Section: New Results
Imaging & Phenomics, Biostatistics
Radiomic analysis to improve diagnosis and therapy in oncology
Participants : Fanny Orlhac [Correspondant] , Nicholas Ayache, Charles Bouveyron, Jacques Darcourt [CAL] , Hervé Delingette, Olivier Humbert [CAL] , Pierre-Alexandre Mattei [Copenhague University] , Thierry Pourcher [CEA] , Fanny Vandenbos [CHU Nice] .
Inria postdoctoral fellowship for 16 months
Radiomics, Statistical learning, Metabolomics
-
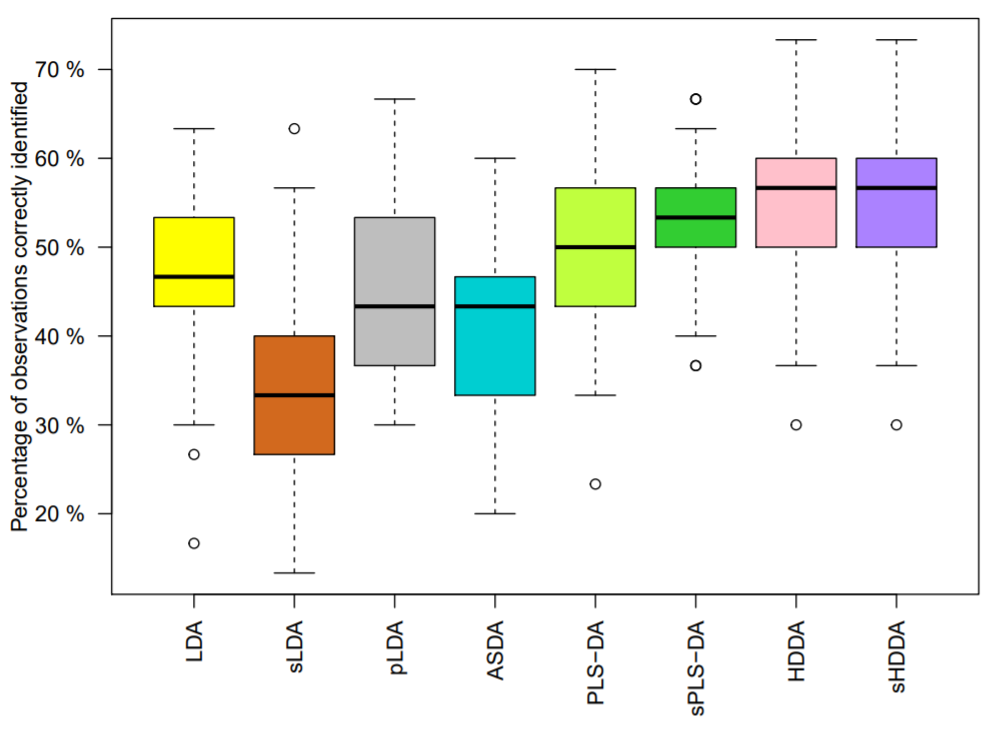
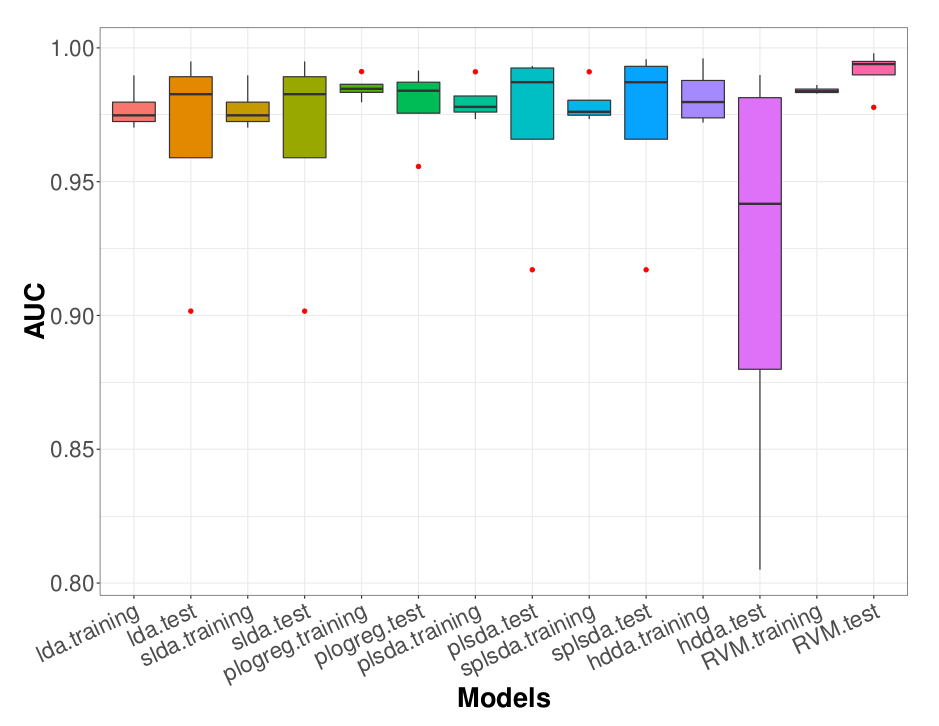
We proposed a modeling which extends the High-Dimensional Discriminant Analysis (HDDA) model by incorporating a sparsity pattern for each class, called sparse HDDA (sHDDA) [21].
