Section: Research Program
Natural and engineered control of growth and gene expression
Participants : Cindy Gomez Balderas, Eugenio Cinquemani, Johannes Geiselmann [Correspondent] , Nils Giordano, Hidde de Jong, Stéphan Lacour, Delphine Ropers, Alberto Soria-Lopéz.
The adaptation of bacterial physiology to changes in the environment, involving changes in the growth rate and a reorganization of gene expression, is fundamentally a resource allocation problem. It notably poses the question how microorganisms redistribute their protein synthesis capacity over different cellular functions when confronted with an environmental challenge. Assuming that resource allocation in microorganisms has been optimized through evolution, for example to allow maximal growth in a variety of environments, this question can be fruitfully formulated as an optimal control problem. We have developed such an optimal control perspective, focusing on the dynamical adaptation of growth and gene expression in response to envrionmental changes, in close collaboration with the BIOCORE project-team.
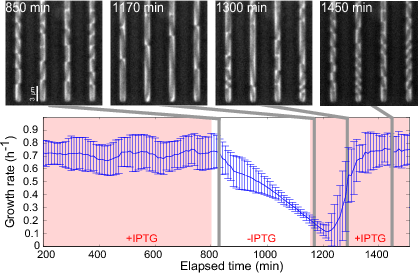
A complementary perspective consists in the use of control-theoretical approaches to modify the functioning of a bacterial cell towards a user-defined objective, by rewiring and selectively perturbing its regulatory networks. The question how regulatory networks in microorganisms can be externally controlled using engineering approaches has a long history in biotechnology and is receiving much attention in the emerging field of synthetic biology. Within a number of on-going projects, IBIS is focusing on two different questions. The first concerns the development of open-loop and closed-loop growth-rate controllers of bacterial cells for both fundamental research and biotechnological applications (Figure 5 ). Second, we are working on the development of methods for the real-time control of gene expression. These methods are obviously capital for the above-mentioned design of growth-rate controllers, but they have also been applied in the context of a platform for real-time control of gene expression in cell population and single cells, developed by the Inria project-team LIFEWARE, in collaboration with a biophysics group at Université Paris Descartes.
|