Section: New Results
Computational Anatomy
Statistical Learning of Heterogeneous Data in Large-Scale Clinical Databases
Participants : Clement Abi Nader [Correspondant] , Nicholas Ayache, Philippe Robert, Marco Lorenzi.
Gaussian Process, Alzheimer's Disease, Disease Progression Modelling
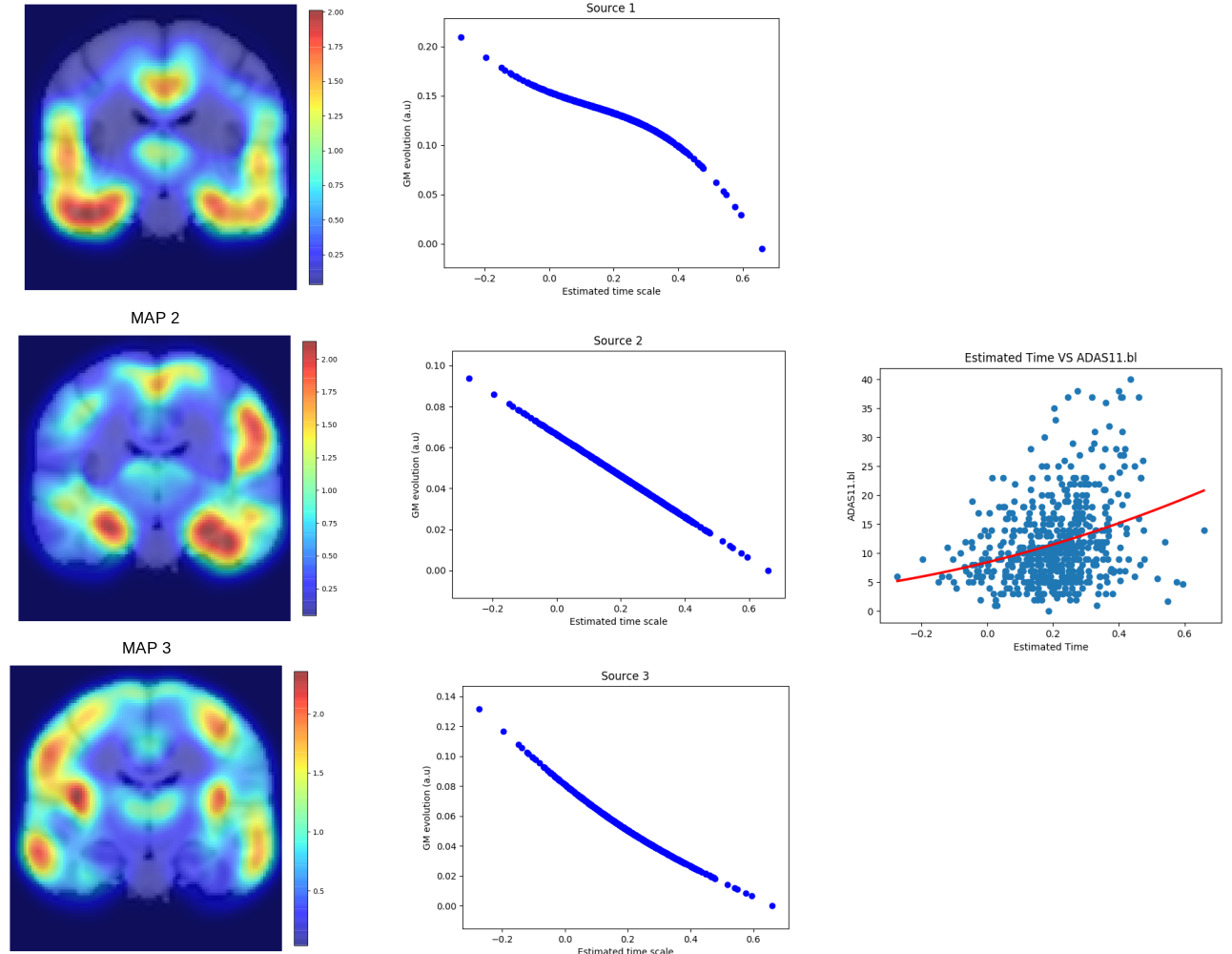
The aim of this project is to develop a spatio-temporal model of Alzheimer's Disease (AD) progession [47]. We assume that the brain progression is characterized by independent spatio-temporal sources that we want to separate. We estimate brain structures involved in the disease progression at different resolutions thus dealing with the non-stationarity of medical images, while assigning to each of them a monotonic temporal progression using monotonic Gaussian processes (Figure 13, left-middle panel). We also compute an individual time-shift parameter to assess the disease stage of each subject (Figure 13, right panel).
