2024Activity reportProject-TeamCOMPO
RNSR: 202124080M- Research center Inria Centre at Université Côte d'Azur
- In partnership with:INSERM, Aix-Marseille Université, CNRS, CAC4 MARSEILLE - Institut Paoli-Calmettes
- Team name: COMPutational pharmacology and clinical Oncology
- In collaboration with:Centre de Recherche en Cancérologie de Marseille
- Domain:Digital Health, Biology and Earth
- Theme:Computational Neuroscience and Medicine
Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.3.2. Data mining
- A3.3.3. Big data analysis
- A3.4.1. Supervised learning
- A3.4.2. Unsupervised learning
- A3.4.5. Bayesian methods
- A6.1.1. Continuous Modeling (PDE, ODE)
- A9.2. Machine learning
Other Research Topics and Application Domains
- B1.1.8. Mathematical biology
- B2.2.3. Cancer
- B2.4.1. Pharmaco kinetics and dynamics
- B2.4.2. Drug resistance
1 Team members, visitors, external collaborators
Research Scientists
- Sebastien Benzekry [Team leader, INRIA, Researcher]
- Elias Ventre [INRIA, Researcher, from Oct 2024]
Faculty Members
- Joseph Ciccolini [Team leader, AMU APHM, Professor, HDR]
- Dominique Barbolosi [AMU, Professor]
- David Boulate [AMU APHM, Professor]
- Raphaelle Fanciullino [AMU APHM, Professor]
- Florence Gattacceca [AMU, Associate Professor]
- Laurent Greillier [AMU APHM, Professor]
- Athur Géraud [Institut Paoli Calmettes, Hospital Staff]
- Athanassios Iliadis [AMU, Emeritus]
- Xavier Muracciole [APHM, Hospital Staff]
- Anne Rodallec [AMU, Associate Professor]
- Sebastien Salas [AMU APHM, Professor]
Post-Doctoral Fellow
- Paul Dufosse [INRIA, Post-Doctoral Fellow, from Mar 2024]
PhD Students
- Salih BENAMARA [AMU]
- Anastasiia Bakhmach [INRIA]
- Celestin Bigarre [AMU, from Feb 2024]
- Celestin Bigarre [INRIA, until Jan 2024]
- Mohamed Boussena [AMU]
- Clarisse Buton [AMU, from Nov 2024]
- Mathilde Dacos [APHM]
- Anthéa Deschamps [AMU, until Oct 2024]
- Hafida Hamdache [INSERM]
- Govind Kallee [APHM, from Sep 2024]
- Clémence Marin [APHM]
- Linh Nguyen Phuong [AMU]
- Loic Osanno [APHM]
- Jessica Ou [AMU]
- Dorian Protzenko [APHM]
- Antonin Ronda [APHM, from Aug 2024]
Technical Staff
- Sarah Giacometti [AMU]
- Andrea Vaglio [INRIA, Engineer, from Nov 2024]
Interns and Apprentices
- Andréa Boniffay [AMU, Intern, from Feb 2024 until Jul 2024]
- Maroua Frikha [INRIA, Intern, from Mar 2024 until Sep 2024]
- Mohamed Ouloum [INRIA, Intern, from Jun 2024 until Aug 2024]
- Ruben Taieb [ENS DE LYON, Intern, until Mar 2024]
- Romain Zakrajsek [INRIA, from Sep 2024]
- Romain Zakrajsek [AMU, Intern, until Jun 2024]
Administrative Assistant
- Sandrine Boute [INRIA]
External Collaborators
- Alice Daumas [AMU]
- Romain Ferrara [AMU, from Aug 2024]
- Quentin Marcou [AMU]
- Caroline Plazy [CHU GRENOBLE-ALPES, from Feb 2024]
- Amelie Pouchin [CHU LA TIMONE]
- Thibaut Reichert [AMU, from Aug 2024]
- Ruben Taieb [ENS DE LYON, from Mar 2024 until Nov 2024]
- Romain Zakrajsek [AMU, from Jul 2024 until Aug 2024]
2 Overall objectives
We aim to optimize therapeutic approaches (i.e., controlling toxicities while ensuring a maximal efficacy) in oncology using mechanistic and statistical modeling (see Figure 1). These therapeutic approaches include immunotherapy, radiotherapy, chemotherapy, targeted therapies and their planification: combinations, sequences, intensification – densification, dosing and scheduling. Specifically, our research will be organized along three main axes:
- Quantitative modeling for personalized clinical oncology.
- Individualizing anticancer drugs regimen.
- Optimizing combinatorial strategies with immune checkpoint inhibitors.
Of note, in the Research Priorities document released by the American Society of Clinical Oncology in February 2021, “Developing and Integrating Artificial Intelligence in Cancer Research”, “Identifying Strategies That Predict Response and Resistance to Immunotherapies” and “Optimizing Multimodality Treatment for Solid Tumors” are listed as top-priorities, which fit quite well with our 3 axes.
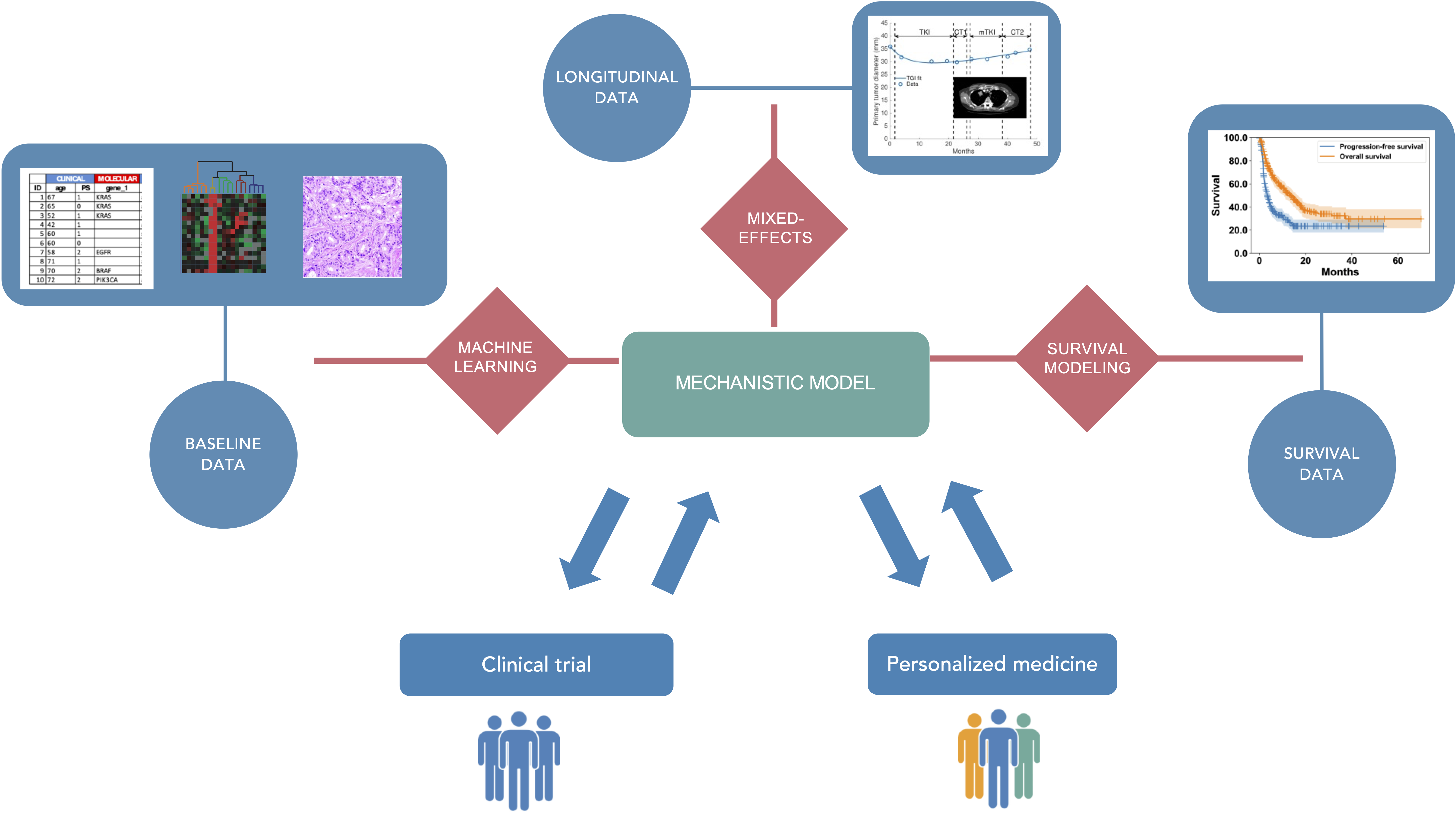
Mechanistic and statistical modeling for pharmacological and clinical oncology.
3 Research program
3.1 Scientific context and motivations
The project-team is based upon the development of model-driven clinical oncology as a means to optimize anticancer therapies. Despite continuous efforts to make available novel drugs beyond traditional cytotoxic chemotherapy (i.e., oral targeted therapies, biologics, immune checkpoint inhibitors), prognosis of outcome remains poor for many cancers. The dosing regimen of anticancer drugs given today remains largely empirical, because dose-finding studies are often performed using outdated, sub-optimal protocols (such as modified-Fibonacci dose-ranging protocols) or because concomitant administration is the rule when combining several drugs. Consequently, clinical oncologists struggle to refine the way they use the anticancer agents made at their disposal. For instance, it took several years of bedside practice to understand that paclitaxel in breast cancer patients should not be administrated using the officially approved 150 mg/m² every 3 weeks scheduling, but rather with an alternate 75 mg/m² weekly dosing 128. Similarly, multi-targets sunitinib is now given on a 50 mg two-weeks on / one-week off basis, rather than the officially approved four-weeks on / two-weeks off schedule 115. Elsewhere, several combinatorial strategies trials have failed to yield convincing results, mostly because of the lack of a strong rationale regarding the best way to sequence treatments 109. Globally, clinical oncology today is still all about finding the best way to treat patients ensuring an optimal efficacy / safety balance.
After having long been limited to cytotoxic chemotherapy (in addition to surgery and radiotherapy), the arsenal of anti-cancer agents has dramatically increased over the last two decades. Indeed, major advances in the understanding of cancer biology that occurred in 1) the discovery and quantification of (epi)genetic alterations leading to targeted therapy and 2) the realization of the importance of the non-cancer cell components of tumors, i.e., the tumor micro-environment and tumor immunity, have helped to identify novel targets. Drugs targeting the tumor vasculature (e.g., first-in-class bevacizumab, approved in the mid-2000’s 97) or tumor immunity (e.g., immune checkpoint inhibitors (ICI) such as first-in-class ipilimumab, approved in the early 2010’s 126) represent groundbreaking innovations in oncology. ICIs in particular are considered as game-changing drugs because diseases with once dismal prognosis (e.g., metastatic melanoma, non-small cell lung cancer (NSCLC), kidney cancer or head and neck cancer 148) now show 20-40% of 5-years survival. Nevertheless, these impressive results are limited to a minority of patients in a limited number of cancers. In addition, no validated biomarker predictive of response has yet been identified, thus highlighting how early prediction of response and probability of future relapse are a critical, unmet medical need. The encouraging yet still insufficient clinical results of ICIs have led current clinical oncology to consider combinations of such immunotherapies with preexisting anti-cancer modalities: radiation therapy 135, cytotoxic 122, targeted 140 or anti-angiogenic therapies 147. However, the near-infinite possibilities of combinations in terms of sequencing, dosing and scheduling challenge the ability of classical trial-and-error methods to find appropriate modes of combination 108.
In addition, day-to-day clinical decisions made by oncologists are based on a large amount of information, coming from: 1) their own knowledge integrating years of clinical practice combined to updated literature and 2) objective data coming from multiple sources (demographic data, radiology, functional imaging, molecular biology, histology, biomarkers, blood counts, etc…). The large amount of clinical and biological data generated now in clinical oncology is not properly analyzed, because of the lack of appropriate models to picture the complexity of longitudinal observations. Oncologists lack a comprehensive framework and numerical software that could support decision of therapeutic strategy (e.g., to treat or not? to what extent? with what treatment (surgery, radiotherapy, systemic therapy)? in what order? etc.), especially when their time dedicated to examination of a given patient case is limited (e.g., in multidisciplinary meetings (RCP), or individual consultations). Furthermore, modeling is the only way to retrieve similar characteristics from very different experimental conditions and clinical protocols.
To address these major issues, our project-team aims at:
- guiding anticancer therapy by developing patient-specific predictive models (individual level);
- better designing clinical trials, in particular regarding combinatorial trials (population level).
3.2 Data
We use non-clinical and clinical data related with the pharmacology of anti-cancer agents and medical monitoring of the disease status. The former includes pharmacokinetics (drug levels in plasma (patients) and full body pharmacokinetics (animal models)), pharmacodynamics (efficacy, safety), pharmacogenetics (i.e., constitutional genetic polymorphisms affecting drug transport and metabolizing enzymes), pharmacogenomics (i.e., molecular and genetic alterations affecting tumor cells). The latter include demographics, anatomical imaging (e.g., tumor sizes derived from CT scan or MRI), functional imaging (e.g., positron emission tomography), histopathology quantifications, biological variables (such as kidney and liver functions or blood counts), immuno-monitoring data (flow cytometry) or cell free DNA. We will especially rely on real-world data (also termed fragmentary data) collected from patient routine monitoring by our members with hospital activity.
Experimental data are generated by the experimental wet-lab group (involving COMPO team members: AR, RF, JC), relying on state-of-the-art experimental pharmacokinetic laboratory fully equipped to perform in vitro and in vivo explorations of drug metabolism, pharmacokinetics and experimental therapeutics in oncology, including bioanalytical support and fluorescence/bioluminescence monitoring in rodents with highly specialized staff. Clinical pharmacokinetics and pharmacogenetics data are generated by the clinical pharmacology group (JC, RF), relying on the expertise of the clinical pharmacokinetics laboratory of the La Timone University Hospital of Marseille, an FDA-labelled, ISO15189-labelled facility with state-of-the-art bioanalytical resources to assay any kind of drugs or drug metabolites in patients. Specific data regarding cancer biology and pharmacodynamics (immunomonitoring, pharmacogenomics) are generated in collaboration with other CRCM teams.
Clinical data and additional biomarker data are collected from either clinical trials or real-world studies performed by hospital pharmacists and oncologists of the joint-team (RF, JC, SS, LG, XM) and their residents, or by other medical oncologist partners. We have strong collaborations and ongoing projects with pediatrics (Pr N. Andre), hematology (Dr G. Venton), nuclear medicine (Pr D. Taïeb) and radiotherapy (Pr L. Padovani). Importantly, the project-team is located near the INCa-labeled center for early clinical trials (CLIP2), thus facilitating the data collection and later, the implementation of modeling approaches in early clinical trials.
In addition, we also rely on publicly available data from online databases such as the TCGA (genomic data), the TCIA (imaging data) or data from clinical trials.