-
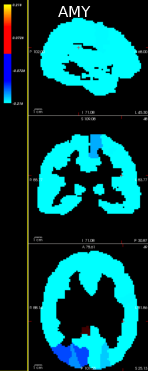
We demonstrated its efficacy in identifying lung lesions based on CT radiomic features (see Figure 8) [21] or triple-negative breast lesions from PET radiomic features and metabolomic data [34], [32], [33]. Thanks to the class-specific variable selection, the final model can be easily interpreted by physicians.
-
We also demonstrated the capacity of the ComBat method to harmonize radiomic features extracted from PET images acquired with different imaging protocols [40], [39].
|
Statistical learning on large databases of heterogeneous imaging, cognitive and behavioral data
Participants : Luigi Antelmi [Correspondent] , Marco Lorenzi, Valeria Manera, Philippe Robert, Nicholas Ayache.
Supported by the French government, through the UCAJEDI Investments in the Future project managed by the National Research Agency (ANR) ref. num. ANR-15-IDEX-01, our research is within the MNC3 initiative (Médecine Numérique: Cerveau, Cognition, Comportement), in collaboration with the Institut Claude Pompidou (CHU of Nice). Computational facilities are funded by the grant AAP Santé 06 2017-260 DGA-DSH, and by the Inria Sophia Antipolis - Méditerranée, "NEF” computation cluster.
statistical learning, joint analysis, neuroimaging
The aim of our work is to build scalable learning models for the joint analysis of heterogeneous biomedical data, to be applied to the investigation of neurological and neuropsychiatric disorders from collections of brain imaging, body sensors, biological and clinical data available in current large-scale databases such as ADNI(http://adni.loni.usc.edu/) and local clinical cohorts.
We developed a computationally efficient formulation of probabilistic latent variable models [37]. This approach is capable to highlight meaningful relationships among biomarkers in the context of Alzheimer's disease (Figure 9) that can be used to develop optimal strategies for disease quantification and prediction.
|
Joint Biological & Imaging markers for the Diagnosis of severe lung diseases
Participants : Benoît Audelan [Correspondant] , Hervé Delingette, Nicholas Ayache.
Lung cancer, Early detection, Sparse Bayesian Learning
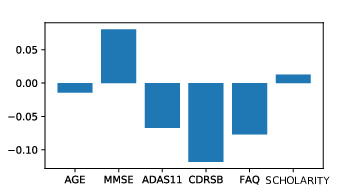
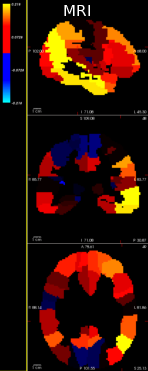
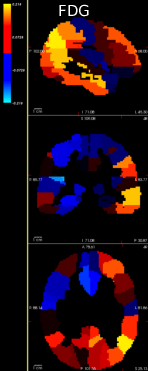
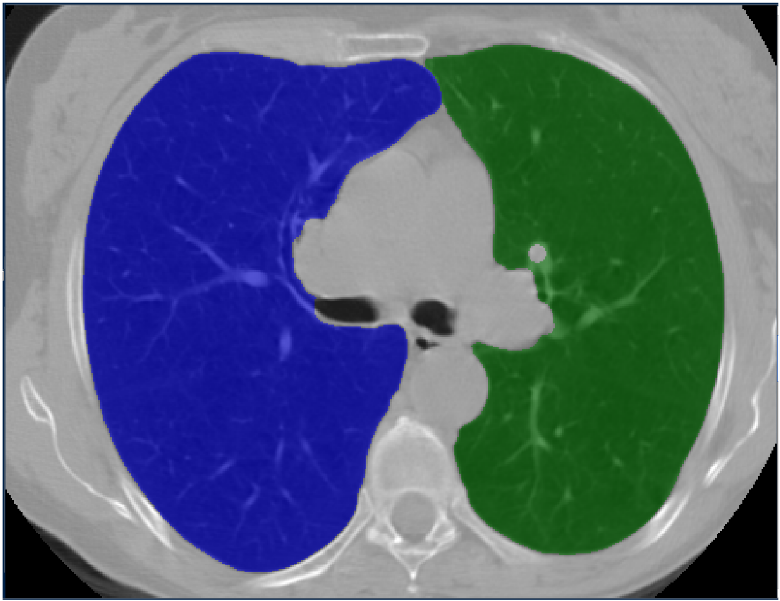
Lung cancer is among the most common cancer and is considered to be one of the most important public health problem. The aim of this work is to improve the detection of lung cancer by combining imaging and biological markers. Exploratory analysis have been conducted to discriminate cancer patients versus controls from circulating miRNAs data using sparse Bayesian learning and to automatically pre-process lung CT images (Fig. 10).
|
A data-driven model of mechanistic brain atrophy propagation in dementia
Participants : Sara Garbarino [Correspondant] , Marco Lorenzi.
