Section: New Results
Medical Image Analysis
Learning a Probabilistic Model for Diffeomorphic Registration
Participants : Julian Krebs [Correspondant] , Hervé Delingette, Tommaso Mansi [Siemens Healthineers, Princeton, NJ, USA] , Nicholas Ayache.
This work is funded by Siemens Healthineers, Princeton, NJ, USA
deformable registration, probabilistic modeling, deep learning, latent variable model, deformation transport, disease clustering
We developed a probabilistic approach for deformable image registration in 3-D using deep learning methods [30]. This method includes:
-
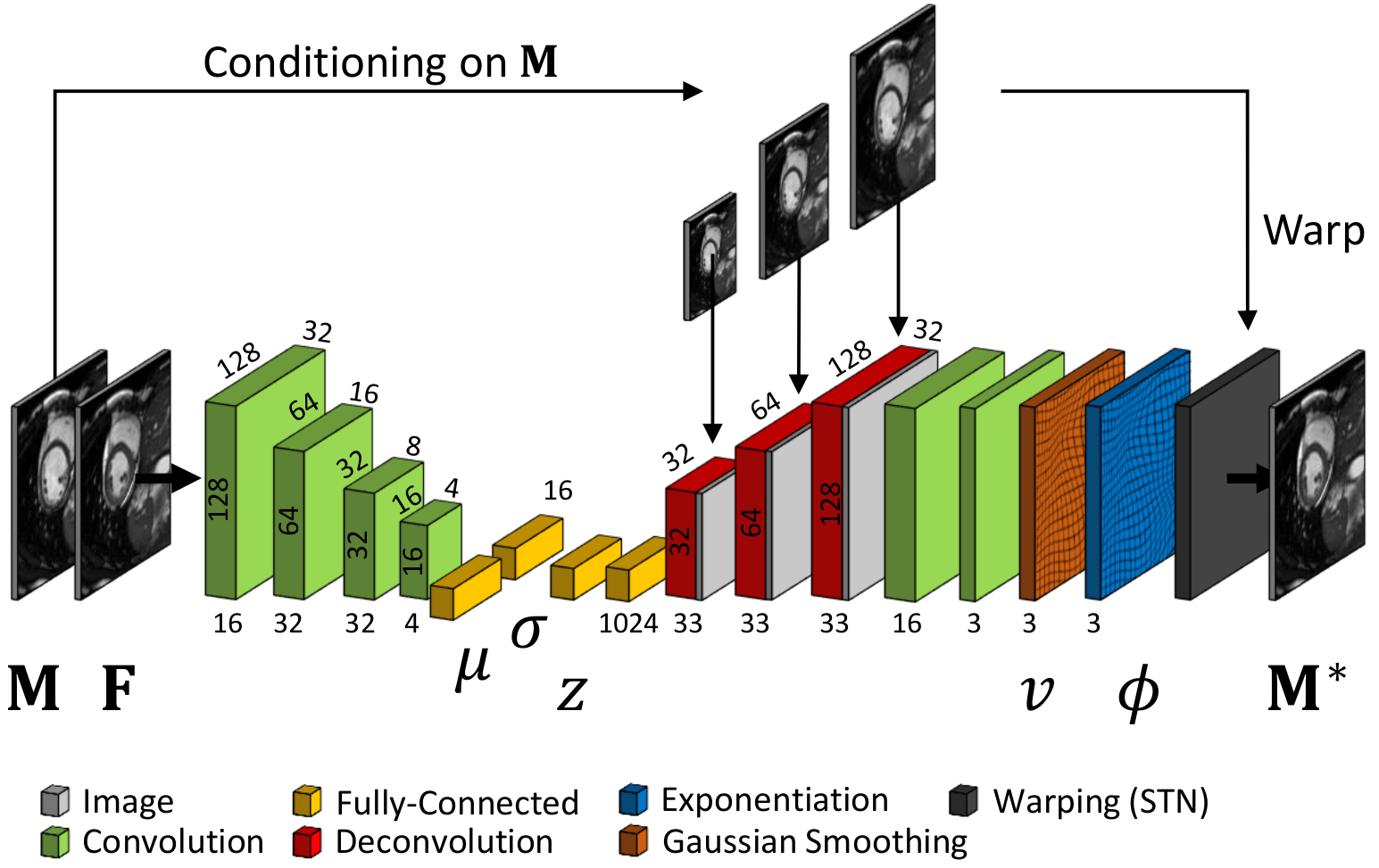
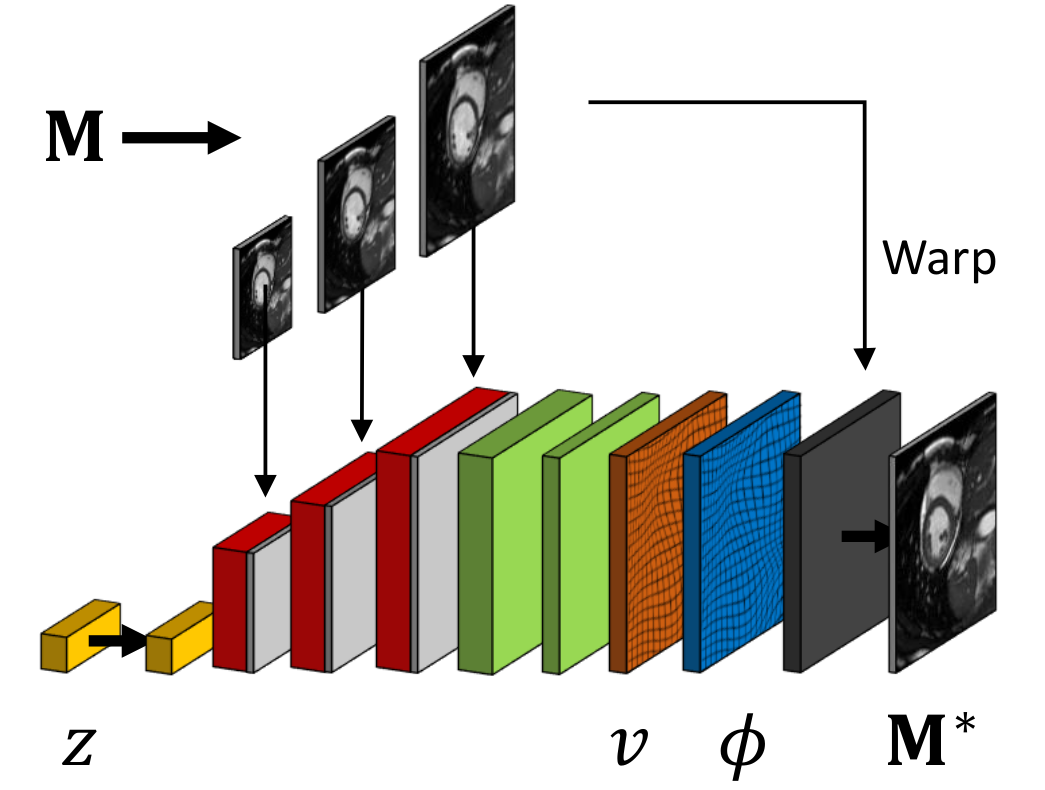
A probabilistic formulation of the registration problem through unsupervised learning of an encoded deformation model (Fig. 4).
-
A differentiable exponentiation layer and an user-adjustable smoothness layer that ensure the outputs of neural networks to be regular and diffeomorphic.