3.3 Mathematical methodology
Our primary objective is centered on the improvement of therapeutic strategies in oncology. Nevertheless, this brings novel methodological challenges requiring developments at the formal level within the generic field of modeling biological and pharmaco-patho-physiological systems. Difficulties to take into account include: the longitudinal profile of the quantities of interest; measurement uncertainty (requiring statistical considerations); difficulties in sampling the real processes leading to scarcity of the observed data and large inter-individual variability. Many specific problems in life science systems are very different from that encountered from physical modeling in industrial applications (e.g., mechanical engineering or energy).
To summarize our methodology, we are interested in modeling the dynamics of pharmaco-oncological processes (mechanistic modeling) and their inter-individual variability (statistical modeling). Our intended methodological contributions are: 1) to invent novel mechanistic models for complex physiological processes able to describe the effect of therapeutic intervention, 2) to design appropriate statistical frameworks for parameter estimation and description of inter-individual variability, 3) to test and validate the models against experimental and clinical data, 4) to combine state-of-the art machine learning (ML) methods with mechanistic models to integrate large dimension data.
3.3.1 Mechanistic modeling
Mechanistic models are defined here as mathematical constructs that describe physiological variables (e.g., plasma drug concentration, tumor size, or biomarkers) and their dynamics based on physical and biological principles (e.g., law of mass conservation). They describe the time profiles of the variables of interest by means of ordinary or partial differential equations (ODEs and PDEs, respectively) and are thus deterministic.
The main challenge of the modeling exercise is to find the appropriate balance between the degree of integration of biological phenomena (model complexity) and granularity of the data available (i.e., sampling time resolution, observed variables, spatio-temporal or only temporal measurements) ensuring the feasibility of parameter estimation. Indeed, cancer biology is extremely complex, involving processes at multiple temporal and spatial scales (intra- and inter-cellular, tissular, organism). It is thus tempting to build intricate models integrating as many phenomena as possible. Along these lines, the last decades have witnessed the proliferation of multiple such complex models. We see two shortcomings to this approach. First, in contrast with models of physical phenomena, the parameters of biological models are often not directly measurable and thus have to be estimated from fitting the models to experimental or clinical data. Therefore, their number has to be commensurate to the available data in order to ensure identifiability. Unfortunately, many complex models from mathematical oncology have too many parameters to be reasonably identified and have thus had a limited application in terms of biological insights or clinical applications. Second, complex, multiscale models are characterized by a reductionist point-of-view whereby general phenomena could be explained by decomposing them into elementary pieces. However, corresponding elementary experiments would not be suitable for quantification of the several homeostatic mechanisms involved in the whole real process. Thereby, we do not adhere to this reductionist vision and for modeling purposes we rather adopt a holistic approach considering the process as an indivisible whole.
To avoid the above-mentioned caveats, our methodology always starts from: 1) a clinically relevant medical problem but more importantly 2) the data available to build models.
In several instances, the mechanistic models are ordinary differential equations (ODEs). This is the case for the simplest type of experimental data that we generate, i.e., tumor growth kinetics. Departing from previous works establishing models for untreated experimental growth, we are now actively engaged into designing pharmacokinetics (PK)/ pharmacodynamics (PD) models of the effect of multiple therapies. These models have to account for the specificities of the drug delivery (e.g., nanoparticles), the biological effect of the treatments (e.g., cytotoxics, antiangiogenics or immunotherapies) and resistance to the therapy (either innate or acquired). The resulting models are novel nonlinear ODEs that need to be validated against the data and, when necessary, theoretically studied for their qualitative behavior. With the advent of immunotherapies, there has been a regain of interest to modeling tumor-immune interactions. Again, despite a wide literature on the subject, very few models have been validated against empirical data. A methodological objective is to establish and validate such models, including effect of immunotherapies.
Description of other phenomena are more adapted to partial differential equations (PDE) models. For instance, following an approach initiated by Iwata et al. 131, structured PDE models can be written for description of a population of metastases (see 4.3.2). Indeed, at the organism scale, cancer diseases are often characterized by a generalized (metastatic) state. However, few modeling efforts are currently focused on this aspect. The only validated models in large cohorts for systemic disease concern the sum of largest diameters as defined by the RECIST criteria 105. We aim to go beyond this state of the art by: 1) providing models of coupled tumor growth with interactions (and quantification of inter-lesion variability) 102, 103 and more importantly 2) developing models accounting not only for growth of the tumors, but also dissemination (birth of new lesions).
To date, most of the available and collectable clinical data about tumor growth and response to therapy consist of scalar data, often even limited to lesion diameters or sum of diameters. This is why we primarily focus our efforts on developing kinetic models of such data, the novelty coming from integrating other longitudinal biological data (e.g., from blood counts). Nevertheless, imaging data are now increasingly accessible and recent advances in image analysis allow the automatic segmentation of lesions make it possible to quantify the spatial shape and texture of tumors without a prohibitive cost for radiologists. This opens the way to develop spatially distributed PDE models of tumor dynamics. The existing models have largely remained unconfronted to data, apart from notable exceptions from the Inria MONC 112 and EPIONE 110 teams, as well as the Swanson 98 and Yankeelov 127 groups. Radiomics approaches quantifying heterogeneity in the images could bring additional information. We will rely on existing or establish collaborations with other dedicated Inria teams (EPIONE, MONC) for such purpose. COMPO would ideally bridge the gap between clinical studies and the Inria ecosystem. Finally, PDE models are also well adapted to describe intra-tumor drug penetration and we have recently developed such models for description of intra-tumor fluid flow and transport of antibody nano-conjugates 149.
3.3.2 Statistical modeling
Statistical models are defined here as mathematical constructs that describe the stochastic sources of variability in the data. They comprise both: 1) classical statistical models defining the functional and probabilistic relationship explicitly and 2) machine learning (ML) algorithms highly based on the data alone (e.g., tree-based models and associated ensemble methods or support vector machines) 124. We use such models for the following purposes: defining appropriate frameworks for parameter estimation; quantitative testing of biological hypothesis; addressing interindividual variability (using nonlinear mixed-effects (NLME) modeling); and building predictive models.
NLME – also termed the population approach in PK/PD modeling, or hierarchical modeling 133 – consists in assuming a statistical distribution of the parameters of the structural (often mechanistic) model, in order to describe longitudinal observations within a population of individuals. Instead of estimating individual parameters on a subject per subject basis – leading to identifiability issues in sparse data situations characteristic of longitudinal measurements in oncology – all data can be pooled together and a joint likelihood is obtained. Likelihood maximization becomes more complex than for classical nonlinear regression, nevertheless this problem has already been addressed by means of algorithms such as the deterministic first-order conditional expansion (FOCE) algorithm 134 or the stochastic approximation of the expectation-maximization algorithm (SAEM) 114. These algorithms are implemented in widely used software in the PMX community such as NONMEM® (Icon) or Monolix® (Lixoft), or R packages (e.g., saemix or nlme). Once the population distribution is estimated, empirical Bayes estimates (EBEs) can be derived for estimation of individual parameters. We also use the language Stan that implements state-of-the art Bayesian methods 106.
Departing from a general distribution of the parameters (often assumed log-normal) with quantified but unexplained interindividual variability, covariates are incorporated to explain this variability and build predictive models. This is traditionally done by means of linear models (possibly up to a functional transformation). However, with the increase in number of such covariates, the traditional tools and algorithms are limited. We thus develop advanced covariate models in NLME incorporating ML algorithms. Such methods require novel contributions. A possible lead is to first identify the EBEs and then use ML algorithms to predict these from the covariates 141. In other cases, ensemble models could be built from the heterogeneous sources of data, integrating one sub-model from EBEs identified from early data. Another, more challenging, avenue would be to adapt the parameter estimation algorithms like SAEM to include ML models in the covariate part.
In addition, because few data have been available longitudinally so far (i.e., small number of quantities measured at each time point), the current use of NLME relies on models with a small number of output variables. In this respect, modern clinical oncology studies bring new modeling and statistical challenges because many more quantitative data are collected at each time point (e.g., hundreds of variables from immuno-monitoring or possibly tens of thousands from circulating DNA, or radiomics features from imaging). Defining high-dimensional ODE models describing all the physiologically meaningful variables becomes intractable, therefore new methods are required. A possible avenue is to have a sequential approach, using first ML methods to reduce the dimension, then model the reduced number of variables. Another, more challenging, avenue would be to perform the two tasks (dimension reduction and temporal modeling) at the same time, and include this in an NLME framework for population estimation. The first part could be done using tools from unsupervised learning such as auto-encoders.
Following the availability of longitudinal tumor measurements, recent developments in the field of NLME have concerned joint modeling 117. This consists in modeling the longitudinal kinetics of a biomarker (e.g., tumor size) together with censored time-to-event data (e.g., overall survival) in a single step. Promising results have been obtained so far and we intend to develop methods beyond the state-of-the art in this area. This includes, in connection with above: 1) extension to models with emergence of new metastases, 2) integration of high-dimensional covariates and 3) high-dimensional longitudinal data. This Bayesian integration of data for updated survival predictions could lead to high impact results, as demonstrated by a recent publication in Cell 132.
Finally, we intend to bring the use of established ML tools to address concrete clinical problems emerging from the data collected in routine or clinical trials. Indeed, such data is so far analyzed using traditional statistical methods. ML algorithms could bring added value for predicting efficacy or toxicity from demographic, clinical and biological data.
3.4 Experimental therapeutics in oncology
The project-team is based upon generating experimental and clinical data to identify and test the models, and to provide proof-of-concept studies so as to validate the model-based dosing and scheduling prior to transposing them in patients. Historically, experimental therapeutics in oncology has relied on a wide variety of in vitro and in vivo models mimicking human cancer disease. In oncology, hundreds of in vitro models using cancer cell lines cultivated following 2D or 3D (spheroids) fashion, plus more sophisticated models with cancer cells enriched with fibroblasts or endothelial cells 142, eventually leading to complex organoids 121. Similarly, almost all kind of tumors can be tested in vivo, mostly in small rodents. In oncology, in vivo models are mostly based upon xenografting human tumors from established cell lines or from patient biopsies (patient-derived xenografts or PDX) so as to better mimic human pharmacology when testing active compounds next. To achieve this, several strains of immune-compromised mice have been successfully developed. Because immune checkpoint inhibitors do not exert direct anti-proliferative activity on cancer cells but are rather expected to harness tumor immunity, human xenografts in immuno-compromised mice is not anymore a suitable model. This has led investigators to shift towards immune-competent syngeneic mice models. Non-clinical experiments with drug candidates in immunotherapy mostly focus on deciphering the pharmacology of the targeted pathways, assessing the cytokine release potential, studying receptor occupancy, by using models the most likely to mimic tumor immunity in human. More sophisticated animal models such as human knock-in mice, immuno-avatar, hemato-lymphoid humanized mice or immune-PDX mice have been developed 145 (i.e., allowing to test immune checkpoint inhibitors in mice models combining human xenograft with relevant, humanized immunity and stroma cells) have been made available as well. Beyond generating data on efficacy such as reduction in primary tumor mass or metastatic spreading, experimental models help providing as well in depth knowledge on human and animal target cells, in vitro and in vivo concentration-effect studies, search for biomarkers, plus the most comprehensive knowledge on animal vs. human differences on dose – exposure – effects relationships and finally drug distribution throughout the body, target expression, affinity of target-binding and intrinsic efficacy, duration and reversibility of the effects. In particular, animal drug metabolism and pharmacokinetics (i.e., exploration of liver metabolism and distribution / absorption processes using in vitro or in vivo dedicated models) help understanding the disposition and distribution of the drug in the body throughout time, especially its ability to target tumor tissues (i.e., in vivo distribution in tumor-bearing mice) and help understanding sources of pharmacokinetic variability. All this information requires state-of-the-art techniques for measuring drugs and drug metabolites into biological fluids in tissues, such as fluorescence-imaging, high-performance liquid chromatography or liquid-chromatography-mass spectrometry bioanalysis. Our team has proven track records in the field of experimental therapeutics in oncology, with two PhDs on developing anticancer nanoparticles in breast and colorectal cancer 120, 144 plus experiments on model-driven way to combine anti-angiogenics with cytotoxics in breast and lung cancers 130, 137, 146, model-driven determination of alternate dosing in neuroblastoma, or methodological studies on monitoring tumor growth 138.
3.5 Axis 1: Quantitative modeling for personalized clinical oncology
The different steps of patient therapeutic management by clinicians consist mostly of: diagnosis, estimation of the extension of the disease, choice of therapy and evaluation of the therapy (efficacy, toxicity). This axis is specifically concerned with such clinical problems, apart from the pharmacological aspects addressed in the other axes.
In this axis, we aim to develop mathematical and statistical models and methods able to process this information to bring added value by inferring hidden parameters and provide simulations and predictions about the past and future behavior of the disease.
In the short-term (4 years), our research projects are: (1) modeling large-scale longitudinal data from immuno-oncology for prediction of response to immune checkpoint inhibition (QUANTIC and TGI-ML projects), (2) developing clinically relevant mathematical models of metastasis and (3) modeling the kinetics of clinical biomarkers.
3.6 Axis 2: Individualizing anticancer drugs dosing regimen
This axis fits within the “population approach” introduced in the 80’s by L. Sheiner 121 and aims at gathering exhaustive information about the multiple sources for variability in response in patients (including but not limited to drug-drug interactions, pharmacogenetics, and cormorbidities affecting renal and liver functions), build specific mechanistic models including relevant covariates and determine the PK/PD relationships of drugs used in oncology-hematology or for treating solid tumors. This covers cytotoxics, oral targeted therapies, biologics or immune checkpoint inhibitors. The overall goal is to achieve precision and personalized drug administration, i.e., the right dosing and scheduling regimen for the right patient.
In the short-term (4 years), our research projects will be focused on the following objectives: 1) predict response or toxicity variability dependent on pharmacogenetic (PGx) and pharmacogenomic data, 2) assess PMX of anticancer agents such as biologics, including ICI, and 3) develop physiologically-based PK models of nanoparticles distribution. Together, these objectives will allow to gain insights in the variability in drug response that depends on PK (1, 2 and 3) or germinal genetic alterations (1).
3.7 Axis 3: Optimizing combinatorial strategies with immune checkpoint inhibitors
Our hypothesis is that so many attempts to combine drugs fail not because the underlying pharmacological concepts are wrong (such as immunogenic cell death triggered by cytotoxics or radiation therapy, or increase in T cells infiltration with anti-angiogenics) but because these combinations probably require fine tuning in terms of dosing, scheduling and sequencing, whereas in practice all the drugs are given the same day. The goal of this second axis is therefore to shift from current empirical and suboptimal combinatorial regimen to model-informed designs to best combine drugs and therapeutic approaches so as to maximize efficacy while controlling toxicities. To do so, we will rely on our pioneering work about model-driven scheduling in early phase trials for combination of cytotoxic agents in metastatic breast cancer (MODEL1 trial) 125, 136 and metronomic vinorelbine in lung cancers (MetroVino trial) 101, 119. Leveraging the unique multidisciplinary aspect of our team, we implement a fully translational approach going from experimental therapeutics, PMX and quantitative systems pharmacology, to clinical trials either in early (phase I/II) or late (phase III) settings. Of note, our group has already an expertise in developing mathematical models determining the best sequencing between chemotherapy and anti-angiogenics.