|
A model of brain morphological evolution
Participants : Raphaël Sivera [Correspondant] , Hervé Delingette, Marco Lorenzi, Xavier Pennec, Nicholas Ayache.
Longitudinal modeling, Deformation framework, Brain morphology, Alzheimer's disease, Aging.
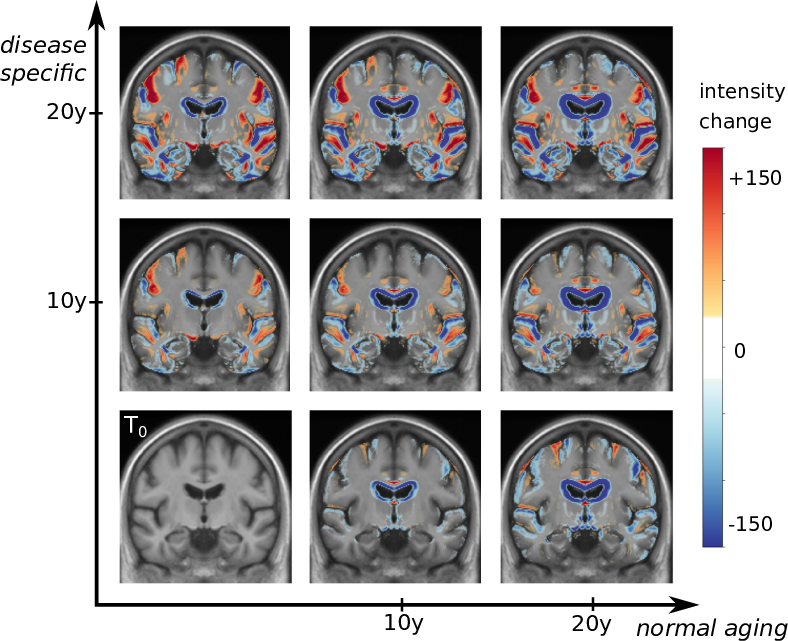
We proposed a deformation-based generative model of the brain morphological evolution that can jointly describes the effect of aging and Alzheimer's disease. It relies on longitudinal description of the aging and disease consequences and can be use to compute image-based cross-sectional progression markers. This approach is able to propose a description of the disease evolution, population and subject-wise(see Figure 14) and open the way to a better modeling of the disease progression.
|
Geometric statistics
Participant : Xavier Pennec [Correspondant] .
This work is partially funded by the ERC-Adv G-Statistics.
Statistics on manifolds, Differential geometry,
Beyond the mean value, Principal Component Analysis (PCA) is often used to describe the main modes of variability and to create low dimensional models of the data. Generalizing these tools to manifolds is a difficult problem. In order to define low dimensional parametric subspaces in manifolds, we proposed in [22] to use the locus of points that are weighted means of a number of reference points. These barycentric subspaces locally define submanifolds which can naturally be nested to provide a hierarchy of properly embedded subspaces of increasing dimension (a flag) approximating the data better and better. This defines a generalization of PCA to manifolds called Barycentric Subspace Analysis (BSA) which provides a new perspective for dimension reduction. It appears to be well suited for implicit manifolds such as the ones defined by multiple registrations in longitudinal or cross-section image analysis. An example of such an application was provided in [23] for 4D cardiac image sequences.
