Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.3. Data and knowledge analysis
- A3.3.3. Big data analysis
- A3.4. Machine learning and statistics
- A3.4.1. Supervised learning
- A3.4.5. Bayesian methods
- A3.4.6. Neural networks
- A3.4.7. Kernel methods
- A3.4.8. Deep learning
- A5.3. Image processing and analysis
- A5.3.2. Sparse modeling and image representation
- A5.3.3. Pattern recognition
- A5.3.4. Registration
- A5.4.1. Object recognition
- A5.4.4. 3D and spatio-temporal reconstruction
- A5.4.5. Object tracking and motion analysis
- A5.4.6. Object localization
- A5.9.1. Sampling, acquisition
- A5.9.2. Estimation, modeling
- A5.9.3. Reconstruction, enhancement
- A5.9.5. Sparsity-aware processing
- A5.9.6. Optimization tools
- A6.1.2. Stochastic Modeling
- A6.1.3. Discrete Modeling (multi-agent, people centered)
- A6.1.4. Multiscale modeling
- A6.1.5. Multiphysics modeling
- A6.2.3. Probabilistic methods
- A6.2.4. Statistical methods
- A6.2.6. Optimization
- A6.3. Computation-data interaction
- A6.3.1. Inverse problems
- A6.3.2. Data assimilation
- A6.3.3. Data processing
- A6.3.4. Model reduction
- A6.3.5. Uncertainty Quantification
- A9.2. Machine learning
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1.1.1. Structural biology
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B2.2.3. Cancer
- B2.6. Biological and medical imaging
1 Team members, visitors, external collaborators
Research Scientists
- Charles Kervrann [Team leader, Inria, Senior Researcher, HDR]
- Patrick Bouthemy [Inria, Senior Researcher, HDR]
- Jean Salamero [CNRS, Senior Researcher, UMR144-CNRS, Insitut Curie, PSL, HDR]
- Cesar Augusto Valades Cruz [Inria, Starting Research Position, until Oct 2020]
Post-Doctoral Fellow
- Anais Badoual [Inria]
PhD Students
- Antonin Deschemps [Inria, from Oct 2020]
- Sebastien Herbreteau [Inria, from Oct 2020]
- Yunjiao Lu [Institut national de recherche pour l'agriculture, l'alimentation et l'environnement]
- Leo Maczyta [Inria]
- Etienne Meunier [Inria, from Sep 2020]
- Antoine Salomon [Inria]
Technical Staff
- Ludovic Leconte [CNRS, Engineer, UMR144-CNRS, Institut Curie, PSL]
- Emmanuel Moebel [Inria, Engineer, until Apr 2020]
- Sylvain Prigent [Inria, Engineer]
Interns and Apprentices
- Leo Maury [Inria, Apprentice]
Administrative Assistant
- Huguette Bechu [Inria]
External Collaborator
- Frédéric Lavancier [Univ de Nantes, HDR]
2 Overall objectives
2.1 Glossary
- LLSM (Lattice Light Sheet Microscopy): high resolution Structured Illumination Microscopy that uses Light Sheet Bessel Beam illumination 38.
- TIRF (Total Internal Reflectance): 2D optical microscopy using evanescent waves and total reflectance 36.
- STORM (Stochastic Optical Reconstruction Microscopy): high-resolution microscopy using stochastic photo-activation of fluorophores and adjustment of point spread functions.
- PALM (Photo-Activated Localization Microscopy): high-resolution microscopy using stochastic photo-activation of fluorophores and adjustment of point spread functions 37.
- Cryo-ET (Cryo-Electron Tomography): 3D representation of sub-cellular and molecular objects of 5-20 nanometers, frozen at very low temperatures, from 2D projections using a transmission electron microscope.
2.2 Scientific context and motivations
During the past two decades, biological imaging has undergone a revolution in the development of new microscopy techniques that allow visualization of tissues, cells, proteins and macromolecular structures at all levels of resolution, physiological states, chemical composition and dynamics. Thanks to recent advances in optics, digital sensors and labeling probes (e.g., Colored Fluorescence Protein), one can now visualize sub-cellular components and organelles at the scale of several hundreds of nanometers to a tens nanometers, in live. As a result, fluorescent microscopy and multimodal imaging (fluorophores at various wavelengths) have become the workhorse of modern biology. As a matter of fact, taking into account all the publications in the 10 most relevant journals in fundamental biology (those with highest IF) for the last 2018-2019 years, the ratio of experimental figures based on BioImage data is close to 70% (GBI EoE.V, Singapour, Sep 2019). All the technological advances in microscopy have created new issues and challenges for researchers in quantitative image processing and analysis. Since the digital processing is now part of the imaging loop, image processing may even drive imaging. A brilliant example of this shift in paradigm is super-resolution localization microscopy (PALM, STED), which was awarded the 2014 Nobel Prize in Chemistry.
2.3 Challenges in biological image processing and quantitative microscopy
In most cases, modern microscopy in biology is characterized by a large number of dimensions that fit perfectly with the complexity of biological features: two or three spatial dimensions, at macro to nano-scales, and one temporal dimension, sometimes spectrally defined and often corresponding to one particular biomolecular species. Dynamic microscopy is also characterized by the nature of the observable objects (cells, organelles, single molecules, ...), by the large number of small size and mobile elements (chromosomes, vesicles, ...), by the complexity of the dynamic processes involving many entities or group of entities sometimes interacting, by particular phenomena of coalescence often linked to image resolution problems, finally by the association, dissociation, recomposition or constitution of those entities (such as membrane fusion and budding). Thus, the corpus of data to be considered for any analysis involving multiple image series acquisitions is massive (up to few GigaBytes per hour). Therefore, it becomes necessary to facilitate and rationalize the production of those multidimensional data, to improve post acquisition analysis, and to favor the organization and the interpretation of the information extracted from this data corpus. It motivates innovative methods and concepts for data fusion, image registration, super-resolution, data mining... More importantly, modern microscopy has led to recent breakthroughs, related to the potential interactions between molecules in the cell. A long-term research consists now in inferring the relationships between the dynamics of macromolecules and their functions. Research on computational biology and quantitative bioimaging lies at the core of the activities of serpico team.
2.4 Objectives of Serpico in cell imaging
In order to tackle the aforementioned challenges, the serpico team aims to develop innovative approaches and paradigms for image reconstruction, 3D molecule tracking and motion estimation, and biophysical parameter estimation to face the huge data volumes acquired with cutting-edge microscopy set-ups. To this end, applied mathematics, image processing and analysis have to be considered in association with biophysics and biology. To be successful, a sustained synergy between all these scientific domains is necessary. To improve state-of-the-art methods and solve important problems in computational bioimaging, the members of serpico especially address the following topics:
- Image restoration/reconstruction motivated by preserving cell integrity (photo-toxicity versus exposure time) and image analysis in multidimensional microscopy;
- Motion analysis and computation of molecule trajectories in live-cell imaging to study molecular interactions in space and time;
- Computational simulation, modeling and estimation of molecule trafficking and interactions at different spatial and temporal scales.
The resulting mathematical models and algorithms will help biologists to decipher molecular processes in fundamental biology and will be exploited for health applications: disease diagnosis, detection of genomic instabilities, deterioration of cell cycle, cancer prevention.
We have successfully developed statistical and variational aggregation methods for image denoising and optical flow, and elaborated powerful methods for image colocalization, diffusion estimation, trajectory estimation-classification, and multimodal registration. An additional issue was the design and distribution of software tools for the biological image analysis and microscopy communities. Finally, the team has focused on the cellular and molecular mechanisms involved in molecule and protein transport and trafficking at the scale of a single cell. Our contributions are detailed in the next sections along three research axes.
2.5 Organization and collaborations
In collaboration with CNRS-UMR 144 Institut Curie (and in cooperation with the “Space Time imaging of Endomembranes and organelles Dynamics” team) and PICT-IBiSA (Cell and Tissue Imaging Facilities), the members of the serpico team have participated in several projects (PhD and post-doc supervision, contracts...) in the field of cell biology and microscopy. We have promoted non-parametric methods since prior knowledge cannot be easily taken into account for extracting unattended but desired information from image data. We have also proposed user-friendly algorithms for processing 2D and 3D image sequences. The projects of serpico were in line with several studies led in the CNRS-UMR 144 Institut Curie Unit. A subset of studies was related to instrumentation in electronic and photonic microscopy (PICT-IBiSA platform) including computational aspects on the reconstruction and enhancement of images related to sub-diffraction light microscopy and multimodal approaches. Serpico projects relied partially on the advances of these instrumental projects and a positive synergy was established.
3 Research program
3.1 Statistics and algorithms for computational microscopy
Fluorescence microscopy limitations are due to the optical aberrations, the resolution of the microscopy system, and the photon budget available for the biological specimen. Hence, new concepts have been defined to address challenging image restoration and molecule detection problems while preserving the integrity of samples. Accordingly, the main stream regarding denoising, deconvolution, registration and detection algorithms advocates appropriate signal processing framework to improve spatial resolution, while at the same time pushing the illumination to extreme low levels in order to limit photo-damages and phototoxicity. As a consequence, the question of adapting cutting-edge signal denoising and deconvolution, object detection, and image registration methods to 3D fluorescence microscopy imaging has retained the attention of several teams over the world.
In this area, the serpico team has developed a strong expertise in key topics in computational imaging including image denoising and deconvolution, object detection and multimodal image registration. Several algorithms proposed by the team outperformed the state-of-the-art results, and some developments are compatible with “high-throughput microscopy” and the processing of several hundreds of cells. We especially promoted non local, non-parametric and patch-based methods to solve well-known inverse problems or more original reconstruction problems. A recent research direction consists in adapting the deep learning concept to solve challenging detection and reconstruction problems in microscopy. We have investigated convolution neural networks to detect small macromolecules in 3D noisy electron images with promising results. The next step consists in proposing smart paradigms and architectures to save memory and computations.
More generally, many inverse problems and image processing become intractable with modern 3D microscopy, because very large temporal series of volumes (200 to 1000 images per second for one 3D stack) are acquired for several hours. Novel strategies are needed for 3D image denoising, deconvolution and reconstruction since computation is extremely heavy. Accordingly, we will adapt the estimator aggregation approach developed for optical flow computation to meet the requirements of 3D image processing. We plan to investigate regularization-based aggregation energy over super-voxels to reduce complexity, combined to modern optimization algorithms. Finally, we will design parallelized algorithms that fast process 3D images, perform energy minimization in few seconds per image, and run on low-cost graphics processor boards (GPU).
3.2 From images to motion descriptors and trajectories
Several particle tracking methods for intracellular analysis have been tailored to cope with different types of cellular and subcellular motion down to Brownian single molecule behavior. Many algorithms were carefully evaluated on the particle tracking challenge dataset published in the Nature Methods journal in 2014. Actually, there is no definitive solution to the particle tracking problem which remains application-dependent in most cases. The work of serpico in particle motion analysis is significant in multiple ways, and inserts within a very active international context. One of the remaining key open issues is the tracking of objects with heterogeneous movements in crowded configurations. Moreover, particle tracking methods are not always adapted for motion analysis, especially when the density of moving features hampers the individual extraction of objects of interest undergoing complex motion. Estimating flow fields can be more appropriate to capture the complex dynamics observed in biological sequences. The existing optical flow methods can be classified into two main categories: i/ local methods impose a parametric motion model (e.g. local translation) in a given neighborhood; ii/ global methods estimate the dense motion field by minimizing a global energy functional composed of a data term and a regularization term.
The serpico team has developed a strong expertise in key topics, especially in object tracking for fluorescence microscopy, optical flow computation and high-level analysis of motion descriptors and trajectories. Several algorithms proposed by the team are very competitive when compared to the state-of-the-art results, and our new paradigms offer promising ways for molecule traffic quantification and analysis. Amongst the problems that we currently address, we can mention: computation of 3D optical flow for large-size images, combination of two frame-based differential methods and sparse sets of trajectories, detection and analysis of unexpected local motion patterns in global coherent collective motion. Development of efficient numerical schemes will be central in the future but visualization methods are also crucial for evaluation and quality assessment. Another direction of research consists in exploiting deep learning to 3D optical flow so as to develop efficient numerical schemes that naturally capture complex motion patterns. Investigation in machine learning and statistics will be actually conducted in the team in the two first research axes to address a large range of inverse problems in bioimaging. Deep learning is an appealing approach since expertise of biologists, via iterative annotation of training data, will be included in the design of image analysis schemes.
3.3 Biological and biophysical models and statistics for quantitative bioimaging
A number of stochastic mathematical models were proposed to describe various intracellular trafficking, where molecules and proteins are transported to their destinations via free diffusion, subdiffusion and ballistic motion representing movements along the cytoskeleton networks assisted by molecular motors. Accordingly, the study of diffusion and stochastic dynamics has known a growing interest in bio-mathematics, biophysics and cell biology with the popularization of fluorescence dynamical microscopy and super-resolution imaging. In this area, the competing teams mainly studied MSD and fluorescence correlation spectroscopy methods.
In the recent period, the serpico team achieved important results for diffusion-related dynamics involved in exocytosis mechanisms. Robustness to noise has been well investigated, but robustness to environmental effects has yet to be effectively achieved. Particular attention has been given to the estimation of particle motion regime changes, but the available results are still limited for analyzing short tracks. The analysis of spatiotemporal molecular interactions from set of 3D computed trajectories or motion vector fields (e.g., co-alignment) must be investigated to fully quantify specific molecular machineries. We have already made efforts in that directions this year (e.g., for colocalization) but important experiments are required to make our preliminary algorithms reliable enough and well adapted to specific transport mechanisms.