In the short-term (4 years), our research projects will be focused on providing model-informed designs for combining ICI with: (1) cytotoxics, (2) an experimental immunoliposome and (3) radiotherapy.
3.8 Mid-term objectives
3.8.1 Modeling
We plan to achieve two main things on the modeling side: 1) the development of effective numerical tools (either as web applications or as part of simulation software) and 2) the empirical validation of the models.
For 1), this includes a tool able to predict response to ICI monotherapy in NSCLC from baseline and early response data . We will work with our industrial partners (in particular, HalioDX from the PIONeeR consortium), to transfer the tool for commercial use. Second, we plan to have a validated numerical tool able to predict metastatic relapse from clinical biomarkers at diagnosis, using our mechanistic model. This model will integrate the effect of adjuvant therapy (hormonotherapy or cytotoxic therapy) and will be able to simulate the long-term impact of alternative treatments (e.g., number of cycles to be administered to prevent distant relapse). It will have been validated from our local databases and will be implemented in a clinically-usable online tool such as PREDICT 150. The main difference with this tool will be the ability to mechanistically simulate the effect of therapy. We plan to have initiated a larger initiative at the national or European level to collect large data bases, validate further the predictive power of the model, and refine its structure if required. We will also extend this tool to other pathologies that share the same problematic (diagnosis at early-stage, important probability of future distant relapse) such as kidney cancer, following our initial work from preclinical data 99, 104, 113.
The pharmacometric models that will have been developed will also be implemented as clinically effective dose adaptation numerical tools directly usable to personalize the dose and scheduling of multiple anti-cancer agents not only by the clinicians and pharmacists from our group, but also by others, at least at a regional level.
For 2), our strategy of validation is the following. First, during the development phase, a proportion of the dataset (usually, 30%) is left aside unused for establishment of the model and initial calibration, and then used as a test set. When the sample size is too small (n 100 patients), only (nested) cross-validation is employed to assess the predictive power of the model. Evaluation metrics will be the classical ones, adapted to the task (classification, regression or survival regression) and include discrimination and calibration. In addition, specific methods will be used when in the context of mixed-effects approach (e.g., visual predictive checks for evaluation at the population level). The second step consists in evaluating the predictive power of the models in retrospective, external data sets. We have for instance initiated a collaboration with Dr C. Scherer (Clermont-Ferrand) to validate our metastatic prediction model on an external database of 3061 patients. The third step is to validate the added value of the model-based approach compared with the standard of care and is a long-term rather than mid-term objective.
3.8.2 Pharmacological and clinical oncology
In axis 2, in addition to standard drugs, developing tools for similarly better understanding the sources of therapeutic and PK variability and understanding the PK/PD relationships of cell therapy in oncology such as CAR-T cell therapies, is a challenging task. The challenge with CAR-T cells is that first, developing bioanalytical tools to monitor them in patients is not trivial, and second, little but nothing is known regarding their PK properties and possible sources impacting on PK/PD relationships such as disease status or immune status of the patient. We aim at developing both a platform to monitor CAR-T cells and future cell therapies in plasma and mechanistic models to describe the disposition of these new therapies in the body.
The nanoPBPK model will be extrapolated to humans and used to determine the specifications of an optimal nanosystem in order to penetrate solid tumors such as pancreatic tumors. The rationally designed nanosystem will be evaluated in vitro and in vivo in order to validate the approach. The nanoPBPK model will be interfaced to become a software and be shared with the scientific community. To achieve this goal, a partnership with ESQlabs, the company developing the opensource PBPK platform PKSim, has already been approved by both sides. The nanoPBPK model will be combined with pharmacodynamic modeling describing the effect of the loaded anticancer drug on tumor growth and metastases spread, on the immune system, and on dose-limiting toxicities.
Our mid-term objective in axis 3 is to assist the design of scheduling regimen for combinatorial treatments in early phase clinical trials, which represent an important clinical challenge of the next 10 years. To do so, we will benefit from our close connection to the INCa-labeled AP-HM's center for early phase clinical trials (CLIP2). Our aim is to design model-based, individualized and adaptive scheduling regimen that depend on the monitoring of the disease evolution. We plan to run phase I/II trials based on the model recommendations. Depending on our achievements and success in phase I/II trials our mid-term goal would be to lead a prospective, randomized, phase III trial comparing a model-based adaptive regimen to the standard of care for combination of immune checkpoint inhibition with chemotherapy and/or anti-angiogenic and targeted therapy. According to our team expertise, the target malignancies would be primarily lung cancer (LG) and head and neck cancer (SS).
3.9 Long-term objectives
At long-term, we globally wish to have established a worldwide leader position in the fields of quantitative mathematical oncology and PMX, as well as the pharmacokinetics of nanoparticles. We hope that this would translate into the achievement of three goals: (1) the development of software effectively used for clinical decision-making and dosing adjustment (estimated achievable), (2) the initiation of prospective, phase III clinical trials comparing model-guided therapy versus standard of care (highly challenging), (3) clinical trials of nanosystems designed by our group (estimated achievable). In addition, we foresee several avenues both in terms of modeling opportunities and applications.
3.9.1 Modeling
Our short-term program is devoted to the development of new models and their confrontation to empirical data. The mid-term program will focus on the validation and refinement of these models. In the long-term, we foresee that this will bring novel questions in terms of mathematical analysis of the models. For instance, metastatic modeling will establish validated models for tumor-tumor interactions, including immune-mediated interactions. In turn, this leads to nonlinear, size-structured, renewal PDEs. Study of the asymptotic behavior of such equations is non-trivial.
More generally, we expect that physiologically structured PDEs (psPDE) can become relevant to practical modeling in oncology, from two types of data: flow cytometry and single-cell sequencing. Flow cytometry is currently becoming of increasing relevance to characterize multiple populations of cells, for instance in the context of immuno-oncology. In the QUANTIC project (10.2), we are starting to interact with such data, only by means of scalar quantities so far. However, the structure of this data is to have, for each cell, a quantitative measure (e.g., a surface marker). Measuring these in a population of millions or billions of cells makes it adapted to modeling by such psPDE. Similarly, single-cell sequencing is a technique by which every cell of a population (e.g., in a tumor) is sequenced, thus having mutation information. In turn, this allows to quantify subclones in the population. Such data has already generated fascinating results, for instance in the study of metastatic development theories 123, 129. Although evolutionary modeling is a wide field with established groups (M. Nowak or F. Michor in Harvard, C. Curtis in Stanford, T. Graham at the CRUK), few groups are modeling dynamical data at single-cell resolution. To this regard, the theoretical work initiated by J. Clairambault and B. Perthame (Inria MAMBA) suggesting to use psPDEs to model evolution in cancer cell populations could be appropriate 107. Parameter estimation in such models is a challenging task 118 and data assimilation from flow cytometry or single-cell sequencing data sets would represent an important avenue. Dynamical data can be provided by circulating tumor DNA and we have already initiated contacts with an important clinical and biological study in Marseille on this topic (the SCHISM study, PIs: SS and F. Fina). The recent developments of technology enabling spatial resolution of single-cell sequencing also paves the way to exciting avenues in terms of modeling 139.
We will also build models that can optimize the effectiveness of treatments incorporating new criteria (other than the evolution of tumor mass) of diagnostic and therapeutic evaluation, especially those we have forged around the information provided by functional imaging (T80) computational algorithm time at which 80% of FDG is metabolized 100, 111, 143.
3.9.2 Pharmacological and clinical oncology
A general, challenging, long-term objective, would be to run prospective clinical trials in which a model-informed arm would be compared to the standard of care. In the model-informed arm, therapeutic decision would be based on the recommendation of the model. This applies to the models developed in all axes. For instance, in breast cancer, the number of cycles of chemotherapy would be adapted based on the model indication (axis 1), decision of the maximum tolerated dose in the treatment of leukemia patients would be based on the PGx/PK/PD model (axis 2) or the combination scheduling regimen would be given by model calculations (axis 3).
Regarding nanosystems initially designed by our group based on the nanoPBPK modeling, they will also be tested in early clinical trials. Our group will drive the design of these based on simulations performed with the nanoPBPK and pharmacodynamic model, in order to guarantee the highest chances of success while ensuring patients safety. In particular, nanoparticles specifically transporting cytoxics could replace standard systemic myelo-ablation in hematopoietic stem cell transplantation, a risky strategy with frequent life-threatening, when not lethal, toxicities. Because of the fully controlled distribution phase in the body, nanoparticles encapsulating several drugs could thus be implemented in the preparative regimens for allogeneic stem cell transplantation in leukemia or myeloid malignancies.
In addition, several groups predict that in addition to standard drugs or biologics, or rising gene therapy and cell therapy strategies, new devices such as nanobots will be developed to treat cancers. Nanobots are entities which are not designed to interact with standard pharmacological targets or genes like current anticancer treatments, but could fix the cancer cell, either by providing a missing protein, or ultimately trigger a mechanic cell-death using radiation, thermal wave, or by disrupting cell membrane. These new entities should exhibit totally new pharmacokinetics, because they are unlikely to be metabolized in the liver or to be cleared by the kidney or the biliary tract. Therefore, new models for PK/PD should be developed, because neither behavior in the body nor intrinsic mechanisms of action are known yet. In addition, the issue of nanosafety with such devices will be particularly critical and will require extensive PMX resources, to predict long-term effects or to keep under control the mechanism of action. Should such nanobots be developed, the COMPO joint-team should develop specific resources to monitor their fate in the body, specific resources to write equations describing nanobots/body, nanobots/immune system, and nanobots/cancer cells interactions, plus new global models encapsulating all the interactions, and pharmacodynamics impact of such devices.
Depending on the expertise we gain in the PK/PD knowledge of those cell therapies as part of the mid-term objectives, optimizing combinatorial strategies to such cell therapies will be another long-term objective.
Last, pharmaco-economic studies including impact of quality of life, increase in both tolerability and efficacy will be performed to determine whether PMX-based dosing is a cost-saving strategy. For instance, by refining the scheduling of immune checkpoint inhibitors, such as determining, using modeling and simulation strategies, when the plasma concentration of the drug reaches the threshold in trough levels necessary to ensure maximal target engagement, it will be possible to customize the frequency of the administrations, with possible strong impact on treatment costs. By using real-life data, we propose to use dedicated models to quickly define alternate and more appropriate dosing and/or scheduling with newly marketed anticancer agents, either chemotherapy, targeted therapies or biologics including immune checkpoint inhibitors.
4 Application domains
The COMPO research team's projects all focus on a serial of complementary and inter-related domains described in an itemized fashion below:
- Health: all the models to be developed within the framework of the COMPO team are related to improving healthcare;
- Cancer: in particular, the models will be developed to address specific issues related to cancerous diseases;
- Precision Medicine: in particular, in cancer the developed models will be part of the implementation of precision medicine in oncology focusing on the following items;
- Combinatorial regimen: developing model-informed strategies to determine the optimal modalities when combining several treatments altogether. With the increasingly diversified arsenal of therapeutic approaches to treat cancers (surgery, radiotherapy, chemotherapy, targeted and anti-angiogenic therapy and immunotherapy), defining optimal combination protocols is highly challenging 108. This spans the issues of sequencing, scheduling and dosing of those therapies, which are to date largely addressed using a trial-and-error approaches. Consequently, too many combinatorial trials fail, and attrition rate with combinatorial immunotherapy is now a rising issue in clinical oncology and we hypothesize that extensive modeling and pharmacometrics could help refining the way anticancer drugs are combined;
- Tools for decision-making: developing model-informed strategies to forecast clinical outcome, i.e., during clinical trials. Assessing the predictive power of markers not only at baseline but also in their change over time is a current challenge. The information available, on the basis of which decision is made, includes clinico-demographic variables, classical biomarkers (e.g., blood counts, thyroglobulin, lactate dehydrogenase levels, etc...) but also an increasing amount of data from other sources (e.g., immuno-monitoring, anatomical functional imaging or genomics) that require state-of-the-art modeling to analyze extremely dense and longitudinal data;
- Adaptive dosing strategies: developing model-informed strategies to customize dosing so as to ensure an optimal toxicity-efficacy ratio. All anticancer agents are approved upon registration trials performed in highly selected patients (i.e., with controlled lifestyle, little comorbidities, controlled polymedication and restricted range of age), thus smoothing the interindividual variability. In real-world practice however, patients are all heavily co-treated with a wide variety of other drugs plus herbal medications, likely to interact with drug metabolism and transport, and are frequently older than in clinical trials. In addition, genetic polymorphisms affecting genes coding for drug transport proteins or drug-metabolizing enzymes in the liver, or transcriptional factors can impact as well on dose-exposure relationships. Consequently, standard dosing may not be suitable in non-standard patients to reach the adequate drug exposure levels associated with optimal toxicity/efficacy balance;
- Nanomedicines: developing model-informed strategies to conceptualize drug-loaded nanoparticles likely to improve the toxicity-efficacy ratio over conventional treatments. As of today, the biodistribution phase of anticancer agents is totally aspecific, making "on-taget off-site" actions an issue because it is associated with drug-related side effects affecting healthy tissues. Nanoparticles present unique features likely to deliver specifically a high amount of payload directly on a tumor site, thus improving the antiproliferative action while sparing healthy tissues. In addition, nanoparticles are expected to reshape the tumor micro-environment, making them good candidates to be further associated with immunotherapy (see Combinatorial Regimen above).
5 Social and environmental responsibility
5.1 Impact of research results
Due to its unique composition including medical oncologists, clinical pharmacologists and mathematical modelers, COMPO is at stake with important social challenges: oncology healthcare and innovation in drug development. The software and results developed by COMPO are devoted to these challenges and aim to be directly used by medical and pharmaceutical oncologists or by the biotech and pharmaceutical industry to help drug development and biomarker discovery.
To give a few examples:
- the KineticsPro software historically developed by Pr Iliadis is used daily by pharmacists to individually adapt the dose of anti-cancer drugs (e.g., for methotrexate, cisplatin or busulfan)
- the compoEDA package is used by physicians to produce automated statistical reports, helping to analyze the data collected for specific medical questions
- the nlml_onco software is developed in collaboration with the industry (Roche / Genentech) to help anticipate the results of phase 3 clinical trials using machine learning models trained on the results of earlier (phase 2) clinical trials 4
- COMPO is in charge of the biostatistical, machine learning and mechanistic modeling analysis of the large-scale PIONeeR RHU project to identify biomarker signatures predictive of the resistance to immunotherapy in lung cancer
- the LUCA-pi RHU, led by COMPO member Pr Boulate, will conduct research to implement lung cancer screening in France (currently not performed)
6 Highlights of the year
6.1 Hiring
This year, two new researchers joined the COMPO team through competitive recruitment: Elias Ventre, hired as an INRIA CRCN researcher and member of COMPO, and Quentin Marcou, hired as a CNRS CRCN researcher and member of the team.
