Section: New Results
Coherent temporal extrapolation of labeled images
Participants : Gaël Michelin, Grégoire Malandain.
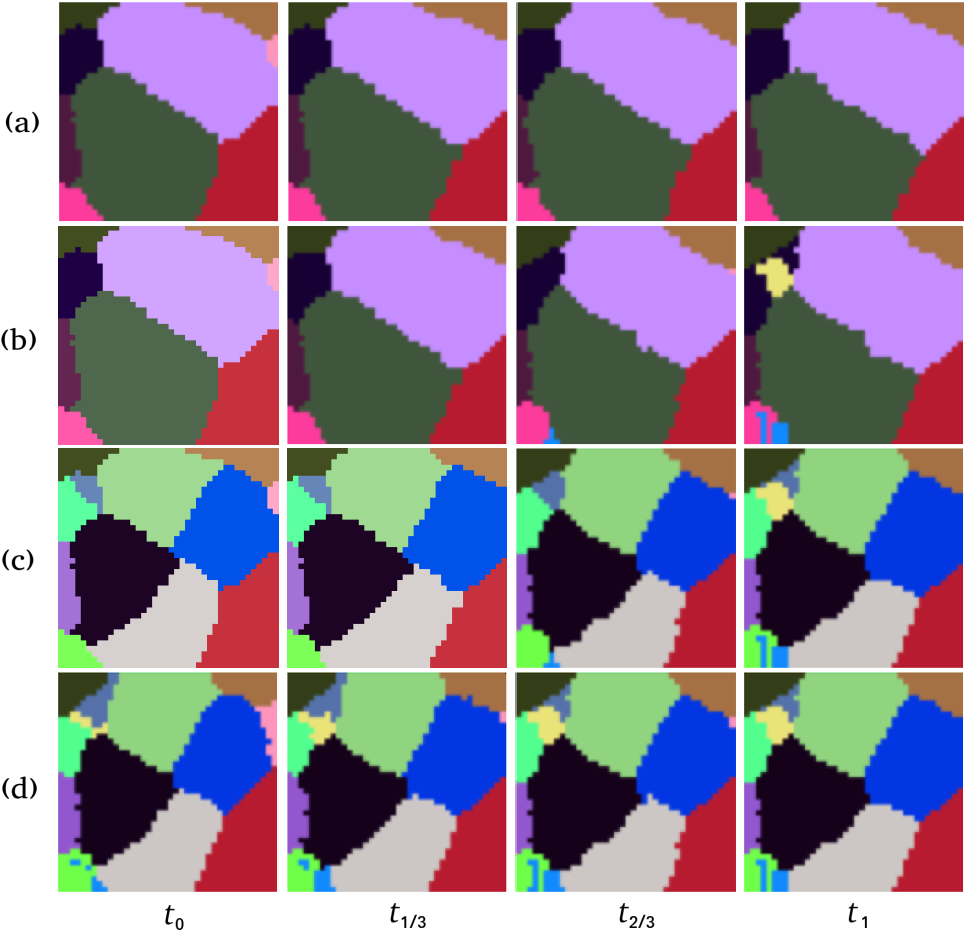
In developmental imaging, 3D+ series of microscopic images allows to follow the organism development at the cell level and has become the standard way of imaging the development of living organs. Dedicated tools for cell segmentation in 3D images as well as cell lineage calculation from 3D+ sequences have been proposed to analyze these data. For some applications (such as section 5.9), it may be desirable to interpolate images at intermediary time-points. However, the known methods do not allow to locally handle the topological changes (ie cell. division).
In the present work, we propose an extrapolation method that coherently deformed the images to be interpolated so that to guarantee a topological continuity of borders (see figure 10).
|