Accordingly, we will study quantification methods to represent interactions between molecules and trafficking around three lines of research. First, we will focus on 3D space-time global and local object-based co-orientation and co-alignment methods, in the line of previous work on colocalization, to quantify interactions between molecular species. In addition, given tracks associated to molecular species, interaction descriptors, dynamics models and stochastic graphical models representing molecular machines will be studied in the statistical data assimilation framework. Second, we will analyse approaches to estimate molecular mobility, active transport and motion regime changes from computed trajectories in the Lagrangian and Eulerian settings. We will focus on the concept of super-resolution to provide spatially high-resolved maps of diffusion and active transport parameters based on stochastic biophysical models and sparse image representation. Third, we plan to extend the aggregation framework dedicated to optical flow to the problem of diffusion-transport estimation. Finally, we will investigate data assimilation methods to better combine algorithms, models, and experiments in an iterative and virtuous circle. The overview of ultrastructural organization will be achieved by additional 3D electron microscopy technologies.
4 Application domains
4.1 Modeling and analysis of membrane transport and molecule trafficking at the single cell scale
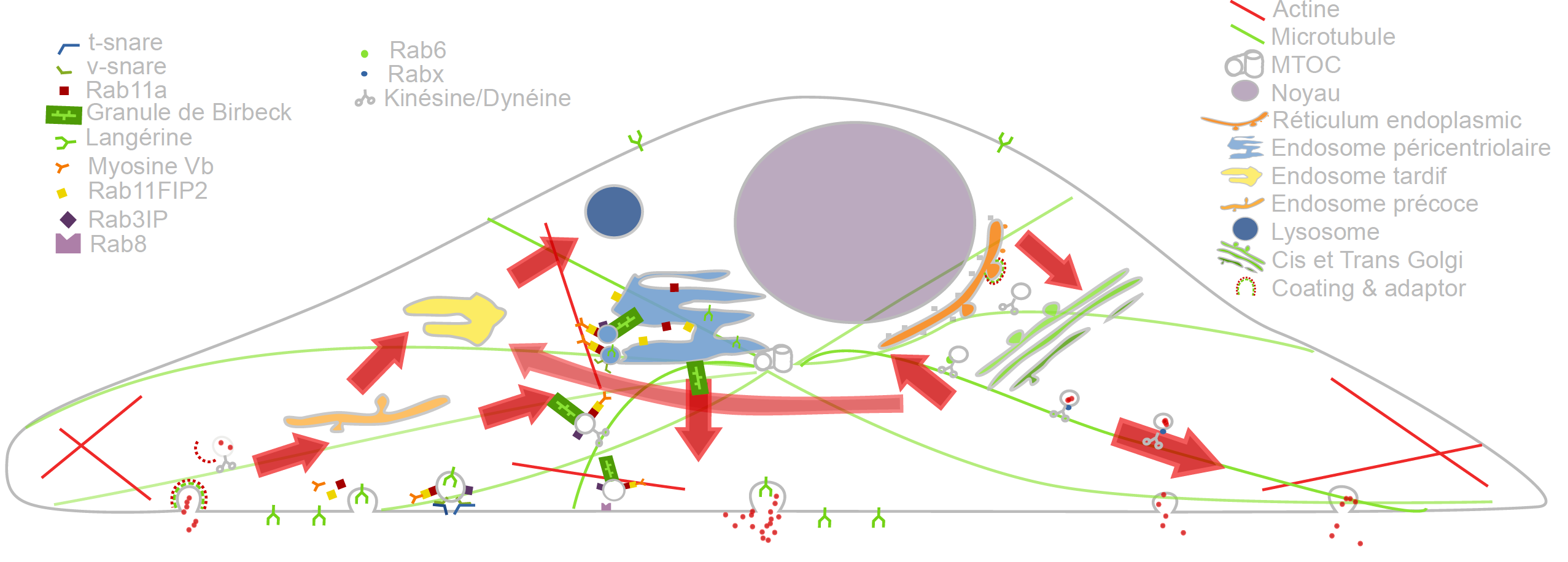
In the past recent years, research carried out out together with the “Space Time imaging of Endomembranes and organelles Dynamics” team at CNRS-UMR 144Institut Curie contributed to a better understanding of the intracellular compartmentation, particularly in specialized model cells such as melanocytes and Langerhans cells of the epidermis, of the components and structural events involved in the biogenesis of their specialized organelles: melanosomes and Birbeck granules, respectively and to the understanding on how the dynamics of those structures relate to their physiological functions. These studies have started to highlight: i/ the measurement of multiple sorting and structural events involved in the biogenesis of these organelles; ii/ complexity of the endo-melanosomal network of these highly specialized cells; iii/ complex molecular architecture organizing and coordinating their dynamics; iv/ intracellular transport steps affected in genetic diseases, among which the Hermansky Pudlak syndrome (HPS) or involved in viral infection (HIV and Langerin in Langerhans cells).
In this context, the central aim of serpico is to understand how the different machineries of molecular components involved are interconnected and coordinated to generate such specialized structures, an issue that become more and more accessible, thanks to improvement in all domains related to live imaging. We need to address the following topics:
- developing new bioimaging approaches to observe and statistically analyze such coordinated dynamics in live material;
- correlating this statistically relevant spatiotemporal organization of protein networks with the biological architectures and at the ultrastructural level;
- modeling intracellular transport of those reference biological complex systems and proposing new experimental plans in an iterative and virtuous circle;
- managing and analyzing the workflow of image data obtained along different multidimensional microscopy modalities.
These studies are essential to unravel the complexity of the endomembrane system, how different machineries evolve together and thus coordinate (e.g. see Fig. 1). They help to decipher cell organization and function at different scales through an integrative workflow of methodological and technological developments. New approaches, such as optogenetics may even help controlling cell functions.
At long term, these studies will shed light on the cellular and molecular mechanisms underlying antigen presentation, viral infection or defense mechanisms, skin pigmentation, the pathogenesis of hereditary genetic disorders (lysosomal diseases, immune disorders) and on the mechanisms underlying cell differentiation and cell transformation. Our methodological goal is also to link dynamics information obtained through diffraction limited light microscopy, at a time regime compatible with live cell imaging and close to biochemical molecular interactions. The overview of ultrastructural organization will be achieved by complementary electron microscopy methods which have also undergone a revolutionary improvement over the last decade. Image visualization and quantitative analysis are of course essential issues in this context.
4.2 Imaging and analysis of cytoskeleton dynamics during cell migration
The ability to migrate in space is among the most fundamental functions of eukaryotic cells and thus is one of the best-studied phenomena in biology. During embryonic development, cell movements result in a massive reorganization of the embryo, from a simple spherical ball of cells into a multi-layered organism; many of the cells at or near the surface of the embryo move to a new, more interior location. Moreover, inadequate or inappropriate migration of immune cells is also critically important for the delivery of protective immune responses to tissues and for wound healing. Finally, cell migration may facilitate the dissemination of tumor cells from primary tumor in blood (extravasation) and eventually the colonization of other organs and the formation of secondary tumors.
It has been established that the cytoskeleton, composed of actin filaments, microtubules and intermediate filaments (elongated structures with a diameter of a few dozens of nanometers), is essential for several cell mechanisms, including cell migration, cell division and molecule trafficking:
- i/ the actin filaments promote cell protrusion, adhesion and retraction;
- ii/ the microtubules are the support of molecule traffic and cell polarization;
- iii/ the intermediate filaments are hypothesized to control microtubule organization.
Nevertheless, the mechanical and chemical states of migrating cells under various external conditions remain largely unknown. In the last decade, high-resolution microscopy methods led to the discovery of novel aspects of cell migration. Most approaches and models are limited to migration in 2D, justified by the flatness of the cell-motile mechanisms. However, the mechanical patterns that govern migration in 2D models are often not essential for efficient migration in 3D. Accordingly, recent very challenging 3D models of cells moving on flat surfaces have begun to emerge. The key challenge, however, is to understand how a 3D motile cell crawls through the 3D extracellular matrix. Another issue is of course to measure and understand how membrane protrusion and retraction keep the cell in homeostasis, which of course relate to membrane traffic.
The objective of serpico is to develop high-end signal processing and computer vision tools to unfold the dynamical coordination of microtubules, actin filaments and intermediate filaments in 3D, involved in cell migration, cell division and how molecular trafficking is coordinated with cytoskeleton changes in these fundamental cellular functions.
5 Social and environmental responsibility
Application of general recommendations related to transport (train, visioconferences). All conferences were conducted in virtual mode in 2020 because of the pandemic crisis. This has lead to a significant reduction of the number of travels, and then a small carbon footprint for the team in 2020.
6 Highlights of the year
The DeepFinder algorithm was ranked 2nd at the international SHREC’20 Challenge: "classification in cryo-electron tomograms".
7 New software and platforms
7.1 New software
7.1.1 GcoPS
- Keywords: Photonic imaging, Fluorescence microscopy, Image processing, Statistic analysis
- Functional Description: The GcoPS (Geo-Co-Positioning System) software is dedicated to the co-localization of fluorescence image pairs for both conventional and super-resolution microscopy. The procedure is only controlled by a p-value and tests whether the Pearson correlation between two binary images is significantly positive. Colocalization amounts here to quantifying the interaction strength by the area/volume of the intersection between the two binary images viewed as random distributions of geometrical objects. Under mild assumptions, it turns out that the appropriately normalized Pearson correlation follows a standard normal distribution under the null hypothesis if the number of image pixels is large. Unlike previous methods, GcoPS handles 2D and 3D images, variable SNRs and any kind of cell shapes. It is able to co-localize large regions with small dots, as it is the case in TIRF-PALM experiments and to detect negative co-localization. The typical processing time is two milliseconds per image pair in 2D and a few seconds in 3D, with no dependence on the number of objects per image. In addition, the method provides maps to geo-co-localize molecule interactions in specific image regions.
-
URL:
http://
icy. bioimageanalysis. org/ plugin/ GcoPS - Publication: hal-02369555
- Contact: Charles Kervrann
- Participants: Thierry Pécot, Frédéric Lavancier, Charles Kervrann, Liu Zengzhen
- Partners: Université de Nantes, UMR 144 CNRS - Institut Curie, Hollings Cancer Center at the Medical University of South Carolina, Charleston SC, USA
7.1.2 THOTH
- Name: Testing HypOtheses for diffusion TricHotomy
- Keywords: Photonic imaging, Fluorescence microscopy, Biomedical imaging, Classification, Statistical categorisation techniques, Statistics, Image sequence, Visual tracking
- Functional Description: The THOTH software classifies biomolecule trajectories of biomolecules (computed with tracking algorithms) into three groups of diffusion: (i) free diffusion, (ii) subdiffusion or (iii) superdiffusion. Brownian motion corresponds to the NULL hypothesis. THOTH is a nonparametric three-decision test whose alternatives are subdiffusion and superdiffusion. Single and multiple testing procedures control respectively the type I error and the false discovery rate. THOTH can be considered as an alternative to the Mean Square Displacement (MSD) method commonly used to address this issue. It gives more reliable results as confirmed by our Monte Carlo simulations and evaluations on real sequences of images depicting protein dynamics acquired with TIRF or SPT-PALM microscopy.
-
URL:
https://
team. inria. fr/ serpico/ software/ thot/ - Publication: hal-01961971
- Contact: Charles Kervrann
- Participants: Vincent Briane, Charles Kervrann, Myriam Vimond
- Partner: ENSAI
7.1.3 SparseVolution
- Name: Sparse Variation for 2D Image Decovolution
- Keywords: Fluorescence microscopy, Image processing, Deconvolution, Inverse problem
- Functional Description: In order to improve the resolution of acquired fluorescence images, we introduced a method of image deconvolution by considering a family of convex regularizers. The considered regularizers are generalized from the concept of Sparse Variation which combines the L1 norm and the first (Total Variation) or second (Hessian Variation) derivatives to favor the colocalization of high-intensity pixels and high-magnitude gradient. The experiments showed that the proposed regularization approach produces competitive deconvolution results on fluorescence images, compared to those obtained with other approaches such as TV or the Schatten norm of Hessian matrix. The final algorithm has been dedicated to deconvolve very large 2D (e.g. 20 000 x 20 000) images or 3D images.
- Contacts: Charles Kervrann, Hoai Nam Nguyen
- Participants: Hoai Nam Nguyen, Charles Kervrann, Sylvain Prigent, Cesar Augusto Valades Cruz
- Partners: Innopsys, UMR 144 CNRS - Institut Curie
7.1.4 AiryscanJ
- Name: Reconstruction of AiryScan microscopy images
- Keywords: Image reconstruction, Fluorescence microscopy, Photonic imaging, Deconvolution, Image analysis
- Functional Description: The AiryscanJ software enables to reconstruct a high resolution image from an array of multiple raw images acquired with the Airyscan technology (32 detectors). Airyscanning is a recent technique based on confocal laser scanning microscopy. The AiryscanJ software gathers four reconstruction methods (ISM, IFED, ISFED, ISM-Deconvolution) to compute a high resolution image: 1/ ISM amounts to summing the preliminarily registered raw images. 2/ IFED is the weighted difference between the inner detectors and the outer detector. 3/ ISFED is the weighted difference between the registered outer detectors and the original outer detectors. AiryscanJ automatically estimates the parameter controlling the IFED and ISFED algorithms. 4/ ISM-Deconvolution allows reconstructing a high resolution image by applying a deconvolution algorithm on the ISM image.