In addition, COMPO has strengthened its medical staff and its link with the Institut Paoli Calmettes (IPC) Cancer Centre with the arrival of Dr. Arthur Géraud, M.D. Ph.D., Medical Oncologist at the IPC.
6.2 PhD Theses Defenses
November, 2024: Jessica Ou, PhD. student at COMPO, succesfully defended her thesis ("Development of PBPK models for optimization of nanomedicines". Supervisor: F. Gattacceca, Co-supervisor: P. Garrigue, Sponsor: Doctoral school 62). 76
December, 2024: Anthea Deschamps, PhD. student at COMPO, succesfully defended her thesis ("Patient-centered population pharmacokinetic approach: proof of concept and application in intensive care". Supervisor: F. Gattacceca, Co-supervisor: R. Guilhaumou, Sponsor: APHM). 74
December, 2024: Clemence Marin, PhD. student at COMPO, succesfully defended her thesis ("PK/PD of cetuximab in head and neck cancer patients: the CetuxiMAX Clinical Trial". NCT 04218136; P.I.: S. Salas, supervisor: J.Ciccolini, Sponsor: Merck Serono). Associated Scientific Production: article 1; article 2 ; article 3 ; article 4 ; article 575
7 New software, platforms, open data
7.1 New software
7.1.1 stats_pioneer
-
Name:
Statistical analysis and reporting of the PIONeeR study
-
Keywords:
PIONeeR, Statistical analysis, Biostatistics
-
Functional Description:
This software was built to analyse the PIONeeR (Precision Immuno-Oncology for advanced Non-small cell lung cancer patients with PD-(L) 1 ICI Resistance) data. PIONeeR is a prospective, multicenter study with primary objective being to validate the existence of a hypothetical immune profile explaining resistance to immunotherapy in non-small cell lung cancer patients.
It initially integrated preprocessing, exploratory data analysis, visualization, statistical analysis, feature selection, machine learning and results generation and reporting. Since, exploratory data analysis, visualization and statistical analysis have been promoted to the COMPO-level `compoEDA` package, feature selection and machine learning to the COMPO-level `ml.tidy` package and .
This software corresponds to the very first step of the data analysis, which is the preprocessing, and the very last: generation of results. Some of its functions aim at:
- preprocessing the data (creation of clinical variables, dictionary, outcome variables, data monitoring and corrections, treatment of the variables types)
- generating the tools to load the data and metadata
- computing statistical tests, logistic or Cox regression, or performing a correlation analysis
- visualising the data (boxplots, barplots, survival curves, ROC curves, volcano plots)
- providing detailed and interactive statistical reports on the data
- displaying statistical results and visualisation to interactive dashboards
- performing supervised and unsupervised machine learning modelling
- providing detailed and interactive machine learning reports
- displaying all reports on two websites (one private, one public)
- automating the production of reports and websites using Gitlab CI/CD
-
News of the Year:
New PK data. New biologically informed groups / subgroups for biomarkers. New documentation about these groups. Large cleaning and refactoring of the repository. Other updates included:
- Preprocess:
- generating new outcomes (primary resistance and secondary resistance)
- complete documentation of all outcomes
- Data exploration:
- new reports on imputation methods performance
- new reports on data correlation
- Feature selection:
- cleaning feature selection general report
- investigation of multiple new signatures
- new report analyzing stability of new signatures
- new report analyzing impact on feature selection methods' stability of data correlation
- Statistical analysis:
- cleaning variable summary dashboard
- Unsupervised machine learning:
- new unsupervised analysis of PIONeeR's dataset taking as input structure of biomarkers (using MFA and HCPC algorithms)
- new reports presenting results
- Supervised machine learning:
- factorization of supervised machine learning reports, then promotion to the "compo.tidyML" COMPO-wise package
- creating generic template report, using compo.tidyML new supervised machine learning pipeline, for performing supervised analysis
- new reports from this template considering multiples outcomes and group of patients (first line, second line, all lines)
- incorporating optimism framework for computing performances of models
- new report presenting learning curves of optimism framework trained models
- new report presenting optimism framework hyperparameter tuning analysis
- documentation about optimism code
- Machine learning:
- new report presenting results for both unsupervised and machine learning models as a companion for ongoing papers
- Continuous integration / continuous development:
- large refactoring
- web solution hosting two PIONeeR's websites (one internal, one as a companion for ongoing papers)
- shiny dashboard added inside both websites
- complete documentation about new CI/CD
- Reporting:
- scripts for reproductible figures of two ongoing papers (methodology, biomarkers analysis)
- Preprocess:
- URL:
- Publications:
-
Contact:
Sebastien Benzekry
-
Partners:
Veracyte, Innate pharma, APHM
7.1.2 compo.EDA
-
Name:
Exploratory Data Analysis for Clinical Oncology Data
-
Keywords:
Data Exploration, Biostatistics
-
Scientific Description:
This library implements as an R package:
- Exploratory analysis:
- Clinical characteristics table
- Kaplan-Meier estimation of the progression-free and overall survival
- Clinical and biological features distribution
- Classification analysis:
- Univariate and multivariate logistic regression
- Odds ratio
- Area under ROC curve
- t test / chi2 test
- Survival analysis:
- Univariate and multivariate Cox regression
- Hazard ratio
- Area under ROC curve
- log-rank test
- Data visualization:
- Correlation plots (Pearson correlation)
- Volcano plots (p-value and adjusted p-value)
- Boxplots (quantitative features) and barplots (qualitative feaures)
- Kaplan-Meier curves
- Automatic comprehensive and customizable statistical reports
- Exploratory analysis:
-
Functional Description:
The package compoEDA aims to provide a comprehensive exploratory analysis of data from clinical studies in oncology. These studies commonly investigate biological markers able to reveal and distinguish different tumor profiles, in order to early adapt the therapeutic strategy for patients.
The objective of this software is to provide a simplified tool for both computational scientists and clinical researchers to easily generate a graphical results and automatic reports containing the following analyses:
- overview and visualization of clinical data and biological markers
- univariate and multivariate classification analysis (logistic regression)
- univariate and multivariate survival analysis (Cox regression, Kaplan-Meier analysis)
- correlation analysis
- statistical tests
- visualization of markers (boxplots, barplots, volcano plots, forest plots ...).
-
Release Contributions:
APP deposit
-
News of the Year:
Aesthetic and cleaning for reports generation. Add the possibility to generate complete statistical reports or smaller, more concise, ones.
Other updates include:
- cleaning functions
- remove hard coding variables
- URL:
-
Contact:
Sebastien Benzekry
-
Participants:
Sebastien Benzekry, Linh Nguyen Phuong, Celestin Bigarre, Paul Dufosse, Melanie Karlsen, Andrea Vaglio
7.1.3 compo.tidyML
-
Name:
Machine learning with tidymodels
-
Keywords:
Survival analysis, Machine learning, Data analysis, Oncology
-
Scientific Description:
This software maximizes the use of the R package tidymodels.
-
Functional Description:
This package provides multiple functions to perform machine learning analysis using the `tidymodels` framework. Tasks include: feature selection, plot feature importances, train, cross-validate, apply supervised machine learning algorithms (classification or survival analyses) and unsupervised machine learning, evaluate metrics of predictive performances, compute learning curves.
Initial development was part of the `stats_pioneer` package (also called `pioneerPackage`) and `ml.tidy` evolved as a standalone package only in February 2023.
-
News of the Year:
A full generic pipeline for generating automatically a complete machine learning reports was added. It can be used to perform both classification or survival analysis. It is composed of multiple parts:
- methods description
- data preprocessing
- feature selection
- analysis of generated signature with SHAP plot and heatmaps
- cross-validation evaluation
- train/test evaluation
- stratified Kaplan-Meier survival analysis
- RADAR significant patients profile displaying
Other updates include:
- adding bobolasso as a feature selection method
- adding xgboost as classifier possible
- adding leave one out cross validation as evaluation method
- adding group-based unsupervised clustering method
- URL:
- Publications:
-
Contact:
Sebastien Benzekry
7.1.4 compo.NLME
-
Name:
R package for fitting and analyzing Non-Linear Mixed Effects (NLME) models using Monolix.
-
Keywords:
Monolix, Nonlinear mixed effects models, Lixoft, Population approach
-
Scientific Description:
Available features:
- Structural models
- constant
- linear
- double exponential
- double exponential with dropout
- hyperbolic
- preprocess blood marker datasets
- preprocess tumor kinetics datasets
- fit NLME models using monolix API
- post-process of results
Available data:
- Tumor Kinetics with dropout data. A simulated dataset of tumor kinetics following the double-exponential model, with parameters obtained from (Benzekry et al., PAGE 20, 2022), which deals with the RECIST-based sum of largest diameters (SLD, in mm) of lung cancer treated with immune-checkpoint blockade (anti-PDL1 drug atezolizumab). Dropout was also simulated using a Weibull survival model.
- Tumor and Blood marker Kinetics with dropout data. A simulated dataset of joint tumor and blood markers (albumin C-reactive protein, lactate dehydrogenase, neutrophils) kinetics following the models and parameters established in (Benzekry et al., PAGE 20, 2022). These are monitoring data during immune-checkpoint blockade (anti-PDL1drug atezolizumab) in lung cancer. Dropout was also simulated using a Weibull survival model.
- Structural models
-
Functional Description:
This R package implements a framework to work with Non-linear Mixed effects models in the context of clinical oncology to predict relapse and survival using longitudinal data.
-
News of the Year:
Integration of postprocessing of Monlix runs:
- computation of ChartsData
- residual plots
- observations VS predictions plots
- individual fits plots
- VPCs
- distribution of individual parameters
- distribution of random effects
- URL:
- Publications:
-
Contact:
Sebastien Benzekry
-
Participants:
Sebastien Benzekry, Celestin Bigarre, Linh Nguyen Phuong, Ruben Taieb
7.1.5 compOC
-
Name:
Optimism correction of machine learning models
-
Keyword:
Feature selection
-
Functional Description:
compOC is a Python implementation of the optimism correction bootstrapping framework for evaluating full machine learning pipelines with or without feature selection. There are 40+ FS methods available, such as Lasso, Stability Selection, Stabl, t-test filtering, hierarchical clustering, recursive feature elimination and others.
-
News of the Year:
Initial development of the package.
- URL:
-
Contact:
Sebastien Benzekry
-
Participants:
Sebastien Benzekry, Anastasiia Bakhmach, Mohamed Boussena
7.1.6 SChISModeling
-
Name:
SChISM modeling
-
Keywords:
SChISM, Statistical analysis, Biostatistics
-
Scientific Description:
- Preprocess
- Exploratory data analysis
- Classification analysis (logistic regression)
- Survival analysis (Cox regression)
- Mixed-effects modeling analysis
- Simulation for ODE models for mechanistic modeling
-
Functional Description:
SChISModeling aims to analyze SChISM data (Size CfDNA Immunotherapies Signature Monitoring). SChISM is a clinical study that introduces an innovative approach to quantify circulating free DNA in cancer patients treated with immunotherapy. The study's objective is to early predict response to immunotherapy in patients at an advanced/metastatic stage according to these quantitative cfDNA data.
This software corresponds to the very first step of the data analysis, which is the statistical analysis. Some of its functions aim at:
- preprocessing the data (creation of clinical variables, dictionary, outcome variables, clinical biomarkers, treatment of the variables types)
- computing statistical tests, logistic or Cox regression, performing a correlation analysis
- visualizing the data (boxplots, barplots, survival curves, ROC curves, volcano plots)
- providing detailed and interactive statistical reports on the data
- simulation for ODE models for mechanistic modeling
-
News of the Year:
- Improved preprocess
- New functions for data visualization
- Simulation for mechanistic model
- New classification outcome added : primary resistance
- Bootstraps
- URL:
-
Contact:
Sebastien Benzekry
-
Participants:
Sebastien Benzekry, Linh Nguyen Phuong, Romain Zakrajsek
7.1.7 metamats
-
Keyword:
Mechanistic modeling
-
Functional Description:
This R package is the implementation of a general framework to build and use models of the metastatic process based on the initial model of Iwata et al. (2000). The family of model that can be built describe the metastatic disease with a partial differential equation (pde) on the size structured distribution of the tumors. These models have three components, a function that characterize the growth of the primary tumor, a function that characterize the growth of the metastases, and a dissemination function that decribes how new metastases are produced.
-
Release Contributions:
Features:
- Model structure
- Direct computation of (C++)
- Individual fit of cumulative size distribution (direct only)
- Many diagnostic plots
-
News of the Year:
Development of the first version of the sofware and several updates.
This sofware was used and will be described in a work to be published in 2025. The application was the use of this model to describe the dynamics of the brain metastasis disease in small cell lung cancer patient with or without prophylactic cranial irradiation (PCI) and study the impact of PCI on the overall survival of the patients as well as the progression of the brain disease.
- URL:
-
Contact:
Celestin Bigarre
-
Participants:
Sebastien Benzekry, Celestin Bigarre
7.1.8 metamatsModels
-
Keyword:
Mechanistic modeling
-
Functional Description:
A collection of models implementation for the metamats R package
-
Release Contributions:
Available models:
- Gompertz growth model (alpha, mu parametrization)
- Gompertz growth model (alpha, K parametrization)
- Gompertz with dormancy growth model (alpha, mu, tau parametrization)
- Proportional dissemination model
- Power dissemination model (mu, gamma parametrization)
- Iwata model object (identical gompertz growth, proportional dissemination)
- Benzekry model object (identical gompertz growth with dormance, power diss)
-
News of the Year:
Development of the first version of the sofware and several updates.
This sofware works in combinaison with the metamats R package and was used and will be described in a work to be published in 2025 (see the BIL entry of metamats R package for more details).
-
Contact:
Celestin Bigarre
-
Participants:
Celestin Bigarre, Sebastien Benzekry
7.2 Open data
Pre- and post-surgical monitoring of experimental primary tumor growth and metastasis under neo-adjuvant treatment
-
Contributors:
Mastri, Michalis (Data collector), Benzekry, Sebastien (Distributor), Ebos, John ML (Contact person)
-
Description:
This comprehensive dataset contains pre-surgical primary tumor volume measurements and pre- and post-surgical records of metastatic burden in 251 mice implanted with human breast cancer cells either untreated or pre-surgically treated with two distinct receptor tyrosine kinase inhibitors (Sunitinib and Axitinib) and multiple dose and scheduling regimen. In addition, the data contains tumor tissue and circulating biomarkers collected at surgery.
- Dataset DOI:
- Publications:
-
Contact:
S. Benzekry
8 New results
Note: Emphasized authors indicate team members.