In classical estimation problems, the number of samples is always finite. The variability that this induces on the estimated empirical mean is a classical result of the law of large numbers in the asymptotic regime. In manifolds, it is not clear how the curvature influences the estimation of the empirical Fréchet mean with a finite number of samples. Preliminary results showed that there is an unexpected bias inversely proportional to the number of samples induced by the gradient of the curvature and a correction term of the same order on the covariance matrix slowering or accelerating the effective convergence rate towards the Fréchet mean of the underlying distribution. These preliminary results were derived using a new simple methodology that is also extending the validity from Riemannian manifolds to affine connection spaces [45].
Brain template as a Fréchet mean in quotient spaces
Participants : Nina Miolane [Correspondant] , Xavier Pennec.
Computational anatomy, Morphological brain template, Hierarchical modeling.
Geometrically, the procedure used to construct the reference anatomy for normalizing the measurements of individual subject in neuroimaging studies can be summarized as the Fréchet mean of the images projected in a quotient space. We have previously shown that this procedure is asymptotically biased, therefore inconsistent. In [15], we presented a methodology that quantifies spatially the brain template's asymptotic bias. We identify the main variables controlling the inconsistency. This leads us to investigate the topology of the template's intensity levels sets, represented by its Morse-Smale complex. We have proposed a topologically constrained adaptation of the template computation that constructs a hierarchical template with bounded bias. We apply our method to the analysis of a brain template of 136 T1 weighted MR images from the Open Access Series of Imaging Studies (OASIS) database.
Cardiac Motion Evolution Modeling from Cross-Sectional Data using Tensor Decomposition
Participants : Kristin Mcleod [Simula Research Laboratory] , Maxime Sermesant, Xavier Pennec.
Cardiac motion tracking, modeling cardiac motion evolution over time
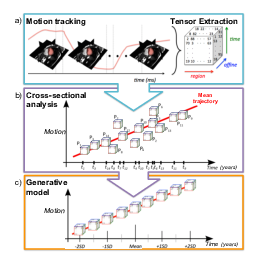
Cardiac disease can reduce the ability of the ventricles to function well enough to sustain long-term pumping efficiency. We proposed in [14] a cardiac motion tracking method to study and model cohort effects related to age with respect to cardiac function. The proposed approach makes use of a Polyaffine model for describing cardiac motion of a given subject, which gives a compact parameterisation that reliably and accurately describes the cardiac motion across populations. Using this method, a data tensor of motion parameters is extracted for a given population. The partial least squares method for higher-order arrays is used to build a model to describe the motion parameters with respect to age, from which a model of motion given age is derived. Based on cross-sectional statistical analysis with the data tensor of each subject treated as an observation along time, the left ventricular motion over time of Tetralogy of Fallot patients is analysed to understand the temporal evolution of functional abnormalities in this population compared to healthy motion dynamics (see Figure 15).
|
Challenging cardiac shape and motion statistics
Participants : Marc-Michel Rohé [correspondant] , Maxime Sermesant, Xavier Pennec.
Shape statistics, Non-rigid registration, Deep learning, Cardiac shape and motion
Two of the methods previously developed by Marc-Michel Rohé in his PhD were benchmarked against other state of the art methods in two successive MICCAI challenges. First, the SVF-net developed to perform a very-fast inter-subject heart registration based on convolutional neural networks was embedded into a multi-atlas segmentation pipeline and tested against other deep learning techniques for the automatic MRI cardiac multi-structures segmentation [2]. Second, a combination of polyaffine cardiac motion tracking and supervised learning was used to predict myocardial infarction [25]. Both challenges demonstrate the good performances of the tested methods.