Sara Garbarino acknowledges financial support from the French government managed by L’Agence Nationale de la Recherche under Investissements d’Avenir UCA JEDI (ANR-15-IDEX-01) through the project “AtroProDem: A data-driven model of mechanistic brain Atrophy Propagation in Dementia”.
Gaussian Processes, Bayesian non–parametric modelling, neuroimaging data, protein dynamics, brain network
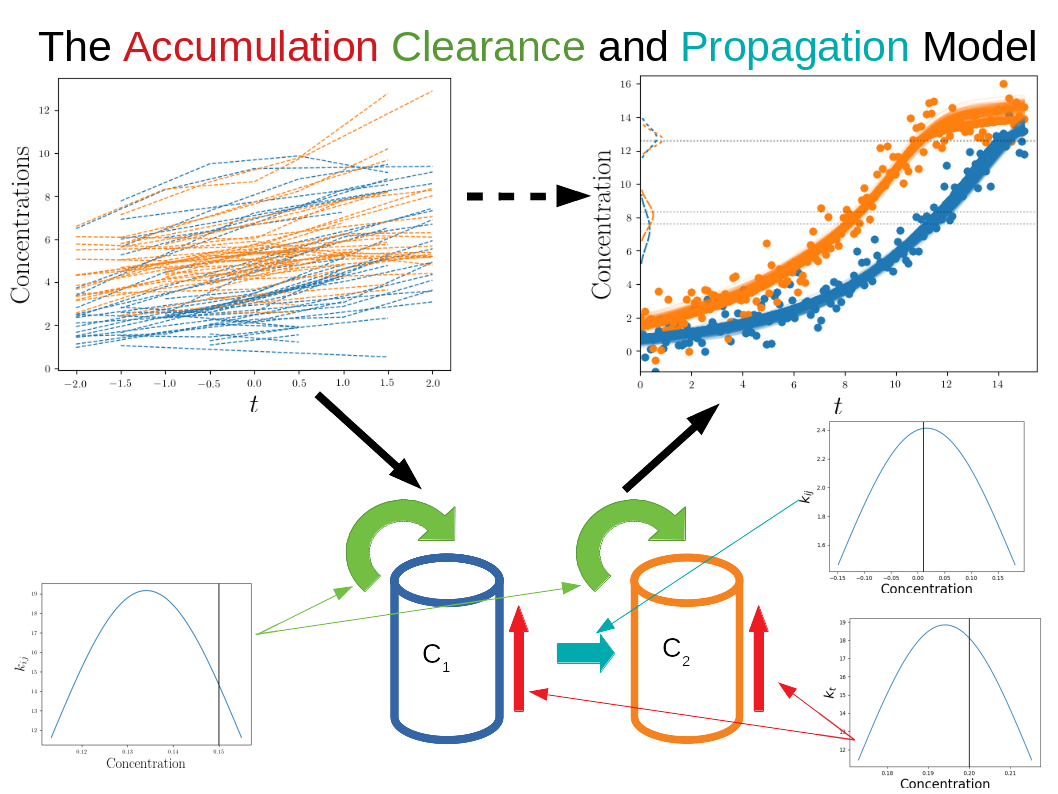
Models of misfolded proteins aim at discovering the bio-mechanical properties of neurological diseases by identifying plausible associated dynamical systems. Solving these systems along the full disease trajectory is usually challenging, due to the lack of a well defined time axis for the pathology. This issue is solved by disease progression models where long-term progression trajectories are estimated via time reparametrization of individual observations. However, due to their loose assumptions on the dynamics, they do not provide insights on the bio-mechanical properties of protein propagation.
In this project we propose a unified model of spatio-temporal protein dynamics based on the joint estimation of long-term protein dynamics and time reparameterization of individuals observations (Figure 11). The model is expressed within a Gaussian Process regression setting, where constraints on the dynamics are imposed through non–linear dynamical systems.
|
Federated Learning in Distributed Medical Databases: Meta-Analysis of Large-Scale Subcortical Brain Data
Participants : Santiago Silva [Correspondant] , Marco Lorenzi, Boris Gutman, Andre Altman, Eduardo Romero, Paul M. Thompson.
This work was supported by the French government, through the UCAJEDI Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-15-IDEX-01 (project Meta- ImaGen).
Federated learning, distributed databases, PCA, SVD, meta-analysis, brain disease.
We proposed a federated learning framework for securely accessing and meta-analyzing any biomedical data without sharing individual information.
-
A frontend pipeline for preprocessing and analyzing data was proposed, including: standardization, confounders correction, and variability analysis via federated PCA.
-
Tested on multi-centric and multi-diagnosis databases (ADNI, PPMI and UK-Biobank) showed a clear differentiation between control and Alzheimer's subjects (Figure 12).
-
Further developments of this study will extend the proposed analysis to large-scale imaging genetics data, such as in the context of the ENIGMA meta-study.