8.1 Predicting survival in patients with advanced NSCLC treated with atezolizumab using pre- and on-treatment prognostic biomarkers
Participants: Sébastien Benzekry, Mélanie Karlsen, Célestin Bigarré, Abdessamad El Kaoutari, Rene Bruno [Roche Genentech], Ales Neubert [Roche pRED], François Mercier [Roche Genentech], Martin Stern [Roche pRED], Bruno Gomes [Roche pRED], Suresh Vatakuti [Roche pRED], Peter Curle [Roche pRED], Candice Jamois [Roche pRED].
Funding and data: Roche pRED
Publication: Published in Clinical Pharmacology and Therapeutics12
Existing survival prediction models rely only on baseline or tumor kinetics data and lack machine learning integration. We introduce a novel kinetics- machine learning (kML) model that integrates baseline markers, tumorkinetics, and four on-treatment simple blood markers (albumin, C- reactive protein, lactate dehydrogenase, andneutrophils). Developed for immune- checkpoint inhibition (ICI) in non- small cell lung cancer on three phase II trials (533 patients), kML was validated on the two arms of a phase III trial (ICI and chemotherapy, 377 and 354 patients). It outperformed the current state- of-the-art for individual predictions with a test set C-index of 0.790, 12-months survival accuracy of 78.7% and hazard ratio of 25.2 (95% CI: 10.4–61.3, P < 0.0001) to identify long-term survivors. Critically, kML predicted the success of the phase III trial using only 25 weeks of on-study data (predictedHR = 0.814 (0.64–0.994) vs. final study HR = 0.778 (0.65–0.931)). Modeling on-treatment blood markers combined with predictive machine learning constitutes a valuable approach to support personalized medicine and drug development. The code is publicly available on gitlab.
8.2 Machine-learning and mechanistic modeling of primary and metastatic breast cancer growth after neoadjuvant targeted therapy
Participants: Sébastien Benzekry, Michalis Mastri, Chiara Nicolò, John ML Ebos.
Data: Preclinical data of primary tumor and metastatic growth in 128 mice.
Publication: published in PLoS Computational Biology13
Background
:
Clinical trials involving systemic neoadjuvant treatments in breast cancer aim to shrink tumors before surgery while simultaneously allowing for controlled evaluation of biomarkers,toxicity, and suppression of distant (occult) metastatic disease. Yet neoadjuvant clinical trials are rarely preceded by preclinical testing involving neoadjuvant treatment, surgery, and post-surgery monitoring of the disease.
Objective
:
Here we used a mouse model of spontaneous metastasis occurring after surgical removal of orthotopically implanted primary tumors to develop a predictive mathematical model of neoadjuvant treatment response to sunitinib, a receptor tyrosine kinase inhibitor (RTKI).
Methods
:
Treatment outcomes were used to validate a novel mathematical kinetics-pharmacodynamics model predictive of perioperative disease progression. Longitudinal measurements of presurgical primary tumor size and postsurgical metastatic burden were compiled using 128 mice receiving variable neoadjuvant treatment doses and schedules (released publicly on Zenodo). A nonlinear mixed-effects modeling approach quantified inter-animal variabilities in metastatic dynamics and survival, and machine-learning algorithms were applied to investigate the significance of several biomarkers at resection as predictors of individual kinetics. Biomarkers included circulating tumor- and immune-based cells (circulating tumor cells and myeloid-derived suppressor cells) as well as immunohistochemical tumor proteins (CD31 and Ki67).
Results
:
Our computational simulations show that neoadjuvant RTKI treatment inhibits primary tumor growth but has little efficacy in preventing (micro)-metastatic disease progression after surgery and treatment cessation. Machine learning algorithms that included support vector machines, random forests, and artificial neural networks, confirmed a lack of definitive biomarkers, which shows the value of preclinical modeling studies to identify potential failures that should be avoided clinically.
Conclusion
:
Mathematical modeling combined with machine learning techniques represent a novel platform for integrating preclinical surgical metastasis models in outcome prediction of neoadjuvant treatment.
8.3 Mechanistic Learning for Predicting Survival Outcomes in Head and Neck Squamous Cell Carcinoma
Participants: Kevin Atsou, Anne Auperin, Joel Guigay, Sebastien Salas, Sebastien Benzekry.
Publication: published in Clinical Pharmacology and Therapeutics: Pharmacometrics and Systems Pharmacology11
We employed a mechanistic learning approach, integrating on‐treatment tumor kinetics (TK) modeling with various machine learning (ML) models to address the challenge of predicting post‐progression survival (PPS)—the duration from the time of documented disease progression to death—and overall survival (OS) in Head and Neck Squamous Cell Carcinoma (HNSCC). We compared the predictive power of model‐derived TK parameters versus RECIST and assessed the efficacy of nine TK‐OS ML models against conventional survival models. Data from 526 advanced HNSCC patients treated with chemotherapy and cetuximab in the TPExtreme trial were analyzed using a double‐exponential model. TK parameters from the first line and maintenance (TKL1) or after four cycles (TK4) were used to predict PPS and post‐cycle 4 OS (OS4), combined with 12 baseline parameters. While ML algorithms underperformed compared to the Cox model for PPS, a random survival forest was superior for OS prediction using TK4 and surpassed RECIST‐based metrics. This model demonstrated unbiased OS4 prediction, suggesting its potential for improving HNSCC treatment evaluation. Trial Registration: ClinicalTrials.gov identifier: NCT02268695.
8.4 Long circulating-free DNA fragments predict early-progression (EP) and progression-free survival (PFS) in advanced carcinoma treated with immune-checkpoint inhibition (ICI): A new biomarker
Publication: communicated at ASCO 94, ESMO 95, PAGE 89 and SMB 71
Participants: Sébastien Salas, Linh Nguyen-Phuong, Frédéric Ginot, Laurent Greillier, Pascale Tomasini, Audrey Boutonnet, Jean-Loup Deville, Frederic Fina, Sébastien Benzekry.
Background
:
Establishing reliable and early predictive biomarkers of response of ICI is essential. Analysis of cfDNA fragmentation profiles (fragmentome) is a promising non-invasive method to do so independently of a specific molecular target, cancer type or treatment. We monitored plasmatic cfDNA concentration and size characteristics of the fragmentome in advanced lung, head and neck, kidney and bladder cancer patients, treated with ICI (n = 111). The aim was to predict EP (defined as progression at the first imaging evaluation) and PFS.
Methods
:
Our novel patented technology made possible to measure accurately cfDNA concentration and size profile directly from tens of microlitres of plasma and without prior DNA extraction (BIABooster system). Statistical association and predictive performances of response from fragmentome-derived metrics (e.g., concentration, size distribution peaks or fragments size ranges) were conducted. The data was split between a training (n=78) and a test (n=33) set. Optimal thresholds were determined through receiver-operator characteristics (ROC) curve analysis, and confidence intervals determined using bootstrap resampling. Classification metrics were assessed in both the training and testing set. The entire process was bootstrapped 100 times to assess the robustness.
Results
:
Quantity of long fragments over 1650 bp (LF) showed the best discriminatory power (AUC = 0.77 (0.65-0.87)) of EP. LF were significantly, strongly and positively associated with non-EP (odd ratio =0.27 (0.14-0.52), p <0.001) and longer PFS (p<0.001, hazard ratio 0.406 (0.274 - 0.599)). The predictive performances of EP were also very high: AUC 0.75 (0.65-0.84), accuracy 71% (95% CI: 63% - 80%), positive predictive value was 0.61 (0.47-0.78), on the test set.
Conclusions
:
These findings highlight a very significant association of cfDNA high-molecular-weight fragments with EP and PFS that outperform the predictive value of the only routinely used marker PDL1.
8.5 Mechanistic modeling of tumor kinetics coupled with biomarker dynamics for survival prediction in non-small cell lung cancer patients
Participants: Ruben Taieb, René Bruno, Jin Jin, Pascal Chanu, Sebastien Benzekry.
Publication: Communicated at PAGE 50
Objectives
:
Simple blood markers derived from routine hematology or biochemistry have been reported to be prognostic factors of overall survival (OS) in cancer patients. However, most studies only use baseline (BSL) values. The only longitudinal biomarker that has been extensively used and modeled to date is tumor size kinetics (TK), linked to OS by parametric survival models (TK-OS).
The few studies investigating blood marker kinetics (BK) used simplified empirical or non-coupled models. These often fail to capture important correlations and lack a systemic view of the processes at stake. Non-trivial, complex dynamic BK profiles, either under monotherapy or combination therapy remain to be quantitatively modeled.
We propose here an analysis of the combined kinetics between tumor size and three BKs: albumin, LDH and neutrophils.
Specifically, our aims were to:
(1) Develop a mechanistic model coupling TK with albumin, LDH and neutrophil counts kinetics (denoted TALN-k)
(2) Integrate TALN-k into a nonlinear mixed-effects (NLME) modeling framework to account for inter-individual variability
(3) Assess its goodness-of-fit and benchmark TALN-k against empirical models
(4) Use TALN-k to disentangle complex TK-BK interactions in non-small cell lung cancer patients (NSCLC).
(5) Integrate the selected model-based TALN-k parameters into a ML model for prediction of individual OS.
Methods
:
Data:
Monotherapy data consisted of the three phase 2 clinical trials POPLAR + FIR + BIRCH (MONO, 862 patients). Combination therapy data consisted of the phase 3 trial IMpower 150 (COMBO, 1115 patients) with 3 arms composed of combinations of atezolizumab (ATZ) and other agents (bevacizumab and carboplatin + paclitaxel).
TALN-k mechanistic model
TLAN-k is a system of coupled ordinary differential equations (with delay), describing the joint tumor, albumin, LDH and neutrophil counts dynamics under immune-checkpoint inhibition either in monotherapy or in combination with other drugs. The model was derived assuming the following pharmaco-biological hypotheses:
TK : Tumor cells were divided into two subpopulations with linear growth rate : one resistant , the other sensitive, both with different linear death rates. For albumin, LDH and neutrophils, production is regulated and elimination is linear in the absence of tumors. Albumin : Albumin production is impaired by tumor-related inflammation, with logistic effect. Inflammation also increases capillary permeability. LDH: In addition to physiological production, LDH levels increase when tumor cells die (Warburg effect), but also following neighboring stroma damage during invasion. Neutrophils: Progenitor cells production occurs with linear due to hematopoiesis. They further undergo a three-compartments chain of maturation (variables ) with linear transition rate. Precursor cell production increases with tumor-related inflammation. Chemotherapy effect on neutrophil production was considered to be constant over time. It was thus implicitly modeled in the precursor production parameter.
Population Model
Individual parameters were all assumed to have log-normally distributed random effects, with a diagonal variance-covariance matrix. The error models were combined (constant + proportional) (S), constant (A), proportional (L and N), which minimized the corrected Bayesian information criterion.
The TALN-k NLME model was fit on the entire tumor, albumin, LDH and neutrophil longitudinal data, on MONO and COMBO separately (total data points: 67,507 COMBO, 44,911 MONO), using the Monolix software and the SAEM algorithm.
Goodness of fit and identifiability were assessed using diagnostic plots, relative standard error (RSE) values and shrinkage of the different estimates.
Definition of the OS prediction problem from on-treatment data
To avoid time-dependent covariate bias for OS prediction, we placed ourselves at cycle 5 day 1 (C5D1, i.e., 3 months after treatment initiation). For each patient, we used only the longitudinal data available before C5D1, to identify their Empirical Bayes estimates (denoted EBEs4 ; 4 as in after 4 cycles). We then discarded the patients who died before C5D1 and computed the shifted OS with C5D1 as baseline for the remaining ones, denoted OS4.
Survival model
The previously defined individual EBEs4 (p = 26 parameters) were adjuncted to baseline covariates (p = 10) to be used as predictors of OS4. Following previous work, a random survival forest machine learning (ML) model was chosen and the complete prediction model denoted TALN-kML4. The ML model performances were assesed using 10-fold cross-validation of each dataset. The C-index was considered as the main (discrimination) predictive metric. The 12-months survival area under the ROC curve was also evaluated.
Results
:
We found the TALN-k model to fit the data very accurately, with interesting, non-trivial and interpretable individual dynamics and trends, such as early and late peaks of LDH, early drops in albumin, and early drops in neutrophil count.
Despite the relatively large number of parameters (p = 26), practical identifiability was also excellent, with minimal correlation between parameters, low RSEs (all < 27%) and eta-shrinkage (< 9%).
Remarkably, this novel mechanistic model outperformed previous modeling attempts using TK + BK empirical models as evidenced by a substantial decrease in the corrected Bayesian information criterion (difference of 5,883 for COMBO, and 1,819 for MONO).
TLAN-kML4 on COMBO yielded a cross-validation test C-index of 0.67± 0.02 and AUC of 0.78±0.03, versus 0.66 ± 0.04 ; 0.74 ± 0.04 and 0.64 ± 0.04 ; 0.7± 0.06, using TK4 or BSL, respectively.
On MONO, the individual predictions were substantially better than on COMBO. TALN-kml4 yielded a 0.74± 0.02 C-index and 0.83± 0.04 AUC; these dropped to 0.71± 0.03 and 0.78± 0.05 with TK4; and 0.66± 0.02 and 0.72± 0.04 using BSL.
Conclusions
:
TALN-k offers interpretability for combined TK-BK dynamics and outperformed previous empirical models. OS prediction was also improved, with better performances for immuno-monotherapy than for immuno-combotherapy.
Such a modeling framework could have three main concrete applications:
To inform indvidual clinical decisions at bedside (personalized healthcare); To integrate previous trials and early data to anticipate the outcome of a phase 3 trial, and optimize the timing for an interim analysis; To help assess in Phase 1b, whether a subsequent Phase 3 would be relevant.
8.6 Mathematical modeling of routine biological parameters kinetics in order to predict durable benefit for first-line immunotherapy in metastatic or unresectable melanoma
Participants: Jessica Forestier, Alice Daumas, Marie-Aleth Richard, Caroline Gaudy-Marqueste, Sebastien Benzekry.
Publication: Communicated at ASCO 84
Background
: Programmed-death 1 (PD1), Cytotoxic T-lymphocyte associated protein 4 (CTLA4) and Lymphocyte-activation gene 3 (LAG3) blockade have shown remarkable efficacy in melanoma patients. Predictive biomarkers are needed to predict response, prevent unneeded toxicity and reduce costs. Despite collective efforts, no biomarker is robust or accessible enough for routine use. Our objective was to evaluate mathematical modeling-based kinetic markers derived from routine biological parameters as early predictors of durable benefit for first-line immunotherapy in advanced melanoma. Our secondary objective was to assess if these modeling could predict progression-free survival (PFS).
