institut Curie, STED team and PICT-IBiSA)
Jérôme Boulanger (UMR 144 CNRS-Institut Curie, STED team)