-
URL:
https://
gitlab. inria. fr/ serpico/ airyscanj_bin - Contact: Charles Kervrann
- Participants: Sylvain Prigent, Stephanie Dutertre, Charles Kervrann
- Partners: Université de Rennes 1, CNRS
7.1.5 DeepFinder
- Name: Deep learning for macromolecule identification within 3D cellular cryo-electron tomograms
- Keywords: Image analysis, Deep learning, Cryo-electron microscopy, Object detection
- Functional Description: DeepFinder is a computational approach that uses artificial neural networks to accurately and jointly localize multiple types and/or states of macromolecules in 3D cellular cryo-electron tomograms. DeepFinder leverages deep learning and outperforms the commonly-used template matching method on ideal data. On synthetic image data (SHREC 2019 challenge), DeepFinder is very fast and produces superior detection results when compared to other competitive deep learning methods, especially on small macromolecules. On experimental cryo-ET data depicting ribosomes, the detection results obtained by DeepFinder are consistent with expert annotations. We have got a high overlap of 86% and a similar structure resolution determined by subtomogram averaging.
-
URL:
https://
gitlab. inria. fr/ serpico/ deep-finder - Contact: Charles Kervrann
- Participants: Emmanuel Moebel, Antonio Martinez, Charles Kervrann
- Partners: Max Planck Institute Martinsried, Fondation Fourmentin-Guilbert
7.1.6 CPAnalysis
- Name: Change point detection algorithm for detecting switches of diffusion along 2D-3D single particle trajectories
- Keywords: Statistical modeling, Statistical physics, Fluorescence microscopy, Motion analysis
- Functional Description: The change point detection algorithm is designed for detecting switches of diffusion along a given 2D-3D particle trajectory. We consider that the particle can switch between three main motion modes: 1/ Superdiffusion which occurs when the particle is transported via molecular motors along the cytoskeleton, 2/ Free diffusion (or Brownian motion) which arises when the particle evolves freely inside the cytosol, 3/ Subdiffusion observed when the particle is confined in a domain or evolves in a crowded area. The algorithm is a sequential nonparametric procedure based on a nonparametric three-decision test computed on local sliding windows along the trajectory.
-
URL:
https://
team. inria. fr/ serpico/ software/ cpanalysis/ - Publication: hal-02424777
- Contact: Charles Kervrann
- Participants: Antoine Salomon, Vincent Briane, Myriam Vimond, Jean Salamero, Charles Kervrann
- Partners: ENSAI, UMR 144 CNRS - Institut Curie
7.1.7 FlowScope
- Name: Optical flow computation for 3D fluorescence microscopy
- Keywords: Motion analysis, Fluorescence microscopy, Image analysis, Data visualization
- Functional Description: The FLOWSCOPE software is able to estimate 3D motion between two fluorescence microscopy volumes. The underlying variational method amounts to minimizing an energy functional made up of two terms: a data term and a regularization term. The data term is derived from the continuous form of Census signature and the smoothness of the flow field is imposed by a L2 regularization term. The method is implemented for a single core CPU. FLOWSCOPE outputs three separate files corresponding to the motion vector components. The flow fields can be visualized with an appropriate color code named 3PHS. The 3PHS map projects the flow field onto the three orthogonal planes selected by the user. The projections of the vector field is color coded in the Hue (direction) and the Saturation (amplitude) spaces.
-
URL:
https://
gitlab. inria. fr/ smanandh/ flowscope - Publication: hal-02308001
- Contact: Sandeep Manandhar
- Participants: Patrick Bouthemy, Charles Kervrann, Sylvain Prigent, Leo Maury, Philippe Roudot
- Partner: Danuser lab, University of Texas Southwestern, Dallas, USA
7.1.8 MWR
- Name: Missing Wedge Restoration (MWR) and Noise Removal in 3D Cryo-Tomography
- Keywords: Image analysis, Inverse problem, Cryo-electron microscopy, Monte-Carlo methods, Bayesian estimation
- Functional Description: The Missing Wedge Restoration (MWR) software enables to reduce the high amount of noise and artifacts observed in 3D cellular cryo-tomograms, induced by the presence of a missing wedge (MW) in the spectral domain. MWR takes as input a 3D tomogram derived from limited-angle tomography, and gives as output a 3D denoised and artifact compensated volume. The artifact compensation is achieved by filling up the MW with meaningful information. A Minimum Mean Square Error (MMSE) estimator is computed by applying a dedicated Markov Chain Monte-Carlo (MCMC) sampling procedure based on the Metropolis-Hasting algorithm. MWR can be used to enhance visualization or as a pre-processing step for image analysis, including segmentation and classification of macromolecules.
-
URL:
https://
gitlab. inria. fr/ serpico/ mwr - Publication: hal-02424804
- Contact: Charles Kervrann
- Participants: Emmanuel Moebel, Antonio Martinez
- Partners: Max Planck Institute Martinsried, Fondation Fourmentin-Guilbert
7.2 New platforms
7.2.1 Bioimage-IT for bioimage management and processing
Participants: Sylvain Prigent, Cesar Augusto Valades Cruz, Ludovic Leconte, Léo Maury, Jean Salamero, Charles Kervrann.
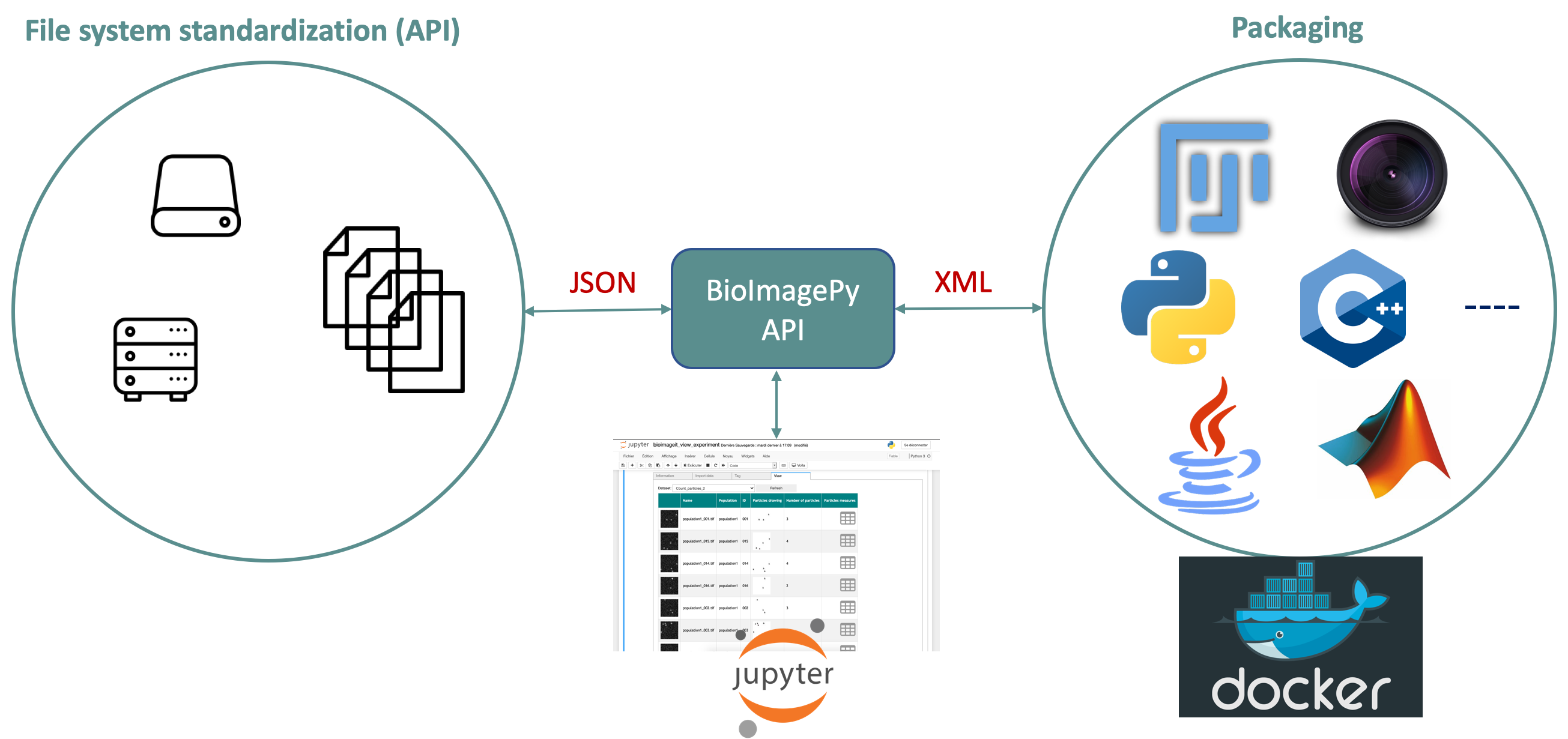
New image acquisition systems generate large number of images and large volume images. Such data sets are hard to store, to process and to analyze for one user in a workstation. Many solutions exist for data management (e.g. Omero, OpenImadis), image analysis (e.g. Fiji, Icy, CellProfiler) and statistics (e.g R). Each of them has its specificities and several bridges have been developed between pieces of software. Nevertheless, in many use-cases, we need to perform analysis using tools that are available in different pieces of software and different languages. It is then tedious to create a workflow that brings the data from one tool to another. It needs programing skills and most of the time, a dedicated script using a dedicated file system for processed data management is developed. The aim of BioImage-IT is to create a “bandmaster” application that allow any scientist to annotate, process, and analyze data using only one single high level application. This BioImage-IT application is based on 3 components:
- an image annotation method based on a json file system,
- an image processing and analysis tools integration method based on Docker and XML commands description,
- an application with a graphical interface to easily annotate data, run processing tools, and visualize data and results.
This software architecture has three main goals. First, data are annotated using a file system. This means that data are not dependent on any software like a SQL database, and each experiment can then be stored in a different directory and can be moved from one server to another or to any drive with a simple copy pasting operation. Second, the processing tools are used as binary packages managed by the Docker technology. Docker enables to gently handle dependencies and several versions of the same tool. Any existing tool can then be integrated in its native programming language. Third, using a single “bandmaster” application allows one to automatically generate metadata for any processed data, improving the traceability and the repeatability of any experimental result.
BioImage-IT (https://
Partners: CNRS-UMR 144 and U1143 INSERM/CNRS-UMR 3666, Institut Curie & France-BioImaging (UMS 3714 CEMIBIO).
8 New results
8.1 Inverse problems and biomolecules dynamics estimation in biological imaging
8.1.1 High-resolution reconstruction and deconvolution of array detector images
Participants: Sylvain Prigent, Charles Kervrann.
Array detector allows a resolution gain for confocal microscopy by combining several images acquired by 32 sub-detectors. A single high resolution image is generally obtained by spatially re-assigning the signal from the 32 sub-detectors and applying a deconvolution procedure. Alternative methods have been proposed to produce high resolution images by linearly combining the signals of sub-detectors. Last year, we proposed a SURE-guided (Stein’s unbiased risk estimation) estimation method for automatically setting the parameter that controls the underlying algorithms, including ISM, IFED, and ISFED, in order to improve spatial resolution. This year, we developed a regularization method that deconvolves directly the stack constituted of 32 spatially re-assigned detectors signals. We demonstrated on both calibration slides and real data that the so-called ISM-SV algorithm allows to produce more homogeneous, well-contrasted, high-resolution images than those obtained with previous deconvolution methods (ISM-RichardsonLucy, ISM-Wiener) (see Fig. 3). All the tested methods have been implemented in an open source software which can be be downloaded for free (Fiji plugin: https://
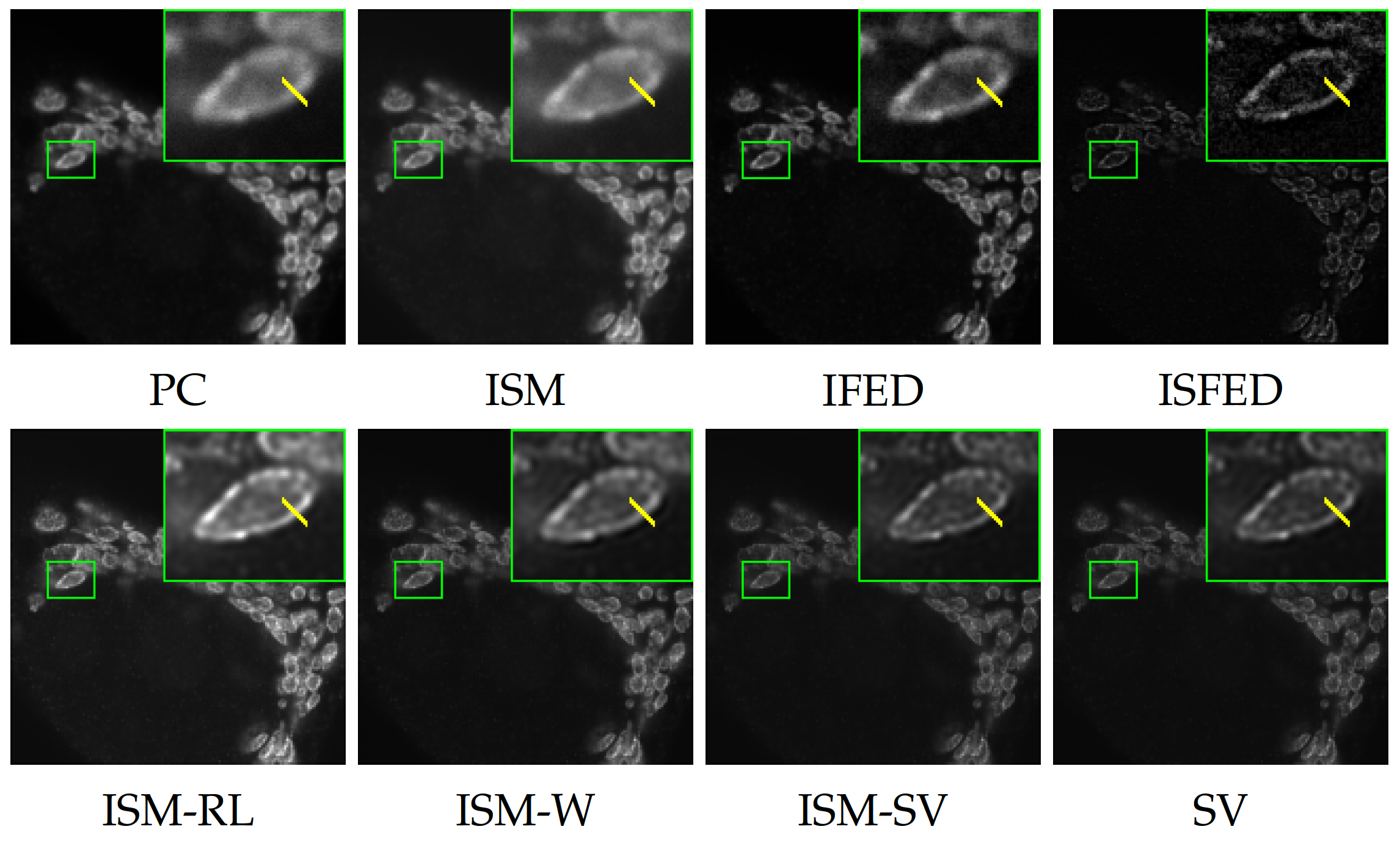
Software: AiryscanJ (see Section 7.1.4).