Methods
: We retrospectively included all consecutive, treatment-naïve, metastatic or unresectable melanoma patients treated with PD1 blockade alone or in combination with CTLA4 or LAG3 blockade between April 2020 and 2022 in our department. Clinical, biological and radiological data were collected. The primary endpoint was durable clinical benefit (DCB) defined as partial or complete response at 6 months or stable disease of at least 6 months. 11 simple blood markers and their on-treatment kinetics were considered. 4 empirical mathematical models (constant, linear, double exponential and hyperbolic) were fitted to the data, allowing to reject the constant null hypothesis for all of them. The association of the best-models coefficients with DCB and PFS was assessed using logistic and Cox regression. To do so, the data was truncated at 1, 2 and 3 months and the model coefficients were retrieved using Bayesian estimation with a population prior identified on the full-kinetics dataset. This study was approved by our local IRB.
Results
: 61 patients were included with 24 months median follow-up. DCB was observed for 57% of them. A low model-based value at baseline using 1 month truncated data was significantly associated with DCB for neutrophils (OR = 4.54[1.69-12.5], p < 0.0001) and - neutrophils/lymphocytes ratio (NLR) (OR = 3.03 [1.47-6.67], p = 0.003). Model-based results increased AUC versus baseline raw data: 0.755 versus 0.699 for neutrophils and 0.76 versus 0.7 for NLR. An elevated model-based at baseline using 1 month truncated data was significantly associated with PFS for neutrophils (HR = 1.56 [1.25-1.96], p < 0.0001), AST (HR = 1.71 [1.24-2.35], p < 0.0001), LDH (HR = 2.0 [1.44-2.77], p < 0.0001), CPR (HR = 1.75 [1.14-2.67], p = 0.01) and ALT (HR = 1.37 [1.05-1.78], p = 0.02). At 1 month, the kinetics parameters on creatinine (HR = 1.38 [1.05-1.82], p = 0.02), albumin (HR = 0.68 [0.48-0.97], p = 0.03) and monocytes (HR = 1.49 [1.02-2.18], p = 0.04) were significantly associated with PFS.
Conclusions
: Modeling the kinetics of routine biological parameters shows potential ability to predict DCB. Perspectives include model improvement, kinetics machine learning integrative model development as well as a toxicity prediction.
8.7 Pharmacokinetic Modeling for drug delivery
Participants: Anne Rodallec, Sophie Marolleau, Sébastien Benzekry.
Publication: communicated at CRS 92 and Pharmacometrics in France 72
Data: PK and PD data on breast cancer bearing mice, Paris-Sud.
Background
Commercial Paclitaxel (PTX) formulations such as Taxol are associated with adverse drug toxicities related to the added emulsionizer. Therefore, PTX formulations have been explored as they could increase efficacy and have improved pharmacokinetic (PK) properties. Recently our partners have developed a new polymer prodrug of PTX for which we proposed that metronomic dosing could result in an effective and tolerable anticancer treatment that is simultaneously convenient for patients as it allows for subcutaneous (SC) administration.
Objective
The objective of this study was to develop a PK/PD model of a novel polymer prodrug subcutaneously injectable to predict the best administration scheme.
Methods
Pharmacokinetics and pharmacodynamics (PD) studies were done on MCF-7 bearing mice using Monolix software. The PK model was developed on both intravenous (IV) Ptx and SC Ptx-PAAm (7mg/kg) data. The PD model was developed on three PD data sets (control, IV Ptx, SC Ptx-PAAm 15mg/kg), and validated on an independent group (SC Ptx-PAAm 60mg/kg). Simulations explored multiple treatment schedules, and the most effective ones were tested in vivo.
Results
The optimal model included a two-compartment PK structure and drug resistance in the PD component. The in vivo results demonstrated excellent agreement with the model predictions. A loading dose followed by daily administration achieved 60% complete response, outperforming previous internal results (tumor volume reduced to 60 versus 1350 mm), without additional toxicity.
Conclusion
Our PK/PD model showed SC Ptx-PAAm with optimized regimens significantly increased efficacy over standard schedules.
8.8 Adaptive dosing of high-dose busulfan in real-world adult patients undergoing haematopoietic cell transplant conditioning
Participants: Dorian Protzenko, Joseph Ciccolini.
Background
To evaluate the effectiveness of a Bayesian adaptive dosing strategy in achieving target busulfan exposure in adult patients undergoing haematopoietic cell transplantation (HCT).
Patients and Methods
This study included 71 adult patients scheduled to receive high-dose busulfan. Busulfan was administered to achieve a cumulative area under the curve (AUC) of 66.0 mg/L/h (16 000 μM/min), 82.60 mg/L/h (20 000 μM/min) or 87.6 mg/L/h (21 200 μM/min) depending on the regimen. Individual pharmacokinetic (PK) parameters of busulfan were estimated from three blood samples using a one-compartment model and Bayesian estimation after the first standard dose. Individual PK parameters were used to adjust subsequent doses to achieve the target exposure.
Results
All patients had their dose adjusted after the first dose administration. The final deviation from the target AUC was significantly improved compared to the initial deviation after standard mg/kg dosing (mean absolute deviation 19.5% vs 11.7%, P ). In addition, the proportion of patients with marked deviation from target exposure (ie, %) decreased significantly from 31% after standard dosing to 10% after PK-guided dosing (P ). Canonical busulfan-related toxicity, specifically veno-occlusive disease, was observed in 5% of patients who achieved successful PK-guided dosing. In contrast, one-third of patients with off-target exposure with poor dosing experienced toxicity.
Conclusion
The Bayesian adaptive dosing strategy significantly improves the accuracy of achieving the target busulfan AUC in patients undergoing HCT. This approach not only reduces marked deviations from target exposure, but also reduces the incidence of busulfan-related toxicity, thereby maintaining a favorable toxicity/efficacy ratio.
8.9 Overcoming immuno-resistance by rescheduling anti-VEGF/cytotoxics/anti-PD-1 combination in lung cancer model
Participants: Guillaume Sicard, Dorian Protzenko, Sarah Giacometti, Joseph Ciccolini, Raphaelle Fanciullino.
Background
Many tumors are refractory to immune checkpoint inhibitors, but their combination with cytotoxics is expected to improve sensitivity. Understanding how and when cytotoxics best re-stimulate tumor immunity could help overcome resistance to immune checkpoint inhibitors.
Methods
In vivo studies were performed in C57BL/6 mice grafted with immune-refractory LL/2 lung cancer model. A longitudinal immunomonitoring study on tumor, spleen, and blood after multiple treatments including Cisplatin, Pemetrexed, and anti-VEGF (either alone or in combination) was performed, spanning a period of up to 21 days, to determine the optimal time window during which immune checkpoint inhibitors should be added. Finally, an efficacy study was conducted comparing the antiproliferative performance of various schedules of anti-VEGF, Pemetrexed-Cisplatin doublet, plus anti-PD-1 (i.e., immunomonitoring-guided scheduling, concurrent dosing or a random sequence), as well as single agent anti-PD1.
Results
Immunomonitoring showed marked differences between treatments, organs, and time points. However, harnessing tumor immunity (i.e., promoting CD8 T cells or increasing the T CD8/Treg ratio) started on D7 and peaked on D14 with the anti-VEGF followed by cytotoxics combination. Therefore, a 14-day delay between anti-VEGF/cytotoxic and anti-PD1 administration was considered the best sequence to test. Efficacy studies then confirmed that this sequence achieved higher antiproliferative efficacy compared to other treatment modalities (i.e., -71% in tumor volume compared to control).
Conclusions
Anti-VEGF and cytotoxic agents show time-dependent immunomodulatory effects, suggesting that sequencing is a critical feature when combining these agents with immune checkpoint inhibitors. An efficacy study confirmed that sequencing treatments further enhance antiproliferative effects in lung cancer models compared to concurrent dosing and partly reverse the resistance to cytotoxics and anti-PD1.
8.10 Pegylated liposome encapsulating docetaxel using microfluidic mixing technique: Process optimization and results in breast cancer models
Participants: Mathilde Dacos, Anne Rodallec, Benoit Immordino, Sarah Giacometti, Guillaume Sicard, Joseph Ciccolini, Raphaelle Fanciullino.
Background
The development of nanoparticles could help to improve the efficacy/toxicity balance of drugs. This project aimed to develop liposomes and immunoliposomes using microfluidic mixing technology.
Patients and Methods
Various formulation tests were carried out to obtain liposomes that met the established specifications. The liposomes were then characterized in terms of size, polydispersity index (PDI), docetaxel encapsulation rate and lamellarity. Antiproliferative activity was tested in human breast cancer models ranging from near-negative (MDA-MB-231), positive (MDA-MB-453) to HER2 positive. Pharmacokinetic studies were performed in C57BL/6 mice. Numerous batches of liposomes were synthesized using identical molar ratios and by varying the microfluidic parameters TFR, FRR and buffer.
Results
All synthesized liposomes have a size < 200 nm, but only Lipo-1, Lipo-6, Lipo-7, Lipo-8 have a PDI < 0.2, which meets our initial requirements. The size of the liposomes was correlated with the total FRR, for a 1:1 FRR the size is 122.2 ± 12.3 nm, whereas for a 1:3 FRR the size obtained is 163.4 ± 34.0 nm (p = 0.019). Three batches of liposomes were obtained with high docetaxel encapsulation rates > 80 %. Furthermore, in vitro studies on breast cancer cell lines demonstrated the efficacy of liposomes obtained by microfluidic mixing technique. These liposomes also showed improved pharmacokinetics compared to free docetaxel, with a longer half-life and higher AUC (3-fold and 3.5-fold increase for the immunoliposome, respectively).
Conclusions
This suggests that switching to the microfluidic process will produce batches of liposomes with the same characteristics in terms of in vitro properties and efficacy, as well as the ability to release the encapsulated drug over time in vivo. This time-efficiency of the microfluidic technique is critical, especially in the early stages of development.
8.11 Body mass index affects imatinib exposure: Real-world evidence from TDM with adaptive dosing
Participants: Paul Maroselli, Joseph Ciccolini, Raphaelle Fanciullino.
Background
Imatinib is the treatment of elderly or frail patients with chronic myeloid leukemia (CML). Trough levels of around 1000 ng/ml are considered as the target exposure.
Methods
We searched for baseline parameters associated with imatinib pharmacokinetics, and studied the clinical impact of subsequent adaptive dosing. We present data from 60 adult CML patients upon imatinib with therapeutic drug monitoring (TDM) and adaptive dosing.
Results
Mean trough levels after treatment initiation were 994.2 ± 560.6 ng/ml (with 56% inter-patient variability). Only 29% of patients were in the therapeutic range. Body weight, height, body surface area, body mass index (BMI), and age were associated with imatinib plasma levels on univariate analysis. Age and BMI remained the only parameters associated with imatinib trough levels on multivariate analysis. As severe toxicities have been previously reported in patients with low BMI treated with standard imatinib, we evaluated the extent to which low BMI may lead to plasma overexposure. We found a statistically significant difference in trough imatinib levels in patients with BMI kg/m2, with exposure +61.5% higher than in patients with 18.5 BMI 24.9 and +76.3% higher than in patients with BMI 25. After TDM with adaptive dosing, a statistically significant difference in dosing between patients was observed, with doses ranging from 200 to 700 mg. No difference in toxicity or efficacy was observed regardless of BMI after adaptive dosing.
Conclusion
Our data suggest that low BMI has a significant impact on imatinib exposure but that pharmacokinetically-guided dosing limits its clinical impact in patients.
8.12 Life-threatening toxicities upon Pembrolizumab intake: could pharmacokinetics be the bad guy?
Participants: Mourad Hamimed, Sophie Marolleau, Joseph Ciccolini.
Background
We report the case of an adult patient diagnosed with Hodgkin's lymphoma who was scheduled for Pembrolizumab after failure of standard therapy. After three well-tolerated courses of Pembrolizumab, a PET scan showed a favorable outcome and a fourth course of Pembrolizumab was started. Unexpectedly, extremely severe toxicities (i.e., autoimmune peripheral hypothyroidism, rhabdomyolysis and severe acute renal failure) occurred after this last course, requiring transfer to the intensive care unit.
Methods
Therapeutic drug monitoring was performed to measure residual Pembrolizumab levels at intervals from the last dose (i.e., 120 and then 170 days), as well as pharmacogenetics investigations on the FCγR gene.
Results
Pembrolizumab plasma concentrations that were still pharmacologically active months after the last administration, suggesting impaired elimination of Pembrolizumab in this patient. Further pharmacokinetic modeling based on the population approach showed that both half-life (47.8 days) and clearance (0.12 L/day) values were significantly different from the standard values usually reported in patients. Further in silico simulations showed that pharmacologically active concentrations of Pembrolizumab were maintained for up to 136 days after the last dose. The search for possible polymorphisms affecting the genes coding for FCγR (i.e., rs1801274 on FCGR2A and rs396991 on FCGR3A gene) was negative. Further TDM showed that Pembrolizumab could be detected up to 263 days after the last administration.
Conclusion
This case report suggests that persistent overexposure in plasma could lead to life-threatening toxicities with Pembrolizumab.
8.13 Poor prognosis of SRSF2 gene mutations in patients treated with VEN-AZA for newly diagnosed acute myeloid leukemia
Participants: Raphaelle Fanciullino.
Background
Mutations in spliceosome genes (SRSF2, SF3B1, U2AF1, ZRSR2) correlate with inferior outcomes in patients treated with intensive chemotherapy for Acute Myeloid Leukemia. However, their prognostic impact in patients treated with less intensive protocols is not well known.
Methods
This study aimed to evaluate the impact of Spliceosome mutations in patients treated with Venetoclax and Azacitidine for newly diagnosed AML. 117 patients treated in 3 different hospitals were included in the analysis.
Results
Thirty-four harbored a mutation in at least one of the spliceosome genes (splice-mut cohort). K/NRAS mutations were more frequent in the splice-mut cohort (47% vs 19%, p=0.0022). Response rates did not differ between splice-mut and splice-wt cohorts. With a median follow-up of 15 months, splice mutations were associated with a lower 18-month LFS (p=0.0045). When analyzing splice mutations separately, we found SRSF2 mutations to be associated with poorer outcomes (p=0.034 and p=0.037 for OS and LFS respectively). This negative prognostic impact remained true in our multivariate analysis.
Conclusion
We believe this finding should warrant further studies aimed at overcoming this negative impact.
9 Bilateral contracts and grants with industry
Participants: Sebastien Benzekry, Joseph Ciccolini, Raphaelle Fanciullino, Laurent Greillier, Sebastien Salas.