-
An analysis of size and structure of a latent variable space for registration.
-
Experiments on deformation transport and disease clustering.
|
Learning Myelin Content in Multiple Sclerosis from Multimodal MRI
Participants : Wen Wei [Correspondent] , Nicholas Ayache, Olivier Colliot [ARAMIS] .
This work is done in collaboration with the Aramis-Project team of Inria in Paris and the researchers at the Brain and Spinal Cord Institute (ICM) located in Paris.
Multiple Sclerosis, MRI, PET, GANs
-
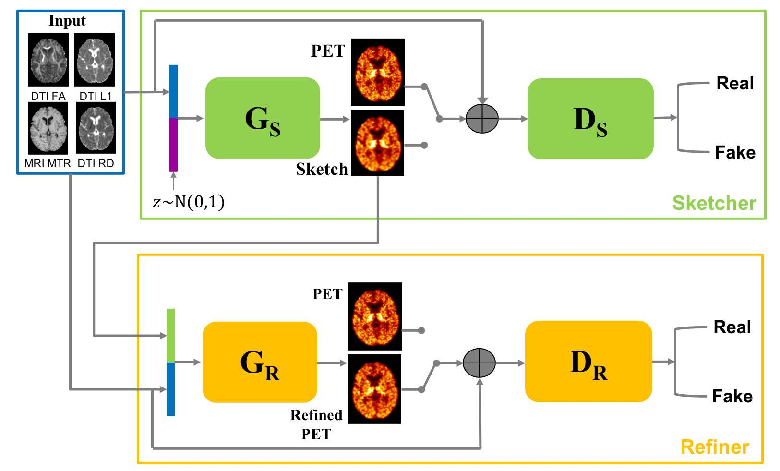
We predict myelin content from multiparametric MRI [36].
-
We design an adaptive loss and a sketch-refinement process for GANs, decomposing the problem into anatomy/physiology and myelin content prediction (Fig. 5).
|
Consistent and Robust Segmentation of Cardiac Images with Propagation
Participants : Qiao Zheng [Correspondant] , Hervé Delingette, Nicolas Duchateau, Nicholas Ayache.