Reference: 23
Collaborators: S. Dutertre, A. Bidaud-Meynard, G. Bertolin, G. Michaux (IGDR – Institute Genetics & Development of Rennes).
8.1.2 Probabilistic reconstruction of truncated particle trajectories on a closed surface: application to membrane-associated molecular dynamics analysis in rod-shaped bacteria
Participants: Yunjiao Lu, Charles Kervrann.
Investigation of dynamic processes in cell biology very often relies on the observation in two dimensions of 3D biological processes. Consequently, the data are partial and statistical methods and models are required to recover the parameters describing the dynamical processes. In the case of molecules moving over the 3D surface, such as proteins on walls of bacteria cell, a large portion of the 3D surface is not observed in 2D-time microscopy. It follows that biomolecules may disappear for a period of time in a region of interest, and then reappear later. Assuming Brownian motion with drift, we addressed the mathematical problem of the reconstruction of biomolecules trajectories on a cylindrical surface (see Fig. 4). A subregion of the cylinder is typically recorded during the observation period, and biomolecules may appear or disappear in any place of the 3D surface. The performance of the resulting algorithm was demonstrated on simulated particle trajectories that mimic MreB protein dynamics observed in 2D time-lapse fluorescence microscopy (TIRFM) in rod-shaped bacteria.
Reference: 18
Collaborators: A. Trubuil and P. Hodara (INRA MaIAGE unit, Jouy-en-Josas); R. Carballido-López and C. Billaudeau (INRA, UR MICALIS, Jouy-en-Josas).
![(a): Several consecutive images from a real TIRFM movie[1]. Tracks are superposed on
the images. (b) Left: Illustration of trajectories observed during recorded time [0,TS][0, T_S] on the surface
of a cylinder. Right: Representation of the dynamics on a 2D unwrapped surface ]-L,0[×[0,H]]-L,0[\times [0,H].](/rapportsactivite/RA2020/serpico/WEB_IMG/IPreconnect.png)
8.1.3 Geo-colocalization of Aurora kinase A/AURKA in the mitochondrial matrix with dSTORM super-resolution microscopy
Participants: Charles Kervrann.
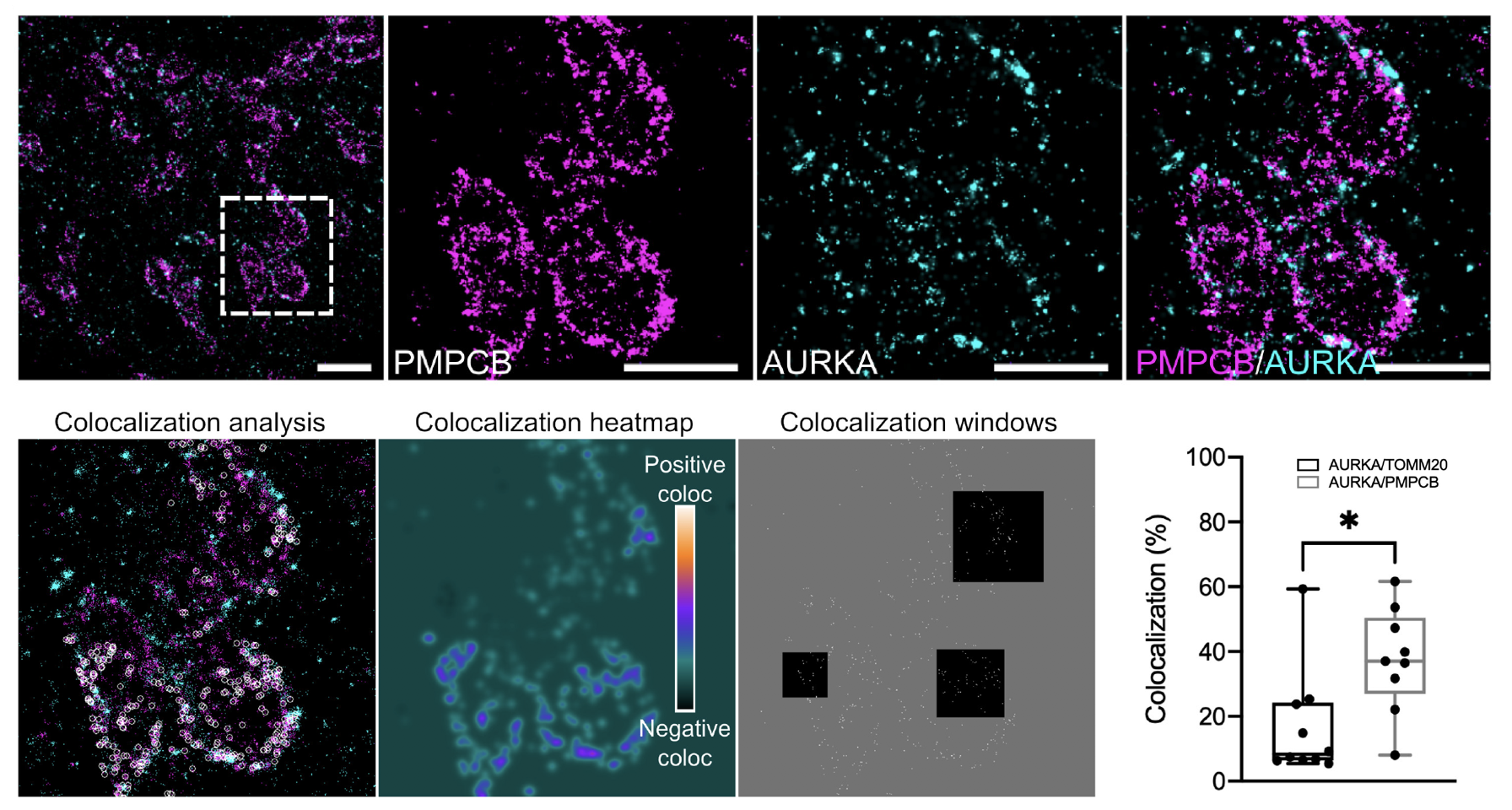
Mitochondria are dynamic organelles playing essential metabolic and signaling functions in cells. Quantifying protein-protein proximities using electron microscopy (EM) is still extremely challenging. Recently, super-resolution microscopy approaches including dSTORM led to valuable advances in our knowledge of mitochondrial ultrastructure, and in linking it with new insights in organelle functions. The cancer-related Aurora kinase A/AURKA is a protein localized at various subcellular locations, including mitochondria. After performing dSTORM, we used the Geo‐coPositioning System (GcoPS) image analysis method to quantify the degree of colocalization of AURKA with compartment-specific mitochondrial markers (see Fig. 5). In this study, we showed that two-color dSTORM provides sufficient spatial resolution to visualize AURKA in the mitochondrial matrix. Our results indicated that dSTORM coupled to GcoPS colocalization analysis is a suitable approach to explore the compartmentalization of non-integral mitochondrial proteins as AURKA, in a qualitative and quantitative manner. This approach also opens up the possibility of analyzing the proximity between AURKA and its multiple mitochondrial partners with exquisite spatial resolution, thereby allowing novel insights into the mitochondrial functions controlled by AURKA.
Software: GcoPS (see Section 7.1.1).
Reference: 33
Collaborators: G. Bertolin (IGDR – Institute of Genetics & Development of Rennes); B. Durel (Cell Imaging Platform, Structure Fédérative de Recherche Necker INSERM US24, CNRS UMS3633, Paris).
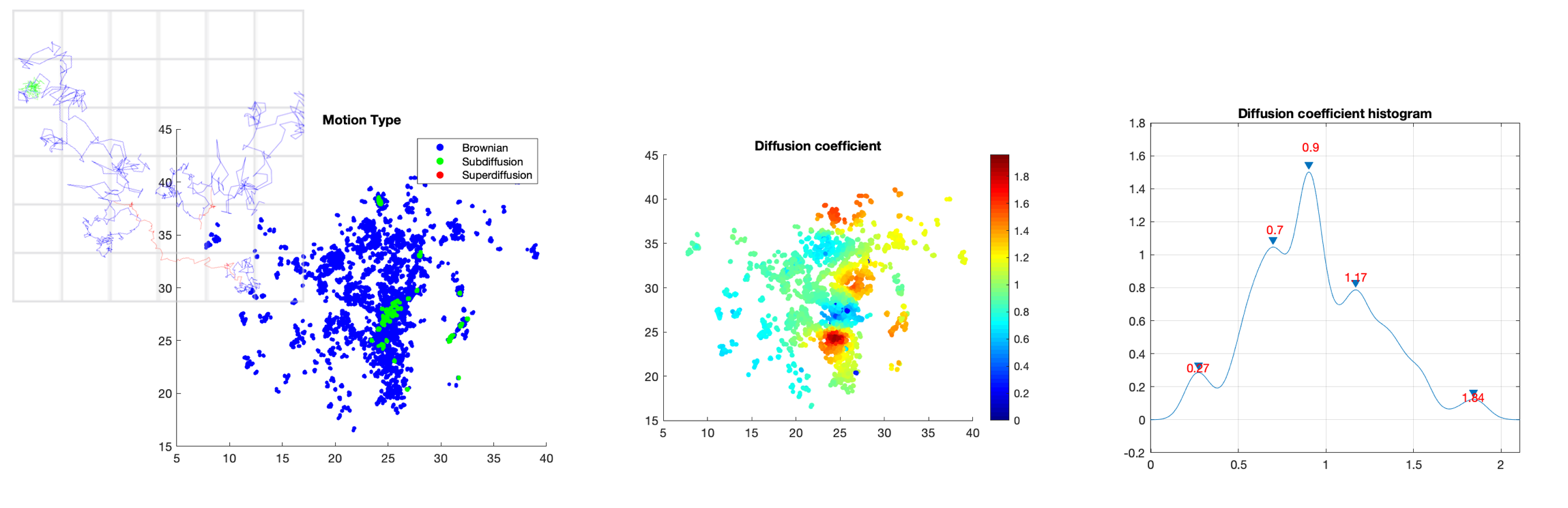
8.1.4 Dense mapping of intracellular diffusion and drift from single-particle tracking data
Participants: Antoine Salomon, Cesar Augusto Valades Cruz, Ludovic Leconte, Jean Salamero, Charles Kervrann.
In this study, we focused on the local estimation diffusion and drift from tracks of biomolecules inside a cell. We assume that particle motion is governed by the following Langevin equation: where denotes the 2D or 3D spatial coordinates of the particle, the drift vector, the diffusion coefficient, and the standard Gaussian white noise. Last year, We proposed a new mapping method to estimate both drift and diffusion (or Kramers-Moyal coefficients) in local neighborhoods centered on trajectory points. This year, we improved the experimental results in terms of resolution by integrating an original data-driven boostrapping technique that increases the number of particle tracks. The method has been tested on a large range of real image sequences and evaluaed in several biological studies, including analysis of the dynamics of transcription factors in the nucleus (see Fig. 6), analysis of biomolecules trafficking of endoplasmic reticulum luminal proteins, as well as analysis of virus dynamics in live cells.
Software: THOTH and CPAnalysis (see Sections 7.1.2 and 7.1.6).
Reference: 24
Collaborators: L. Johannes (U1143 INSERM / CNRS-UMR 3666, Institut Curie, PSL Research University); L. Muresan (Cambridge Advanced Imaging Centre, UK).
8.1.5 Spatial birth-death-move processes : basic properties and estimation of their intensity functions
Participants: Frédéric Lavancier, Charles Kervrann.
Spatial birth-death processes are generalisations of simple birth-death processes, where the birth and death dynamics depend on the spatial locations of individuals. In this study, we further let individuals move during their life time according to a continuous Markov process. This generalisation,that we call a spatial birth-death-move process, finds natural applications in bioimaging and individual-based modeling in ecology. In the first part of the study, we verified that birth-death-move processes are well-defined homogeneous Markov processes, studied their convergence to an invariant measure, and established their underlying martingale properties. In the second part, we addressed the non-parametric estimation of their birth, death and total intensity functions, in presence of continuous-time or discrete-time observations. We introduced a kernel estimator that we proved to be consistent under fairly simple conditions, in both settings. We also discussed how we can take advantage of structural assumptions made on the intensity functions, and we explained how bandwidth selection by likelihood cross-validation can be conducted. A simulation study completed the theoretical results. We finally applied our model to the analysis of the spatio-temporal dynamics of proteins involved in exocytosis mechanisms in live cells.