9.1 Research contracts
kML 2.0
- Partner: Genentech
- Title: Kinetics - machine learning modeling for prediction of outcome in oncology
- Funding: 25 k€
- Duration: Oct 2023 - Apr 2024
- Principal investigator: S. Benzekry
9.2 Clinical trials
CetuxiMAX
- Registration: NCT4218136
- Partner: Merck Serono
- Title: Maximizing Cetuximab efficacy in head and neck cancer patients through PK/PD modeling
- Funding: 40 k€
- Duration: 2020 - 2024
- Principal investigator: Sébastien Salas
IMHOTEP
- Registration: NCT04795661
- Partner: INCa
- Title: Immunotherapy in MSI/dMMR Tumors in Perioperative Setting
- Funding: 800 k€
- Duration: 2023 - 2027
- COMPO investigator: Joseph Ciccolini
MOIO
- Registration: NCT05078047
- Partner: BMS and INCa
- Title: Study Comparing the Standard Administration of IO Versus the Same IO Administered Each 3 Months in Patients With Metastatic Cancer in Response After 6 Months of Standard IO
- Funding: 500 k€
- Duration: 2020-2024
- Principal investigator: Gwenaelle Gravis (IPC)
- COMPO investigator: Joseph Ciccolini
PEMBOV
- Registration: NCT03596281
- Partner: INCa
- Title: Pembrolizumab in Combination With Bevacizumab and Pegylated Liposomal Doxorubicin in Patients With Ovarian Cancer (PEMBOV)
- Funding: 700 k€
- Duration: 2020-2025
- Principal investigator: Judith Mitchels (IGR)
- COMPO investigator: Joseph Ciccolini
REZOLVE
- Registration: ANZGOG-1101
- Partner: Sydney Medical Center Australia
- Title: Pembrolizumab in Combination With Bevacizumab and Pegylated Liposomal Doxorubicin in Patients With Ovarian Cancer (PEMBOV)
- Funding: Aus$ 800k
- Duration: 2018-2024
- Principal investigator: Sonia Yip (Sydney University)
- COMPO investigator: Joseph Ciccolini
VENETACIBLE
- Registration: AORC 2023-AO1546-39
- Partner: CHU Nice
- Title: Personalizing Venetoclax dosing in AML patients non-eligible for HD chemotherapy as first line treatment.
- Funding: 30 k€ (Nice University Hospitals)
- Duration: 2023-2024
- COMPO Investigator: Raphaelle Fanciullino
PDC-LUNG-101
- Registration: Eudract 2018-002382-19 / NCT03970746
- Partner: PDC*line Pharma SAS
- Title: Safety, immunogenicity and preliminary clinical activity study of PDC*lung01 cancer vaccine in NSCLC
- Duration: 2022-2024
- Principal investigator: Johan Vansteenkiste, Prof KU Leuven
- COMPO investigator: Laurent Greillier
PROPEL
- Registration: Eudract 2019-003474-35 / NCT03138889
- Partner: Nektar Therapeutics
- Title: Bempegaldesleukin and Pembrolizumab With or Without Chemotherapy in Locally Advanced or Metastatic Solid Tumors
- Duration: 2017-2024
- COMPO investigator: Laurent Greillier
NCT04721015
- Registration: Eudract 2020-004953-57 / NCT04721015
- Partner: AbbVie
- Title: Study of Intravenous (IV) ABBV-637 Alone or in Combination With IV Docetaxel/Osimertinib to Assess Adverse Events and Change in Disease Activity in Adult Participants With Relapsed/Refractory (R/R) Solid Tumors
- Duration: 2021-2026
- COMPO investigator: Laurent Greillier
PIONeeR
- Registration: NCT03493581
- Partner: APHM, HalioDx, Innate Pharma, AMU
- Title: Precision Immuno-Oncology for Advanced Non-small Cell Lung Cancer Patients With PD-1 ICI Resistance
- Duration: 2018-2024
- Principal investigator: Laurent Greillier, Jean-Olivier Arnaud
NCT01817192
- Registration: NCT01817192
- Partner: Razor Genomics
- Title: Adjuvant Chemotherapy in Patients With Intermediate or High Risk Stage I or Stage IIA Non-squamous Non-Small Cell Lung Cancer
- Duration: 2020-2025
- Principal investigator: David R Spigel, Sarah Cannon
- COMPO investigator: Laurent Greillier
NCT04042558
- Registration: NCT04042558
- Partner: Centre Francois Baclesse
- Title: A Study Evaluating Platinum-Pemetrexed-Atezolizumab (+/-Bevacizumab) for Patients With Stage IIIB/IV Non-squamous Non-small Cell Lung Cancer With EGFR Mutations, ALK Rearrangement or ROS1 Fusion Progressing After Targeted Therapies
- Duration: 2019-2024
- COMPO investigator: Laurent Greillier
Canopy-A
- Registration: NCT03447769
- Partner: Novartis Pharmaceuticals
- Title: Study of Efficacy and Safety of Canakinumab as Adjuvant Therapy in Adult Subjects With Stages AJCC/UICC v. 8 II-IIIA and IIIB (T>5cm N2) Completely Resected Non-small Cell Lung
- Duration: 2018-2027
- COMPO investigator: Laurent Greillier
NCT04350463
- Registration: NCT04350463
- Partner: Celgene
- Title: A Safety and Efficacy Study of CC-90011 in Combination With Nivolumab in Subjects With Advanced Cancers
- Duration: 2020-2024
- COMPO investigator: Laurent Greillier
TROPION-LUNG01
- Registration: NCT04656652
- Partner: Daiichi Sankyo, Inc., AstraZeneca
- Title: Study of DS-1062a Versus Docetaxel in Previously Treated Advanced or Metastatic Non-small Cell Lung Cancer Without Actionable Genomic Alterations
- Duration: 2020-2024
- COMPO investigator: Laurent Greillier
MERMAID-1
- Registration: NCT04385368
- Partner: AstraZeneca
- Title: Phase III Study to Determine the Efficacy of Durvalumab in Combination With Chemotherapy in Completely Resected Stage II-III Non-small Cell Lung Cancer (NSCLC)
- Duration: 2020-2026
- Principal investigator: Solange Peters, Charles Swanton
- COMPO investigator: Laurent Greillier
CARMEN-LC03
- Registration: NCT04154956
- Partner: Sanofi
- Title: SAR408701 Versus Docetaxel in Previously Treated, Carcinoembryonic Antigen-related Cell Adhesion Molecule 5 (CEACAM5) Positive Metastatic Non-squamous Non-small Cell Lung Cancer Patients
- Duration: 2020-2024
- COMPO investigator: Laurent Greillier
SKYSCRAPER-03
- Registration: NCT04513925
- Partner: Hoffmann-La Roche
- Title: A Study of Atezolizumab and Tiragolumab Compared With Durvalumab in Participants With Locally Advanced, Unresectable Stage III Non-Small Cell Lung Cancer (NSCLC)
- Duration: 2020-2027
- COMPO investigator: Laurent Greillier
NCT03899155
- Registration: NCT03899155
- Partner: Bristol-Myers Squibb
- Title: Pan Tumor Nivolumab Rollover Study
- Duration: 2019-2025
IMbrella A
- Registration: NCT03148418
- Partner: Hoffmann-La Roche
- Title: A Study in Participants Previously Enrolled in a Genentech- and/or F. Hoffmann-La Roche Ltd-Sponsored Atezolizumab Study
- Duration: 2017-2030
- COMPO investigator: Laurent Greillier
BEAT-meso
- Registration: NCT03762018
- Partner: European Thoracic Oncology Platform, Hoffmann-La Roche
- Title: Bevacizumab and Atezolizumab in Malignant Pleural Mesothelioma
- Duration: 2019-2024
- Principal investigator: Enriqueta Felip, Sanjay Popat
- COMPO investigator: Laurent Greillier
SPECTA
- Registration: NCT02834884
- Partner: European Organisation for Research and Treatment of Cancer - EORTC
- Title: Screening Cancer Patients for Efficient Clinical Trial Access
- Duration: 2017-2026
- Principal investigator: Vassilis Golfinopoulos
- COMPO investigator: Laurent Greillier
TROPION-Lung05
- Registration: EudraCT 2020-002774-27
- Partner: Daiichi Sankyo, Inc.
- Title: Phase 2 Study of DS-1062a in Advanced or Metastatic Non-small Cell Lung Cancer with Actionable Genomic Alterations
- Starting year: 2021
- COMPO investigator: Laurent Greillier
SKYSCRAPER-06
- Registration: NCT04619797
- Partner: Hoffmann-La Roche
- Title: A Study of Tiragolumab in Combination With Atezolizumab Plus Pemetrexed and Carboplatin/Cisplatin Versus Pembrolizumab Plus Pemetrexed and Carboplatin/Cisplatin in Participants With Previously Untreated Advanced Non-Squamous Non-Small Cell Lung Cancer
- Duration: 2020 - 2025
- COMPO investigator: Laurent Greillier
PERSEE
- Registration: EudraCT 2020-002626-86
- Partner: CHRU de Brest
- Title: A trial comparing the pembrolizumab platinum based chemotherapy combination with pembrolizumab monotherapy in first line treatment of non small-cell lung cancer (NSCLC) patients
- Starting year: 2020
- Principal investigator: Renaud Descourt, Chantal Decroisette, Christos Chouaid
- COMPO investigator: Laurent Greillier
SAPPHIRE
- Registration: EudraCT 2019-001043-41
- Partner: Mirati Therapeutics, Inc.
- Title: A Randomized Phase 3 Study of Sitravatinib in Combination with Nivolumab Versus Docetaxel in Patients with Advanced Non-Squamous Non-Small Cell Lung Cancer with Disease Progression On or After Platinum-Based Chemotherapy and Checkpoint Inhibitor Therapy
- Starting year: 2020
- COMPO investigator: Laurent Greillier
Nivothym
- Registration: EudraCT 2015-005504-28
- Partner: European Organisation for Research and Treatment of Cancer
- Title: Single-arm, multicenter, phase II study of immunotherapy in patients with type B3 thymoma and thymic carcinoma previously treated with chemotherapy
- Starting year: 2017
- Principal investigator: Nicolas Girard, Solange Peters
- COMPO investigator: Laurent Greillier
ELEVATE HNSCC
- Registration: NCT04854499
- Partner: Gilead Sciences
- Title: Study of Magrolimab Combination Therapy in Patients With Head and Neck Squamous Cell Carcinoma
- Duration: 2021-2025
- COMPO investigator: Sébastien Salas
ROMANE
- Registration: ML43492
- Partner: ROCHE
- Title: Retrospective observational study of patients with locally advanced squamous cell carcinoma of the head and neck
- Starting year: 2023
- COMPO investigator: Sébastien Salas
XRAY VISION
- Registration: NCT05386550
- Partner: EMD Serono Research & Development Institute, Inc.
- Title: Phase III Xevinapant (Debio 1143) and Radiotherapy in Resected LA SCCHN, High Risk, Cisplatin-ineligible Participants
- Duration: 2022-2030
- COMPO investigator: Sébastien Salas
Iintune-1
- Registration: NCT04420884
- Partner: Takeda
- Title: A Study of Dazostinag as Single Agent and Dazostinag in Combination With Pembrolizumab in Adults With Advanced or Metastatic Solid Tumors
- Duration: 2020-2026
- COMPO investigator: Sébastien Salas
AHEAD-MERIT
- Registration: NCT04534205
- Partner: BioNTech SE
- Title: A Clinical Trial Investigating the Safety, Tolerability, and Therapeutic Effects of BNT113 in Combination With Pembrolizumab Versus Pembrolizumab Alone for Patients With a Form of Head and Neck Cancer Positive for Human Papilloma Virus 16 and Expressing the Protein PD-L1
- Duration: 2021-2028
- COMPO investigator: Sébastien Salas
CODEBREAK IGR
- Registration: NCT05631249
- Partner: Gustave Roussy, Cancer Campus, Grand Paris
- Title: Sotorasib in Previously Treated Locally Advanced or Metastatic NSCLC Subjects With Mutated KRAS p.G12C
- Duration: 2022-2026
- COMPO investigator: Laurent Greillier
ADAPTABLE
- Registration: NCT05781308
- Partner: Intergroupe Francophone de Cancerologie Thoracique
- Title: Combination of Paclitaxel-bevacizumab ± Atezolizumab in Patients With Advanced NSCLC Progressing After Immunotherapy & Chemotherapy
- Duration: 2023-2026
- COMPO investigator: Laurent Greillier
MATISSE
- Registration: NCT05742607
- Partner: Innate Pharma
- Title: IPH5201 and Durvalumab in Patients With Resectable Non-Small Cell Lung Cancer
- Duration: 2023-2026
- COMPO investigator: Laurent Greillier
DESTINY-Lung04
- Registration: NCT05048797
- Partner: AstraZeneca
- Title: A Study to Investigate the Efficacy and Safety of Trastuzumab Deruxtecan as the First Treatment Option for Unresectable, Locally Advanced/Metastatic Non-Small Cell Lung Cancer With HER2 Mutations
- Duration: 2021-2027
- COMPO investigator: Laurent Greillier
KRYSTAL-12
- Registration: NCT04685135
- Partner: Mirati Therapeutics Inc.
- Title: Phase 3 Study of MRTX849 (Adagrasib) vs Docetaxel in Patients With Advanced Non-Small Cell Lung Cancer With KRAS G12C Mutation
- Duration: 2021-2024
- COMPO investigator: Laurent Greillier
PECATI
- Registration: NCT04710628
- Partner: MedSIR
- Title: Combination of Pembrolizumab and Lenvatinib, in Pre-treated Thymic CArcinoma paTIents
- Duration: 2021-2026
- COMPO investigator: Laurent Greillier
A2A-005
- Registration: NCT05403385
- Partner: iTeos Belgium SA
- Title: Study of Inupadenant (EOS100850) With Chemotherapy as Second Line Treatment for Nonsquamous Non-small Cell Lung Cancer
- Duration: 2022-2025
- COMPO investigator: Laurent Greillier
Delivir
- Registration: NCT05788926
- Partner: Transgene
- Title: A Clinical Trial of TG6050 in Patients With Metastatic Non-Small Cell Lung Cancer
- Duration: 2023-2025
- COMPO investigator: Laurent Greillier
BEAMION-Lung 1
- Registration: NCT04886804
- Partner: Boehringer Ingelheim
- Title: A Study to Test Different Doses of BI 1810631 in People With Different Types of Advanced Cancer (Solid Tumors With Changes in the HER2 Gene)
- Duration: 2021-2027
- COMPO investigator: Laurent Greillier
ELEVATE Lung & UC
- Registration: NCT04827576
- Partner: Gilead Sciences
- Title: Study of Magrolimab in Patients With Solid Tumors
- Duration: 2021-2025
- COMPO investigator: Laurent Greillier
IDE397-001
- Registration: NCT04794699
- Partner: IDEAYA Biosciences
- Title: Study of IDE397 in Participants With Solid Tumors Harboring MTAP Deletion
- Duration: 2021-2024
- COMPO investigator: Laurent Greillier
CA099-003
- Registration: NCT05407675
- Partner: Bristol-Myers Squibb
- Title: A Study to Evaluate the Safety and Tolerability of BMS-986408 Alone and in Combination With Nivolumab or Nivolumab and Ipilimumab in Participants With Advanced Solid Tumors
- Duration: 2022-2027
- COMPO investigator: Laurent Greillier
10 Partnerships and cooperations
Participants: Dominique Barbolosi, Sebastien Benzekry, Elias Ventre, David Boulate, Joseph Ciccolini, Raphaelle Fanciullino, Florence Gattacceca, Laurent Greillier, Xavier Muracciole, Anne Rodallec, Sebastien Salas.