This project is funded by European Research Council (MedYMA ERC-AdG-2011-291080).
Cardiac segmentation, deep learning, neural network, 3D consistency, spatial propagation
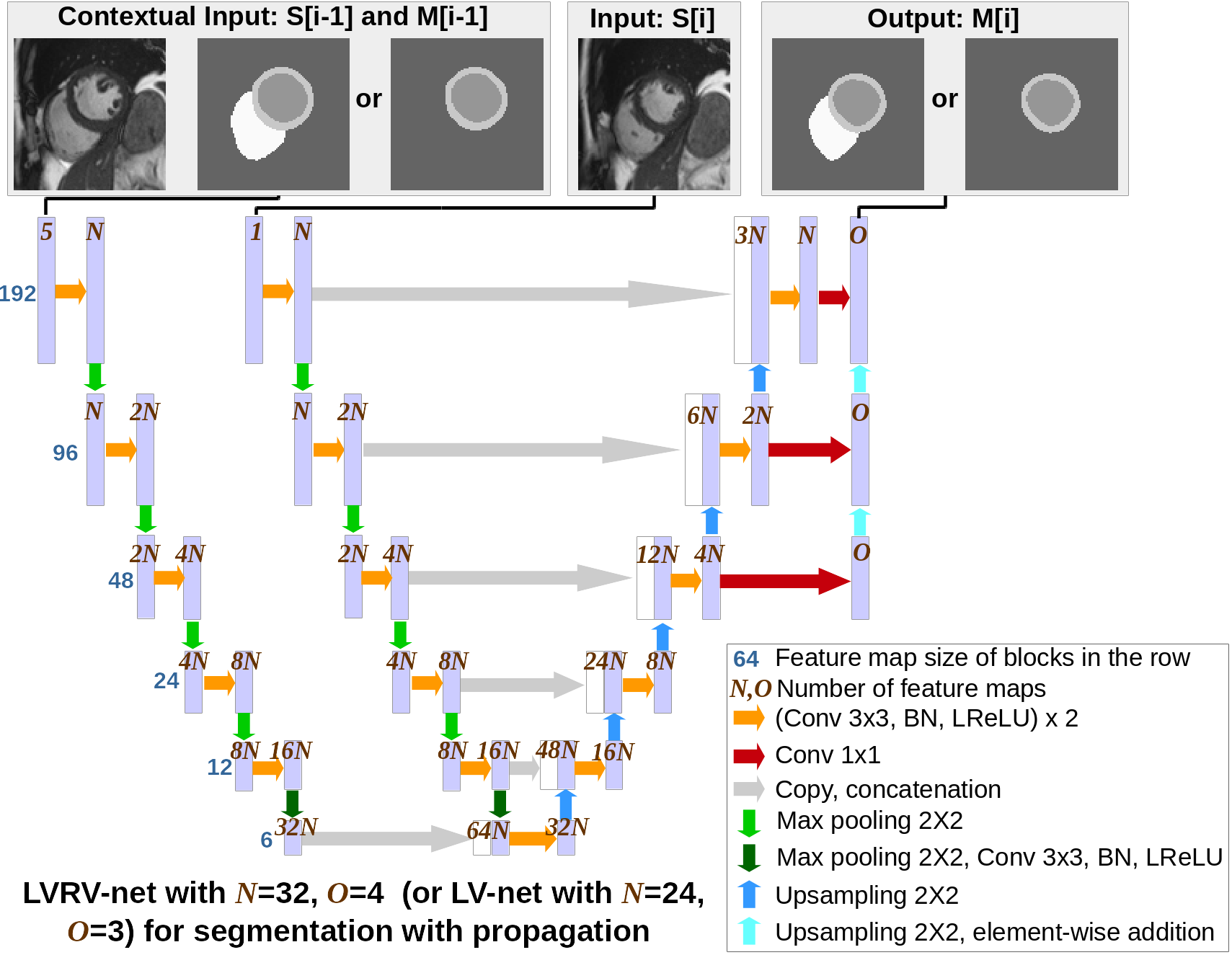
We propose a method based on deep learning to perform cardiac segmentation on short axis MRI image stacks iteratively from the top slice (around the base) to the bottom slice (around the apex) [26][62]. At each iteration, a novel variant of U-net is applied to propagate the segmentation of a slice to the adjacent slice below it (Fig. 6).
-
Robustness and generalization ability to unseen cases are demonstrated.
-
Results comparable or even better than the state-of-the-art are achieved.
The corresponding open source software, CardiacSegmentationPropagation, is available in https://team.inria.fr/epione/en/software/.