Collaborators: R. Le Guevel (Univ Rennes, CNRS, IRMAR - UMR 6625, Rennes); J. Salamero (CNRS-UMR 144, Institut Curie, PSL Research University).
8.1.6 Detection and tracking of Tau/microtubules interactions in neural cells
Participants: Léo Maury, Sylvain Prigent, Charles Kervrann.
The microtubule-associated phosphoprotein Tau regulates microtubule dynamics and is known to be involved in neurodegenerative diseases. The majority of Tau molecules decorate axonal microtubules, thereby stabilizing them. However, it is an open question how the Tau protein regulates microtubule dynamics and interact with other molecular partners. In this study, we proposed a pipeline based on GFP-Tau proteins and microtubules detection and tracking in Total Internal Reflection Fluorescence (TIRF) to better estimate the Tau oligomerization state and residency time on microtubules (see Fig. 7). A generic plug-in ImageJ for proteins detection 19 and tracking has been designed for end-users.
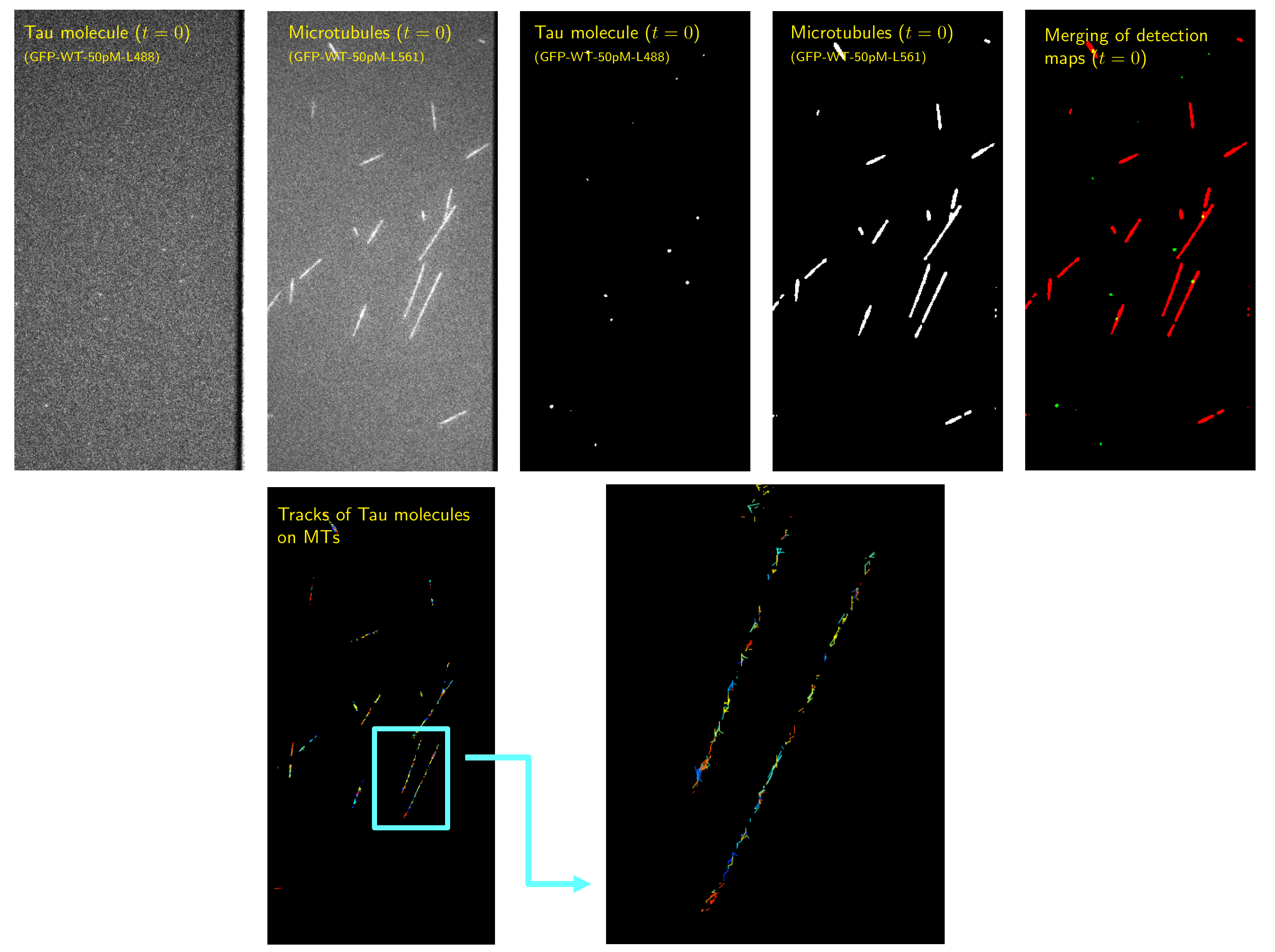
Collaborators: I. Arnal and V. Stoppin-Mellet (Institut of Neuroscience Grenoble).
8.2 Deep learning for object and event localization and classification in microscopy and image sequence analysis
8.2.1 Deep Learning for macromolecule identification in 3D cellular cryo-electron tomograms
Participants: Emmanuel Moebel, Charles Kervrann.
Cryo-electron tomography (cryo-ET) visualizes the 3D spatial distribution of macromolecules at nanometer resolution inside native cells. While this label-free cryogenic imaging technology produces data containing rich structural information, automated identification of macromolecules inside cellular tomograms is challenged by noise and reconstruction artifacts, as well as the presence of many molecular species in the crowded volumes. Last year, we developed a computational procedure that uses artificial neural networks to accurately localize several macromolecular species in cellular cryo-electron tomograms. The DeepFinder algorithm (see Fig. 8) is significantly faster than template matching, and performs better than other competitive deep learning methods at identifying macromolecules of various sizes in both synthetic and experimental datasets. This year, we demonstrated that, on cellular cryo-ET data, DeepFinder was able to localize membrane-bound and cytosolic ribosomes (3 MDa), rubisco (500 kDa soluble complex), and photosystem II (500 kDa membrane complex) with comparable accuracy to expert-supervised ground truth annotations. DeepFinder is currently used to detect smaller macromolecules such as 200 kDa disk-shaped nucleosomes.
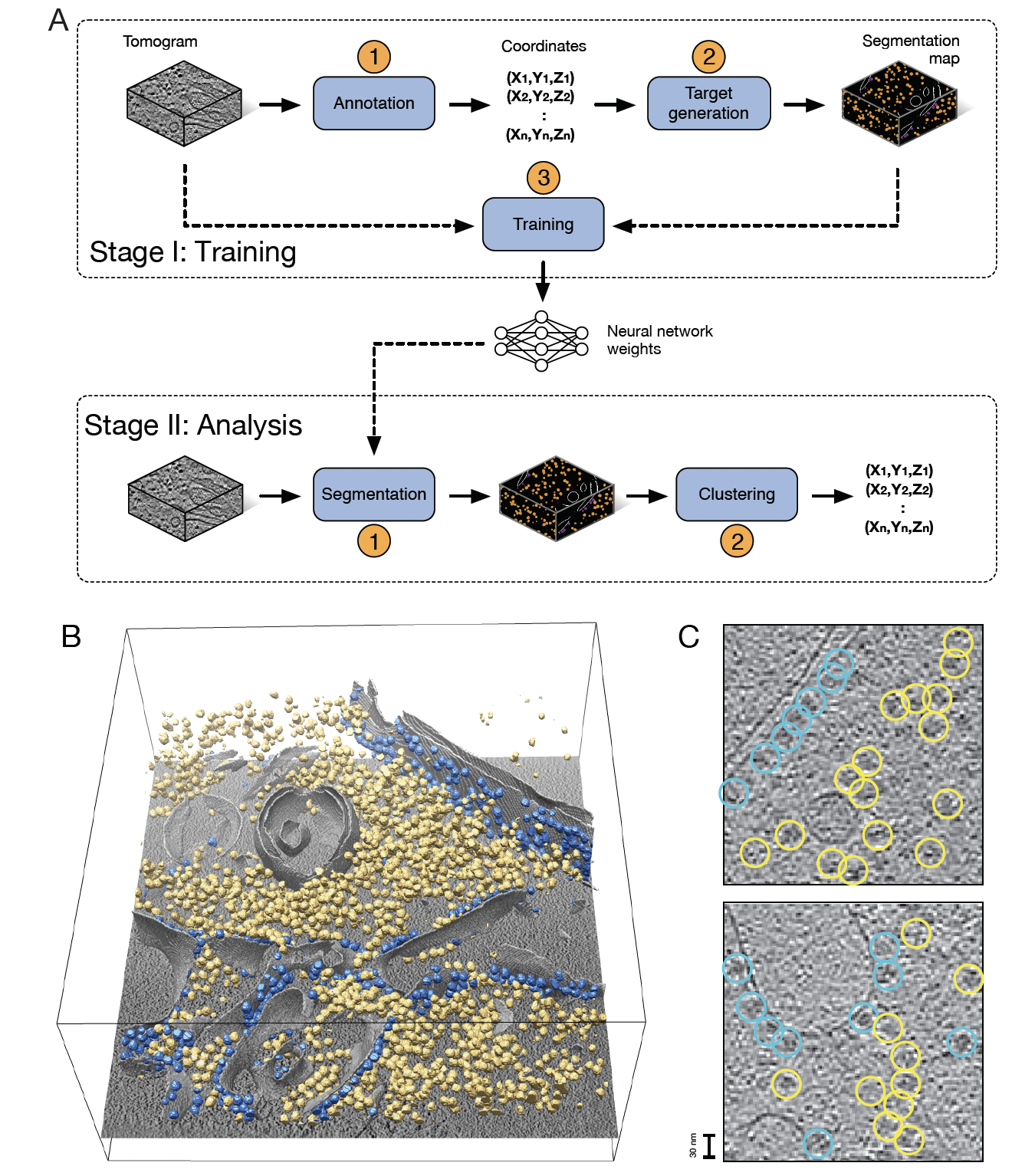
Collaborators: B. Engel, S. Pfeffer and W. Baumeister (Max Planck Institute, Biochemistry Department, Martinsried, Germany); C.O Sorzano (Biocomputing Unit, CSIC, Madrid, Spain); P. Schultz and M. Eltsov (IGBMC Strasbourg); D. Levy and A. Bertin (UMR168 CNRS, Institut Curie, Paris).
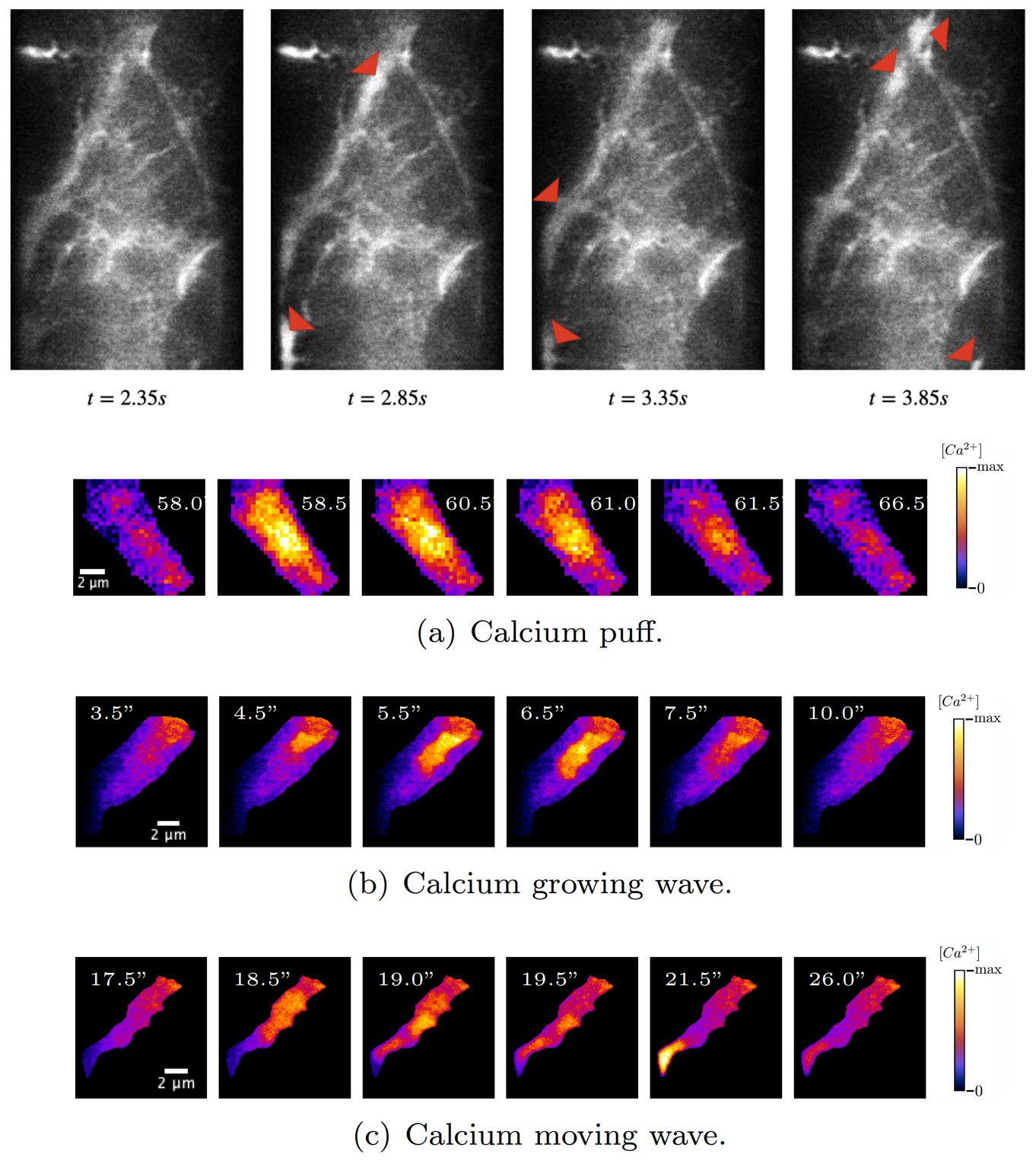
8.2.2 Convolutional Neural Networks algorithms for calcium signal segmentation in astrocytes observed with 3D lattice light sheet fluorescence microscopy
Participants: Anais Badoual, Charles Kervrann.
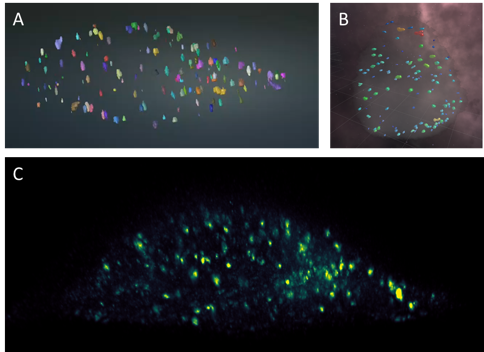
Astrocytes are glial cells in the central nervous system that recently emerged as key partners of neurons for the processing of information. Astrocytic calcium signals are involved in many key brain functions (e.g., memory and learning), and their alterations can lead to brain diseases. These signals also exhibit an important spatiotemporal diversity, and it is still unknown whether this variability relates to their role in distinct neurobiological functions. Not surprisingly, decoding this calcium code is a leading topic in neuroscience. The challenge is that the recording of astrocytic calcium activity is an intricate task that requires high resolution in both space and time. Indeed, most signals occur in fine astrocyte ramifications (50 - 200nm) and last between milliseconds to tens of seconds. Until recently, the quantification of intracellular propagation of calcium signal in astrocytes with fluorescent calcium indicators has been restricted to two dimensions, either 2D cell cultures or 2D slicing of a 3D setup. Unfortunately, key information can be lost by ignoring the third dimension, such as the number of calcium signals orchestrated in a single astrocyte and the location from which they originated. The recent emergence of lattice light sheet microscopy (LLSM) now enables a high spatio-temporal resolution with low phototoxicity (see Fig. 9). This finally provides a refined 3D imaging of the dynamical behavior of calcium signals in astrocytes inside living brain slices and in the intact mouse brain in vivo. Unfortunately, the community is currently lacking of image analysis tools to detect, segment and quantify these signals in LLSM images. In this context, we developed an image processing tool for neurobiologists which 1) detects and segments calcium signals in 3D+time LLSM images, and 2) classifies these signals based on their 3D space-time morphological characterization. To do so, we focused on unsupervised 3D convolutional network and machine learning techniques.
Collaborators: M. Arizono and V. Nägerl (Interdisciplinary Institute for Neuroscience, UMR5297 CNRS, Bordeaux); A. Denizot (Computational Neuroscience Unit, Okinawa Institute of Science and Technology, Onna, Japan); H. Berry (EPC beagle, Inria Rhone-Alpes, Villeurbanne).
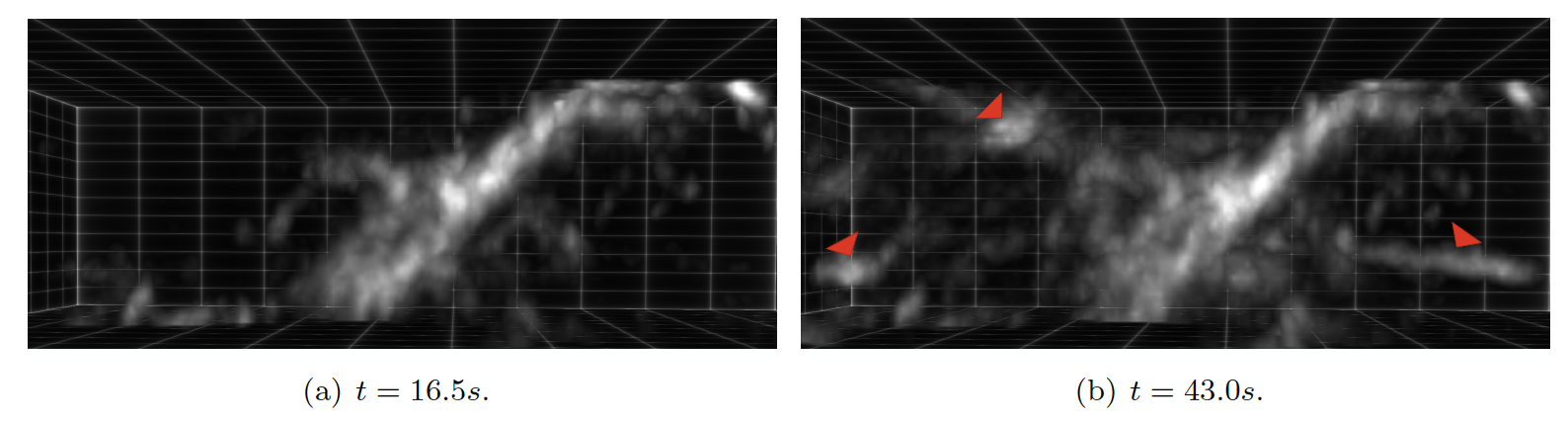
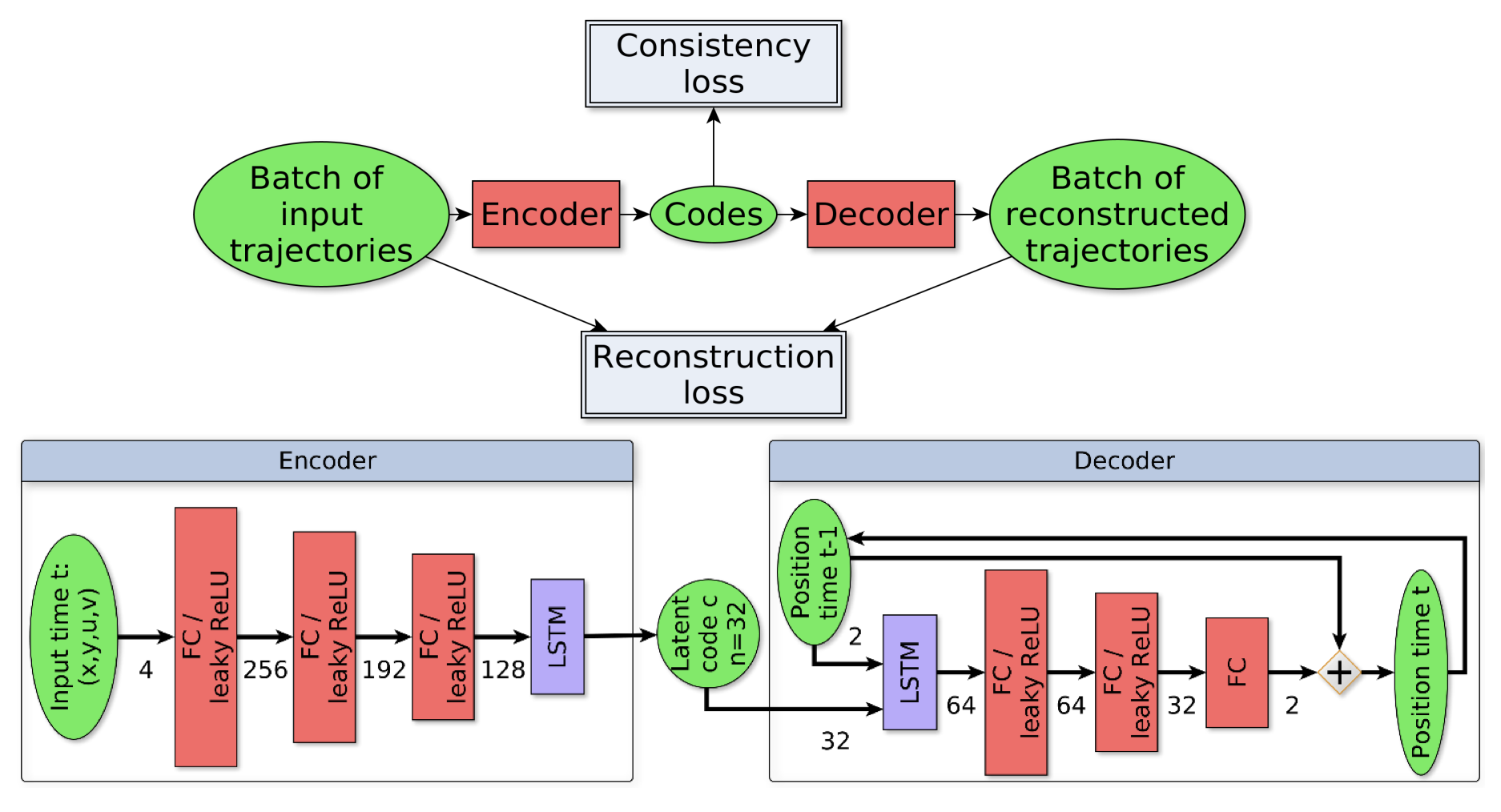
8.2.3 Trajectory saliency detection using consistency-oriented latent codes
Participants: Léo Maczyta, Patrick Bouthemy.
In this work, we are concerned with the detection of progressive dynamic saliency from video sequences. More precisely, we are interested in saliency related to motion and likely to appear progressively over time. It can be relevant to trigger alarms, to dedicate additional processing or to detect specific events. Trajectories represent the best way to support progressive dynamic saliency detection. Accordingly, we talk about trajectory saliency detection. A trajectory will be qualified as salient if it deviates from normal trajectories that share a common motion pattern related to a given context. First, we need a compact while discriminative representation of trajectories. We adopt a (nearly) unsupervised learning-based approach, summarized in Fig. 10. The latent code estimated by the recurrent LSTM-based auto-encoder provides the desired representation. In addition, we enforce code consistency for normal (similar) trajectories through the auto-encoder loss function. The distance of the trajectory code to a prototype code accounting for normality is the means to detect salient trajectories. We have validated our trajectory saliency detection method on synthetic and real trajectory datasets. Our method outperforms existing methods on several scenarios drawn from the publicly available dataset of pedestrian trajectories acquired in a railway station 35. We carried out additional experiments on particle trajectories computed from 2D fluorescence TIRF microcopy sequences. Trajectories corresponding to particles undergoing directed motion were acting as salient trajectories among Brownian and confined motions.
Reference: 34
Collaborators: O. Le Meur (Percept team, IRISA, Rennes).
8.2.4 Attention mechanisms and dynamic saliency in image sequences
Participants: Etienne Meunier, Patrick Bouthemy.
In this work, we aim to define attention mechanisms in image sequences driven by motion information. The goal is to highlight moving elements of interest within an overall motion in the image or among other individual motions exhibiting no specificity. This can also be referred to as dynamic or video saliency paradigms. The first step is to leverage the convolutional neural network we described in 39 for detecting the presence of motion saliency in every frame of a video. The idea is to predict the image location of salient moving objects by finding image areas contributing to the network decision. Then, a motion-based partition of the image is used to delineate precisely their location. To this end, we have designed a network that segments the optical flow map into several regions that have coherent motions, while estimating a quadratic motion model per region and determining the number of moving regions. Moreover, salient apparent motion may arise in two situations: due to independently moving objects (the one we are interested in) or due to static objects in the scene foreground if the camera is moving. We can predict if such a salient region is actually related to an independent motion in the scene or not by leveraging geometric properties of the flow linked to the camera motion after canceling the camera rotation. This method does not require training, and again exploits the segmentation of the flow.
8.3 Simulation and visualization of 3D+time microscopy data
8.3.1 Simulation of astrocytic calcium dynamics in Lattice Light Sheet Microscopy images
Participants: Anais Badoual, Charles Kervrann.
Astrocytes regulate neuronal information processing through a variety of spatio-temporal calcium signals. Recent advances in calcium imaging have started to shine light on astrocytic activity, but the complexity and size of the recorded data strongly call for more advanced computational analysis tools. Their development is currently hindered by the lack of reliable, labeled annotations that are essential for the evaluation of algorithms and the training of learning-based methods. To solve this labeling problem, we have designed a generator of 2D/3D lattice light sheet microscopy (LLSM) sequences which realistically depict the calcium dynamics of astrocytes (see Fig. 11). By closely modeling calcium kinetics in real astrocytic ramifications, the generated datasets open the door for the deployment of convolutional neural networks in LLSM.
Reference: 21
Collaborators: M. Arizono and V. Nägerl (Interdisciplinary Institute for Neuroscience, UMR5297 CNRS, Bordeaux); A. Denizot (Computational Neuroscience Unit, Okinawa Institute of Science and Technology, Onna, Japan); M. Ducros (Bordeaux Imaging Center, UMS3420 CNRS, US04 INSERM, Bordeaux, France); H. Berry (EPC beagle, Inria Rhone-Alpes, Villeurbanne).
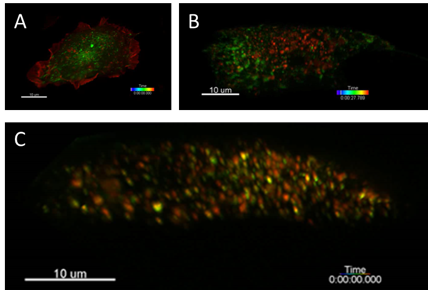
8.3.2 Monitoring and visualization of intracellular (endocytosis, exocytosis) processes
Participants: Cesar Augusto Valades Cruz, Ludovic Leconte, Jean Salamero.