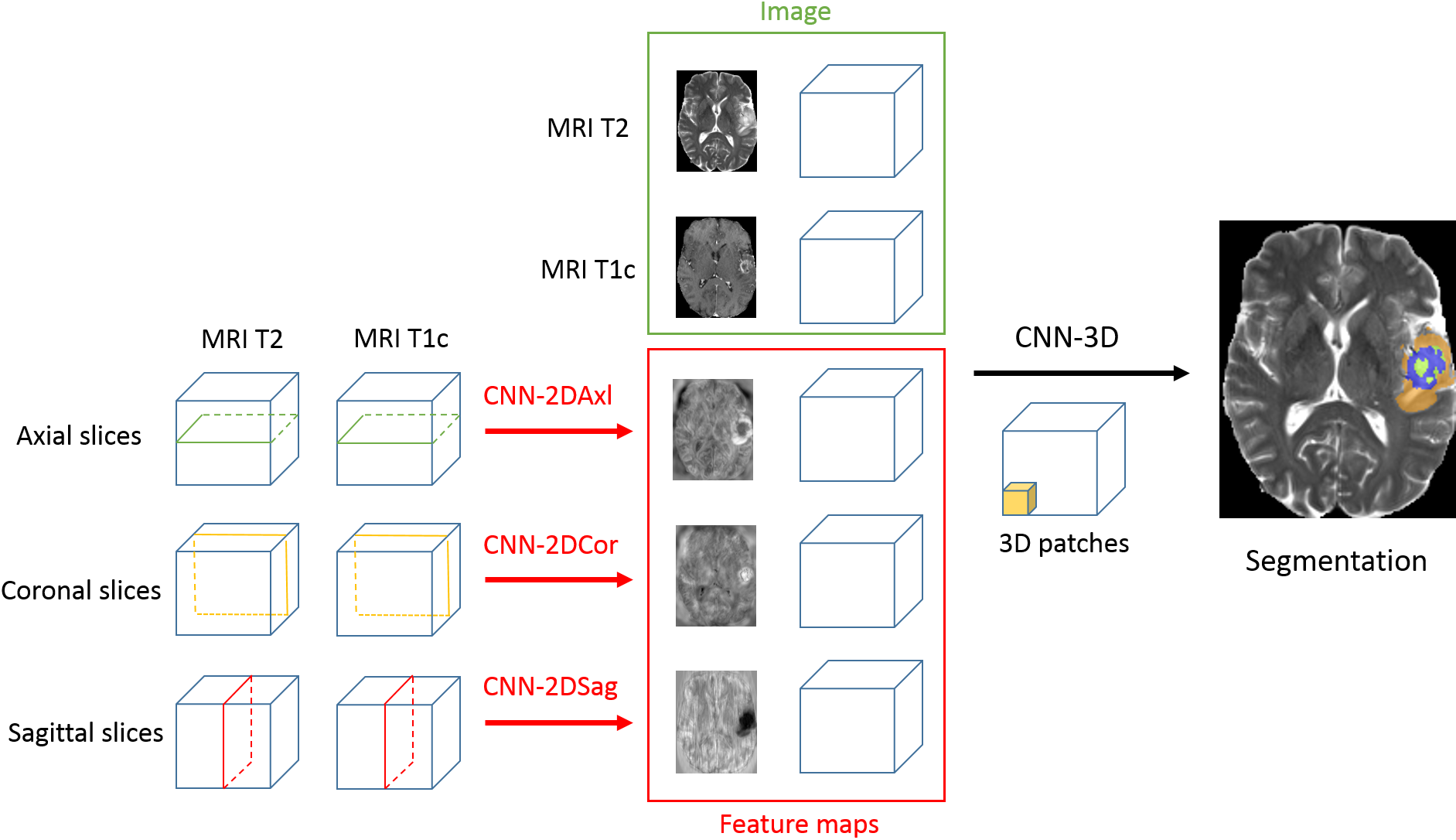
Deep Learning for Tumor Segmentation
Participants : Pawel Mlynarski [Correspondant] , Nicholas Ayache, Hervé Delingette, Antonio Criminisi [MSR] .
This work is funded by Inria-Microsoft Joint Center and is done in cooperation with Microsoft Research in Cambridge.
deep learning, semi-supervised learning, segmentation, MRI, tumors
-
We proposed a model for tumor segmentation which is able to analyze a very large spatial context by combining 2D and 3D CNNs [56] (Fig. 7). Top-3 performance was obtained on BRATS 2017 challenge.
-
We proposed an approach to train CNNs for tumor segmentation with a mixed level of supervision [55]. Our approach significantly improves segmentation accuracy compared to standard supervised learning.
-
We designed a system for segmentation of organs at risk for protontherapy. Promising preliminary results were obtained.