The study of the whole cell dynamics of endocytic/exocytic-recycling events has proven difficult until recently because of lack of sensitivity, limited speed, photobleaching and phototoxicity associated with conventional imaging modalities. The Lattice Light Sheet Microscope (LLSM) allows overcoming these difficulties yet reaching high spatial resolution. In addition, this imaging technique and 3D-tracking will allow to look at molecular machinery throughout the full sequence of events that lead to exocytic fusion event or endocytic carrier formation, from initial membrane recruitment and budding and intracellular trafficking throughout the entire endocytic membrane system. An integrated framework based in 3D single particle tracking to study the coordination of vesicle recycling from the endosomal recycling compartment up to the plasma membrane has been developed. This method is based on U-Track and cmeAnalysis from Danuser Team. This method has been applied to characterize a cell adhesion molecule, CD166, endocytosis and its interplay with bar-domain proteins (see Fig. 12).
Moreover, we have got preliminary results of the coordination of vesicle recycling from the endosomal recycling compartment up to the plasma membrane using LLSM imaging and 3D tracking. These include the imaging of exocytosis events such as Transferrin Receptor recycling, coincidences of diverse actors involved in one particular mechanism (such rab proteins) or in the coordination of multiple mechanisms such recycling of endosomes and cytoskeleton remodeling in moving cells (see Fig. 13).
Reference: 20
Collaborators: L. Johannes (U1143 INSERM / CNRS-UMR 3666, Institut Curie, PSL Research University).
![: CD 166 endocytosis in U2OS cells [17]. (A) Maximum Intensity projection of CD166 (red) and clathrin (green). (B) Detected spots of CD166. (C) Average endocytic intensity traces per lifetime and classification of endocytic events for CD166 and Tf uptake. (D) Representative 2D projections of acquired 3D stacks of cargo (red) and clathrin (green) and overlay (yellow).](/rapportsactivite/RA2020/serpico/WEB_IMG/LLSMendocytosis.png)
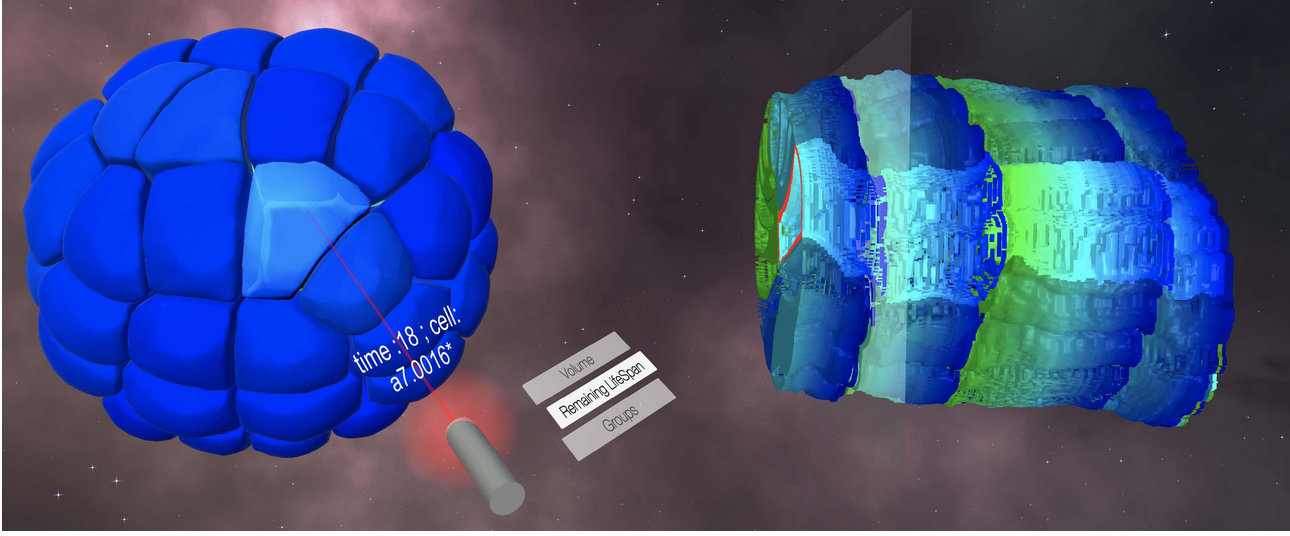
8.3.3 Immersive interaction and visualization of temporal 3D data
Participants: Gwendal Fouché, Cesar Augusto Valades Cruz, Ludovic Leconte, Jean Salamero, Charles Kervrann.
Nowadays, the detection and visualization of important localized events and process in multidimensional and multi-valued images, especially in cell and tissue imaging, is tedious and inefficient. Methods from the field of Immersive Analytics could be beneficial for biological imaging, where large time-varying datasets are more and more present, and have emerging constraints from the growing spatio-temporal resolutions.
In this context, we have proposed an extension of the Space-Time Cube (STC) visualization technique to spatio-temporal 4D data, named Space-Time Hypercube (STH). The STH approach considers that the data to visualize lays in a 4D hypercube with three spatial dimensions and one temporal dimension. We then project this 4D volume into a 3D STC relying on a user-defined cross-section on the spatial dimension. This cross-section is computed along the temporal dimension and the results are stacked, yielding a 3D volume which can be directly rendered and enriched with numerical and categorical data. The improved depth perception thanks to stereoscopic rendering improves the visual extraction of meaningful structures, and the 3D user interfaces and interaction methods allow the user to explore and manipulate naturally the STC from different perspectives in the Virtual Environment.
Experimental results suggested that this visualization method could help identifying behaviors of multiple cells and events with a large temporal component (see Fig. 14). Meanwhile, ). This VR system allows an immersive interaction and visualization of 3D single particle tracking of Rab5 endosomes in the whole cell (see Fig 15). Our objective here was to follow and manipulate fission-fusion events of early endosomes, a function led by Rab5 and its effectors, to geolocalize these events in the whole cell, and further annotate them in conjunction with the behavior of "mother/daughter" endosomes tracks at the whole cell level, thanks to the quasi-isotropic nature of LLSM images.
Collaborators: F. Argelaguet (EPC hybrid, Inria Rennes); E. Faure (Laboratory of Computer Science, Robotics and Microelectronics of Montpellier).
9 Bilateral contracts and grants with industry
9.1 Bilateral contracts with industry
9.1.1 Contract with Fourmentin-Guilbert Foundation: Macromolecule detection in 3D cellular cryo-electron tomograms
Participants: Emmanuel Moebel, Charles Kervrann.
Funding: Fourmentin-Guilbert Foundation
Duration: 5 months (Dec 2019 – Apr 2020)
Collaborators: D. Larivière & E. Fourmentin (Fourmentin-Guilbert Foundation), B. Engel, A. Martinez and W. Baumeister (Max Planck Institute, Martinsried, Germany).
The objective of the project is to improve the DeepFinder software dedicated to the detection and identification of macromolecules within 3D cellular cryo-electron tomograms. In collaboration with Fourmentin-Guilbert Foundation, the goal is to build cellular atlases of several organisms from localizations of macromolecules (see Section 8.2.1 and Software DeepFinder in Section 7.1.5).
9.1.2 Contract with GATACA Systems: Super-resolution microscopy and in live cell imaging
Participants: Jean Salamero, Ludovic Leconte, Charles Kervrann.
Funding: GATACA Systems company
Duration: 48 months (2017 – 2020)
Collaborator: C. Gueudry (GATACA Systems)
The objective of the project is to transfer innovations for Multi-Angle TIRFM (using Azymuthal TIRFM from Ilas2/3) and collaborate as “β-Test site” for 3D-SIM in Nipkow disk microscopy.
9.1.3 Contract with CryoCapCell SA: 3D LIVE CLEM (Correlative Light and Electron Microscopy) to decipher fates and functions of exosomes in vivo
Participant: Jean Salamero.
Funding: DIM–ELICIT Empowering LIfe sCiences with Innovative Technologies (Région Ile de France)
Duration: 24 months (Oct 2018 – Sep 2020)
Collaborators: G. Van Niel (coordinator, Institute of Psychiatry and Neuroscience of Paris), G. Raposo (CNRS-UMR 144 Institut Curie PSL Research), X. Heiligenstein (CryoCapCell SA).
The objective of the project is to link dynamic biogenesis of intracellular membrane compartments with their ultrastructures. It combines fast high resolution photonic imaging ( MA-TIRFM and fast high pressure freezing for 3D cryoEM. It requires adapted registration methods in 3D, in order to navigate through the multiple scales.
9.2 Bilateral grants with industry
9.2.1 Contract with DGA: Motion saliency analysis in videos
Participants: Léo Maczyta, Patrick Bouthemy.
Funding: DGA (National Defense Agency) and Région-Bretagne
Duration: 36 months (Oct 2017 – Sep 2020)
This project funded by the DGA (Ministry of defense) and Région-Betagne concerns the PhD thesis (co-funding) carried out by Léo Maczyta. The goal is to develop motion saliency methods along three axes: temporal motion saliency detection, saliency map estimation, trajectory-based saliency detection (see Section 8.2.3).
9.2.2 Contract with IRSN: DeepSuN – Localization of chromosomal aberrations induced by nuclear radiation dose excess
Participants: Antonin Deschemps, Charles Kervrann.
Funding: IRSN (Institut de Radioprotection et Sureté Nucléaire) and Région-Bretagne
Duration: 36 months (Oct 2020 – Sep 2023)
Collaborator: Mohamedamine Benadjaoud (IRSN, Fontenay-aux-Roses)
This project funded by the IRSN (Institut de Radioprotection et Sureté Nucléaire) and Région-Betagne concerns the PhD thesis (co-funding) carried out by Antonin Deschemps. The goal is to develop statistical and deep learning and methods for localizing and classifying chromosomal aberrations observed in 2D microscopy images (blood test) and estimating radiation dose following a postulated nuclear reactor accident.
9.2.3 Contract with AIRBUS Defence and Space SAS: LION Chaine Image Elargie (LiChIE)
Participants: Sébastien Herbreteau, Etienne Meunier, Patrick Bouthemy, Charles Kervrann.
Funding: Bpifrance / Projets Structurants pour la Compétitivité (PSPC)
Duration: 54 months (Aug 2019 – Jan 2024)
Collaborators: M. Ortner and R. Fraisse (Defence and Space SAS)
This project funded by Bpifrance concerns the PhD theses carried out by Sébastien Herbreteau and Etienne Meunier. The goal is to develop statistical and deep learning and methods for image restoration in night conditions and saliency detection in image sequences, respectively. The resulting algorithms will be embedded in hardware plateforms for a next generation of observation satellites.
10 Partnerships and cooperations
10.1 International initiatives
10.1.1 Inria International Labs
Informal international partners
- Collaboration with Max-Planck Institute, Martinsried, Germany (with B. Engel, A. Martinez and W. Baumeister): Detection and segmentation of macromolecules in cryo-electron tomography (project in progress with E. Moebel and C. Kervrann).
- Collaboration with Biocomputing Unit, CSIC, Madrid, Spain (with C.O. Sorzano): Integration of DeepFinder software into the Scipion platform dedicated to cryo-electron microscopy image analysis (project in progress with E. Moebel and C. Kervrann).
- Collaboration with the Cambridge Advanced Imaging Centre, Cambridge, UK (with L. Muresan): dense mapping for transcription factors analysis in single molecule localization microscopy (project in progress with A. Salomon and C. Kervrann).
- Collaboration with the PKU University, Institute of Molecular Medecine, Beijing, Peoples Republic of China. (with L. Chen and YM Liu): 3D reconstitution of the biogeneisis of Endoplasmic Reticulum-Plasma Membrane Contact Sites- ER-PM MSCs upon Ca2+ store depletion or replenishment (collaboration funded by a contract between PSL and PKU for exchange of students, in progress with C.A. Valades Cruz and J. Salamero)
10.2 European initiatives
10.2.1 Collaborations in European programs, except FP7 and H2020
ESFRI initiative program: EuroBioImaging
Participants: Charles Kervrann, Jean Salamero.
Coordinator: J. Eriksson (Turku University, Finland).
Funding: Member states of the European Union.
Partners: 15 European countries in 2020.
As a member of the National Research Infrastructures (RI) France BioImaging, serpico is involved in the ESFRI Euro-BioImaging (https://
EIT Digital program: Photodermatoses
Participants: Sylvain Prigent, Charles Kervrann.
Duration: 12 months (Nov 2019 – Oct 2020)
Funding: EIT Digital
Partners: UVisio and Nobleo Projects B.V., Eindhoven, The Netherlands.
serpico is involved in a European project which aims at developing a connected wearable device for diagnosis and treatment of photodermatoses. Using the data on skin sun sensitivity and UV exposure habits with machine learning algorithms (e.g. SVM) will enable to make more precise optimal sun exposure predictions for patients. The wearable device will be useful for a larger population to increase awareness around overexposure to UV as a main cause of sun damage and worst-case skin cancer.
10.3 National initiatives
10.3.1 France-BioImaging project
Participants: Sylvain Prigent, Cesar Augusto Valades Cruz, Patrick Bouthemy, Charles Kervrann, Ludovic Leconte, Jean Salamero.
Duration: 2011 – 2024
Funding: Investissement d'Avenir, ANR INBS-PIA1 2011 and “FBI Next Generation” (ANR program 2020-2024).
Coordinator: J. Salamero (UMS 3714 / CNRS-UMR 144, Institut Curie) (until June 2020).
Partners: CNRS, University of Paris-Diderot-Paris 7, Aix-Marseille University, University of Bordeaux, University of Montpellier, Institut Pasteur, Institut Curie, Inria, ENS Paris, University of Paris, UPMC, Ecole Polytechnique, Inserm.
serpico is a member of the French initiative, the so-called “France-BioImaging” (FBI) National Research Infrastructure which gathers several outstanding cellular imaging centers (microscopy, spectroscopy, probe engineering and signal processing). FBI is on the French Roadmap of Research Infrastructure (https://
10.3.2 ANR DALLISH project: Data Assimilation and Lattice LIght SHeet imaging for endocytosis / exocytosis pathway modeling in the whole cell
Participants: Antoine Salomon, Cesar Augusto Valades Cruz, Patrick Bouthemy, Frédéric Lavancier, Ludovic Leconte, Jean Salamero, Charles Kervrann.
Duration: 48 months (Oct 2016 – Sep 2020).
Funding: ANR (Agence Nationale de la Recherche) PRC (Collaborative Research Project).
Coordinator: C. Kervrann.
Partners: Inria (serpico, beagle, fluminance teams), INRA MaIAGE Unit Jouy-en-Josas, Institut Curie (CNRS-UMR 144 & U1143 INSERM / UMR 3666) Paris.
Cutting-edge Light Lattice Sheet microscopy represents the novel generation of 3D fluorescence microscopes dedicated to single cell analysis, generating extraordinarily high resolved and sharp, but huge 3D images and videos. One single live cell experiment in one single biological condition can result into up to one terabyte of data. The goal of the project is to develop new paradigms and computational strategies for image reconstruction and 3D molecule motion estimation and tracking. Furthermore, establishing correspondences between image-based measurements and features, stochastic motion models, and underlying biological and biophysical information remains a challenging task. In a larger perspective, the quantitative description of image data corresponding to protein transport will be a prerequisite for understanding the functioning of a cell in normal and pathological situations including cancer, viral infection and neurodegenerative diseases (see Sections 8.1.4, 8.1.5, and 8.3.2).
10.3.3 Inria DEFI NAVISCOPE: image-guided NAvigation and VISualization of large data sets in live cell imaging and microCOPy
Participants: Gwendal Fouché, Cesar Augusto Valades Cruz, Ludovic Leconte, Anais Badoual, Jean Salamero, Charles Kervrann.
Duration: 60 months (2018 – 2022)
Funding: Inria
Coordinator: C. Kervrann.
Partners:aviz Inria team (Saclay); beagle Inria team (Lyon), hybrid Inria team (Rennes), morpheme Inria team (Sophia-Antipolis); mosaic Inria team (Lyon), parietal Inria team (Saclay), serpico Inria team (Rennes); MaIAGE INRA Unit (Jouy-en-Josas); CNRS-UMR 144, Institut Curie, PSL Research University (Paris).
In the frame of the "Naviscope" IPL project (https://
- Novel machine learning methods able to detect the main regions of interest, and automatic quantification of sparse sets of molecular interactions and cell processes during navigation to save memory and computational resources.
- Novel visualization methods able to encode 3D motion and deformation vectors and dynamics features with color and texture-based and non-sub-resolved representations, abstractions, and discretization, as used to display 2D motion and deformation vectors and patterns.
- Effective machine learning-driven navigation and interaction techniques for complex functional 3D+Time data enabling the analysis of sparse sets of localized intra-cellular events and cell processes (migration, division, etc.).
Meanwhile, we address the technological challenge of gathering up the software developed in each team to provide a unique original tool for users in biological imaging, and potentially in medical imaging.
10.4 Regional initiatives
10.4.1 Motion saliency analysis in videos
Participants: Léo Maczyta, Patrick Bouthemy.
Funding: DGA (National Defense Agency) and Région-Bretagne
Duration: 36 months (Oct 2017 – Sep 2020)
See Section 9.2.1.
10.4.2 Deep-SuN – Localization of chromosomal aberrations induced by nuclear radiation dose excess
Participants: Antonin Deschemps, Charles Kervrann.
Funding: IRSN (Institut de Radioprotection et Sureté Nucléaire) and Région-Bretagne
Duration: 36 months (Oct 2020 – Sep 2023)
See Section 9.2.2.
11 Dissemination
11.1 Promoting scientific activities
11.1.1 Scientific events: organization
Member of the organizing committees
-
Anais
Badoual was organizer of the minisymposium “Bio-imaging: new opportunities for mathematical and computational advances”
for the workshop on “Mathematical Signal and Image Analysis”, Germany, March 30 - April 2, 2020 (canceled two weeks before due to Covid-19) (http://
www-m15. ma. tum. de/ Allgemeines/ MSIA20) - Patrick Bouthemy was member of the organizing committee of the BioImage Computing (BIC) workshop in conjunction with ECCV 2020 on-line, Aug 23, 2020.
- Jean Salamero was co-organizer of the ReTUBI (Twin H2020) Super Resolution meeting and Imaging “BioImaging Course”, Lisbon, Portugal (Virtual Meeting), Oct 20-23, 2020.
- Jean Salamero was co-organizer of the Global BioImaging 2020 “EOE.V”. Global BioImaging – Exchange of Experience V – Virtual Meeting, Sep 8-9, 2020 (Time zone Japan).
11.1.2 Scientific events: selection
Reviewer
- Anais Badoual was reviewer for IEEE ISBI'2020.
- Charles Kervrann was reviewer for IEEE ISBI'2020.
11.1.3 Journal
Member of the editorial boards
- Patrick Bouthemy is co-editor in chief of the open access journal "Frontiers in Computer Science", section "Computer Vision and Image Analysis" (until July 2020).
Reviewer - reviewing activities
- Charles Kervrann was reviewer for Acta Crystallographica (Section D).
- Jean Salamero was reviewer for Nature Methods, Scientific Reports, PLoS Computational Biology, and J. Microscopy.
- Cesar Augusto Valades Cruz was reviewer for Bioinformatics, PLoS Computational Biology, and J. Physical Chemistry Letters.
11.1.4 Invited talks
Charles Kervrann:
- – "Statistical and computational methods for intracellular trajectory analysis in fluorescence microscopy", Key-Note Talk, Quantitative BioIamging (QBI) conference, Jan 6-9 2020, Oxford, UK.
Jean Salamero:
- – "Live cell imaging in High Space-time Resolution", ReTUBI (Twin H2020) Super Resolution meeting and Imaging, Oct 20-23 2020, Lisbon, Portugal. (virtual meeting)
- – "Open access to Bioimaging infrastructures: Requirements, opportunities and challenges", Spanish and Portugese Advanced Optical Microscopy Meeting (SPAOM2020), Nov 24-27 2020. (virtual meeting)
11.1.5 Leadership within the scientific community
-
Charles
Kervrann is member of the executive board of the CNRS GdR ImaBio (2588 - Microscopie Fonctionnelle du Vivant).
- Patrick Bouthemy is member of the board of AFRIF (Association Française pour la Reconnaissance et l'Interprétation des Formes).
11.1.6 Scientific expertise
- Patrick Bouthemy was head of the jury for the AFRIF thesis prize (2019, 2020).
- Jean Salamero was was reviewer for European Research Council, FNS (Switzerland), NOW (Netherlands), ANR, INCA, ARC, Regional councils, and ITMO Cancer program.
11.1.7 Research administration
Charles Kervrann:
- – Member of the executive board of the project committee "Bureau du Comité des Projets" of the Inria Rennes - Bretagne Atlantique centre since 2010.
-
–
Co-head of the "BioImage Informatics" node (ANR France-BioImaging project http://
france-bioimaging. org/), National Research Infrastructure"for Biology and Health) since 2011.
Patrick Bouthemy:
-
–
Head of Excellence Lab (Labex) CominLabs (http://
www. cominlabs. ueb. eu) since April 2014. -
–
Deputy member of the board of directors and member of the Selection and Validation Committee of the Images & Réseaux competitivity cluster (http://
images-et-reseaux. com/). - – Member of the Research Committee of IMT Atlantique.
- – Head of the ANR evaluation committee for the 2020 call for proposals regarding the Artificial Intelligence field.
Jean Salamero:
-
–
Director of the National Research Infrastructure in Biology and Health, France BioImaging (2015-2020) (https://
france-bioimaging. org/) (until June 2020). - – Director of the Service Unit UMS 3714 (2015-2020) CEMIBIO CNRS-Institut Curie (until June 2020).
- – Member of the Advisory Committee for the creation of the National Biomedical Imaging Center (NBIC@Beijing, Republic of China) since 2017.
- – Member of the Advisory Board of EMBL Core Facilities.
- – Member of the Steering Committee of the DIM-ELICIT program (Région Ile de France) since 2016.
- – Member of the Qlife “Institut Convergences” since 2018.
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
Anais Badoual:
- – PhD degree: Image analysis (supervision and correction of exercise sessions), 6 hours, Gothenburg University, Sweden.
Charles Kervrann:
- – Master: From Bioimage Processing to BioImage Informatics, 5 hours, coordinator of the module (30 hours), Master 2 Research IRIV, Telecom-Physique Strasbourg and University of Strasbourg.
- – Master: Analysis of Image Sequences, 9 hours, Master 2 Research SISEA, University of Rennes 1.
- – Engineer Degree and Master 2 Statistics and Mathematics: Statistical Models and Image Analysis, 37 hours + 15 hours (TP, Yunjiao Lu), 3rd year, Ecole Nationale de la Statistique et de l'Analyse de l'Information (ENSAI), Rennes.
11.2.2 Supervision
- Léo Maczyta (PhD defended in Nov 2020): Motion saliency in video sequences, supervised by P. Bouthemy and O. Lemeur (Team Percept, IRISA, Rennes)).
- Yunjiao Lu (PhD in progress): Intracellular dynamics and super-resolution imaging: analysis of bacteria wall at the molecular scale (started in October 2017, supervised by C. Kervrann, A. Trubuil and R. Carballido-Lopez (INRAE, Jouy-en-Josas)).
- Antoine Salomon (PhD in progress): Statistical aggregation for image analysis in fluorescence microscopy and super-resolution (started in November 2017, supervised by C. Kervrann).
- Gwendal Fouché (PhD in progress): Immersive interaction and visualization of temporal 3D data (started in October 2019, supervised by C. Kervrann, F. Argelaguet (EPC hybrid, Inria, Rennes), and E. Faure (LIRMM, Montpellier)).
- Sébastien Herbreteau (PhD in progress): Deep Learning for image restoration in night conditions (started in october 2020, supervised by C. Kervrann).
- Antonin Deschemps (PhD in progress): Statisical leanring for localization and classification of chromosomal aberrations induced by nuclear radiation dose excess (started in october 2020, supervised by C. Kervrann).
- Etienne Meunier (PhD in progress): Deep Learning for saliency detection in image sequences (started in September 2020, supervised by P. Bouthemy).
- Claire-Jing Rouchet (PhD in progress): Role of membrane microdomains in bacterial morphogenesis (started in October 2020, supervised by R. Carballido-Lopez, C. Billaudeau (INRAE, Jouy-en-Josas), C. Kervrann).
11.2.3 Juries
Charles Kervrann:
- – Reviewer of the HdR of T. Walter (University PSL, MINES-ParisTech).
- – Reviewer of the PhDs of C. Jaques (EPFL, Lausanne, Switzerland, supervised by J.-M. Odobez and M. Liebling), J. Boyd (University PSL, MINES-ParisTech, supervised by T. Walter).
Patrick Bouthemy:
- – Reviewer and President of the PhD thesis committee of T. Champetier (University PSL, ENS Paris, supervised by A. Genovesio).
12 Scientific production
12.1 Major publications
- 1 articleAdaptive spot detection with optimal scale selection in fluorescence microscopy imagesIEEE Transactions on Image Processing2411November 2015, 16
- 2 articleAn extended model of vesicle fusion at the plasma membrane to estimate protein lateral diffusion from TIRF microscopy imagesBMC Bioinformatics1812017, 352
- 3 article A patch-based method for repetitive and transient event detection in fluorescence imaging PLoS ONE 5 10 Oct 2010
- 4 articlePatch-based nonlocal functional for denoising fluorescence microscopy image sequencesIEEE Transactions on Medical Imaging292Feb 2010, 442-453
- 5 articleStatistical analysis of particle trajectories in living cellsPhysical Review E 976June 2018, 1-20
- 6 article Fast live simultaneous multiwavelength four-dimensional optical microscopyProc Natl Acad Sci USA10737Sep 2010, 16016-16022
- 7 articleMotion Textures: Modeling, Classification, and Segmentation Using Mixed-StateSIAM Journal on Imaging Sciences64December 2013, 2484-2520
- 8 inproceedings PEWA: Patch-based Exponentially Weighted Aggregation for image denoising NIPS - Neural Information Processing Systems Neural Information Processing Systems Foundation Montreal, Canada December 2014
- 9 articleBackground Fluorescence Estimation and Vesicle Segmentation in Live Cell Imaging with Conditional Random FieldsIEEE Transactions on Image Processing242February 2015, 14
- 10 article Counting-based particle flux estimation for traffic analysis in live cell imaging IEEE Journal of Selected Topics in Signal Processing September 2015
12.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Doctoral dissertations and habilitation theses
Reports & preprints
12.3 Cited publications
- 35 inproceedingsSocially-aware large-scale crowd forecastingIEEE Conference on Computer Vision and Pattern Recognition (CVPR)Columbus, OH, USA,June 2014, 2211-2218
- 36 articleTotal Internal Reflection Fluorescent Microscopy in cell biologyTraffic22004, 4658--4668
- 37 articleImaging intracelluar fluorescent proteins at nanometer resolutionScience3132006, 1642--1645
- 38 articleLattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolutionScience34662082014, 1257998
- 39 articleCNN-based temporal detection of motion saliency in videosPattern Recognition Letters41282019, 298--305