10.1 International research visitors
10.1.1 Visits of international scientists
Marija Jovanovic; Masa Roganovic; Milena Kovacevic
-
Status:
Researcher (Clin PK)
-
Institution of origin:
University of Belgrade
-
Country:
Serbia
-
Dates:
December 2024
-
Context of the visit: :
Joint Program on clinical pharmacokinetics of immune checkpoint inhibitors
-
Mobility Program:
Pavle Savic bilateral grant
10.1.2 Visits to international teams
Research stays abroad
Anne Rodallec
-
Visited institution:
Pharmaceutical Institut of the University of Bonn
-
Country:
Germany
-
Dates:
September 2023 - August 2024
-
Context of the visit:
This exchange was funded by the Deutscher Akademischer Austauschdienst (DAAD) and the fondation ARC to develop our skills on immunomonitoring in vitro and ex vivo for several projects involving Immunotherapies, including COMPLICITY project
-
Mobility program/type of mobility:
Research stay + Lectures
Joseph Ciccolini
-
Visited institution:
School of Pharmacy University of Belgrade
-
Country:
Serbia
-
Dates:
November 2024
-
Context of the visit:
Exchange based upon the Pavle Savic bilateral grant at Campus France supporing a clinical research project on the PK/PD of nivolumab in patientswith cancer
-
Mobility program/type of mobility:
Research stay + Lectures
10.2 National initiatives
PIONeeR - biomarkers
-
Title:
Precision Immuno-Oncology for advanced Non-small cell lung cancer patients with PD-1 ICI Resistance
-
Partner Institutions:
- AP-HM, Marseille
- 13 national cancer centers, including Centre Lyon Bérard and IUCT in Toulouse
- Marseille Immmunopôle (immuno-monitoring platform)
- Vasculo-monitoring platform of AP-HM
- Inserm
- Aix-Marseille University
- Veracyte, Marseille
- InnatePharma, Marseille
-
Date/Duration:
2018 - 2024
-
Funding:
8 M€, ANR
-
Principal investigator:
F. Barlesi (IGR)
-
COMPO members involved:
S. Benzekry, J. Ciccolini, L. Greillier, P. Dufosse, M. Boussena, M. Hamimed, M. Karlsen, S. Marolleau, C. Marin, A. Vaglio.
LUCA-pi RHU
-
Title:
Lung cancer prevention and interception
-
Partner Institutions:
- Gustave Roussy Institute (G. Kroemer, L. Zitvogel)
- Therapanacea (N. Paragios)
- CIML (P. Milpied)
-
Date/Duration:
2023 - 2028
-
Funding:
10 M€, ANR
-
Principal investigator:
D. Boulate (COMPO)
-
COMPO seniors:
D. Barbolosi, S. Benzekry
AML
-
Title:
Prédiction de la Toxicité du Venetoclax dans la Population de Patients Agés traités pour une LAM
-
Partner Institutions:
- AP-HM, Marseille
-
Date/Duration:
2023 - 2026
-
Funding:
30k€, GIRCI
-
Principal investigator:
Sylvain Garciaz (IPC, Marseille)
-
COMPO members involved:
S. Benzekry, R. Fanciullino, A. Bakhmach
DIGPHAT PEPR Digital Health
-
Title:
Digital pharmacological twin
-
Partner Institutions:
- JB Woillard (CHU Limoges)
- C. Battail (CEA Grenoble)
- M. Ursino + S. Zohar (HEKA, Inria, Paris)
- J. Josse (PREMEDICAL, Inria, Montpellier)
- E. Chatelut + M. White-Koning (IUCT, Toulouse)
-
Date/Duration:
2023 - 2028
-
Funding:
Total 1.8 M€, COMPO 251k€
-
Principal investigator:
JB Woillard (CHU Limoges)
-
COMPO senior:
S. Benzekry.
-
COMPO junior:
A. Bakhmach
COPYCAT
-
Title:
Combining Organoid technology with Mathematics to develop innovative models mimicking tumor cellular heterogeneity and plasticity for pediatric oncology
-
Partner Institutions:
- L. Broutier (CRCL, Inserm, CNRS, UCBL, Lyon)
- E. Pasquier (CRCM)
- R. Mounier (INMG, Inserm, CNRS, UCBL, Lyon)
-
Date/Duration:
2023 - 2027
-
Funding:
Total 922k€, COMPO 115k€ (INCa)
-
Principal investigator:
L. Broutier (CRCL, Inserm, CNRS, UCBL, Lyon)
-
COMPO members involved:
S. Benzekry, E. Ventre
SChISM
-
Title:
Size CfDNA Immunotherapies Signature Monitoring
-
Partner Institutions:
- APHM
- M. Lavielle (XPOP – Inria)
- Adelis,
- F. Fina (ID-Solutions Oncology)
-
Date/Duration:
2022 - 2025
-
Funding:
120k€, APHM + PhD grant ICI - Laennec
-
Principal investigator:
S. Benzekry, S. Salas
-
COMPO junior:
L. Nguyen Phuong
METAMATS
-
Title:
Mechanistic modeling for the prediction of metastatic relapse in breast cancer
-
Partner Institutions:
- F. Bertucci (IPC, Marseille)
- G. MacGrogan (Institut Bergonié , Bordeaux)
-
Date/Duration:
2020 - 2024
-
Funding:
100k€, Inria-Inserm PhD grant
-
Principal investigator:
S. Benzekry, X. Muracciole
-
COMPO members involved:
C. Bigarre
PhD H. Hamdache
-
Title:
Improving therapeutic efficacy and managing side effects and sequelae in pediatric cancer through improved and personalized nutritional programs using computer simulations
-
Partner Institutions:
- V. Pancaldi (IUCT, Inserm, Toulouse)
-
Date/Duration:
2023 - 2026
-
Funding:
120k€, Inria-Inserm PhD grant
-
Principal investigator:
V. Pancaldi (IUCT, Inserm, Toulouse)
-
COMPO members involved:
S. Benzekry (co-supervisor).
South-ROCK
-
Title:
South-research on cancer for kids
-
Partner Institutions:
28 constitutive teams
- P. Mehlen (Centre Léon Bérard + Hospices Civils de Lyon)
- E. Pasquier (CRCM, Marseille)
- M. Castets (CRCL, Lyon)
-
Date/Duration:
2023 - 2028
-
Funding:
Total 2 M€
-
Principal investigators:
P. Mehlen (CLB + HCL, Lyon), E. Pasquier (CRCM, Marseille), M. Castets (CRCL, Lyon)
-
COMPO seniors:
S. Benzekry, F. Gattacceca, J.Ciccolini
THERMONANO
-
Title:
Nanoassemblies for the subcutaneous self-administration of anticancer drugs
-
Partner Institution:
- Institut Galien Paris-Saclay (UMR CNRS 8612)
-
Date/Duration:
2019 - 2024
-
Funding:
1.8 M€, ERC
-
Principal investigator:
J. Nicolas (Institut Galien, Paris-Sud)
-
COMPO members involved:
A. Rodallec, S. Benzekry, S. Marolleau.
TORNADO
-
Title:
TOols for Rational NAnoDrugs Optimization: development of a physiologically-based pharmacokinetic model for nanomedicines
-
Partner Institution:
- P. Garrigue (CERIMED, Marseille)
- S. Legeay (MINT, Univ Angers)
- E. Roger (MINT, Univ Angers)
-
Date/Duration:
2021-2024
-
Funding:
120k€ (Doctoral grant AMU ED62)
-
Principal investigator:
F Gattacceca
-
COMPO junior:
J. Ou (PhD student), M. Mahdjoub (M2 intern)
PEMBOV
-
Title:
Pembrolizumab in Combination With Bevacizumab and Pegylated Liposomal Doxorubicin in Patients With Ovarian Cancer
-
Partner Institution:
- Institut Gustave Roussy (IGR)
-
Date/Duration:
2021-2024
-
Funding:
300 K€, INCa
-
Principal investigator:
J. Michels (IGR)
-
COMPO members involved:
J. Ciccolini, M. Hamimed
REZOLVE
-
Title:
A phase 2 trial of intraperitoneal bevacizumab to treat symptomatic ascites in patients with chemotherapy-resistant, epithelial ovarian cancer
-
Date/Duration:
2020-2025
-
Funding:
total funding undisclosed, COMPO funding 40 K€
-
Principal investigator:
S. Yip (Sydney Medical Center Australia), J.Ciccolini
-
COMPO senior:
J.Ciccolini
-
COMPO junior:
C. Marin
COMPLICITY
-
Title:
COMPutationaL tools for NanoBooster In Cancer ImmunoTherapY
-
Partner Institution:
- Pharmaceutical Institute, Bonn University, GERMANY: A Lamprecht M Shetab Boushehri
-
Date/Duration:
2023-2026
-
Funding:
25k€ (Amidex Pepiniere), 30K€ (ARC), 2K€ (DAAD)
-
Principal investigator:
A. Rodallec
-
COMPO senior:
A. Rodallec, F. Gattacceca
-
COMPO junior:
C. Perez
Paris Saclay Cancer Cluster
-
Title:
Clinical Pharmacokinetics Platform
-
Partner Institution:
- Paris Saclay
-
Date/Duration:
2025-2029
-
Funding:
Paris Saclay
-
Principal investigator:
E. Vivier (PSCC)
-
COMPO members involved:
J. Ciccolini
Prevalung
-
Title:
Epidemiological Study to Assess the Prevalence of Lung Cancer in patients with smoking-associated atherosclerotic cardiovascular diseases
-
Partner Institution:
- APHM, Gustave Roussy Institute
-
Date/Duration:
2019-2025
-
Funding:
7M€ Horizon Europe
-
Principal investigator:
D. Boulate
-
COMPO members involved:
D. Boulate, D. Barbolosi, C. Buton
10.3 Regional initiatives
PETRA Network
-
Title:
PE“TRANSLA” : translational research in neuro-oncology
-
Partner Institution:
- Assistance Publique Hôpitaux de Marseille (APHM)
-
Date/Duration:
2023-2027
-
Funding:
Canceropole PACA
-
Principal investigator:
E. Tabouret (APHM)
-
COMPO members involved:
J. Ciccolini
VENETACIBLE
-
Title:
Etude PK/PD du Venetoclax dans les leucémies aigues myéloides
-
Date/Duration:
2023-2025
-
Funding:
to be determined
-
Principal investigator:
G. Venton
-
COMPO senior:
R. Fanciullino, J.Ciccolini
-
COMPO junior:
L. Osanno
CEREAL
-
Title:
Allogreffe de cellules souches hématopoïétiques après conditionnement à toxicité réduite associant cladribine, fludarabine et busulfan chez les patients atteints d'hémopathies malignes réfractaires (Phase I)
-
Date/Duration:
2020-2025
-
Funding:
to be determined
-
Principal investigator:
R. Devillier (IPC), J.Ciccolini
-
COMPO senior:
J.Ciccolini
-
COMPO junior:
D. Protzenko
MOIO
-
Title:
A non-inferiority randomized phase III trial of standard immunotherapy by checkpoint inhibitors vs. reduced dose intensity in responding patients with metastatic cancer: the MOIO protocol study
-
Date/Duration:
2022-2027
-
Funding:
1M€ total (INCA, PHRC + BMS grant), COMPO funding undetermined yet
-
Principal investigator:
G. Gravis (IPC Marseille), J.Ciccolini
-
COMPO members involved:
J.Ciccolini
IMHOTEP
-
Title:
étude de phase 2, avec 4 cohortes évaluant l’efficacité du pembrolizumab MK-3475 chez des patient(e)s atteints d'une tumeur MSI/dMMR ou d'un cancer gastrique EBV+, résécable non prétraité.
-
Date/Duration:
2024-2027
-
Funding:
1M€ total (INCA, PHRC), COMPO funding undetermined yet
-
Principal investigator:
C. de la Fouchardière (IPC Marseille), J.Ciccolini
-
COMPO members involved:
J.Ciccolini
PKID
-
Title:
Population pharmacokinetic approach centered on the patient: a patient pharmacokinetic ID from one drug to predict the PK of the next one
-
Partner Institution:
- R. Guilhaumou (APHM, Marseille)
-
Date/Duration:
2021-2024
-
Funding:
APHM (hospital intern)
-
Principal investigator:
F Gattacceca
-
COMPO junior:
A. Deschamps
PhD M. Boussena
-
Title:
Machine learning methods for clinical oncology data: application to the prediction of immunotherapy response in lung cancer
-
Partner Institutions:
- J. Josse (PREMEDICAL, Inria)
- GFPC
- CHU Brest
- Inserm Brest
-
Date/Duration:
2023 - 2026
-
Funding:
120k€, Institut Laennec
-
Principal investigator:
S. Benzekry, L. Greillier
-
COMPO members involved:
M. Boussena
PICOMALE
-
Title:
Phase I Clinical trials Oncology & Machine Learning
-
Partner Institutions:
- CLIP: N. Andre, P. Tomasini
- LIS (Laboratoire d'informatique et des systèmes): C. Roman, S. Sellami, A. Bouchra
-
Date/Duration:
2023 - 2026
-
Funding:
Total 100k€, COMPO 66k€
-
Principal investigator:
N. Andre
-
COMPO senior:
S. Benzekry.
-
COMPO junior:
C. Bigarre
COALA
-
Title:
Cure Oncogene-Addicted Lung Adenocarcinoma
-
Date/Duration:
2024 - 2029
-
Funding:
3M total, 130k€ COMPO
-
Principal investigator:
J. Mazières (CRCT)
-
COMPO members involved:
S. S. Benzekry, D. Barbolosi
Brain meta SCLC
-
Title:
Modeling the impact of prophylactic whole brain radiotherapy to prevent brain metastases in SCLC
-
Date/Duration:
2022 - 2023
-
Funding:
Inria – Inserm PhD + APHM for AD
-
Principal investigator:
S. Benzekry, X. Muracciole, L.Padovani
-
COMPO members involved:
C. Bigarre, A. Daumas
11 Dissemination
Participants: Dominique Barbolosi, Sebastien Benzekry, Elias Ventre, David Boulate, Joseph Ciccolini, Raphaelle Fanciullino, Florence Gattacceca, Laurent Greillier, Xavier Muracciole, Anne Rodallec, Sebastien Salas.
11.1 Promoting scientific activities
11.1.1 Scientific events: organisation
General chair, scientific chair
- J. Ciccolini
- Chair, Pharmacology and Molecular Mechanism (PAMM) group -EORTC, Brussels Belgium.
- F. Gattacceca
- President of GEPK (French Group of Lecturers in PK), organized the 2024 annual meeting, Poitiers, France, July 4-5, 2024.
- PK Docs: Creation and organization of the international online monthly event for PK PhD students.
Member of the organizing committees
- A. Rodallec
- Member of the YSC : organization of the meet up station for the CRS in Bologna, Italy 2024
- Member of the ECS of the CRS Local Chapter France and Benelux: Organizing comitee of the 1st meeting in Ghent, Belgium 2024
- J. Ciccolini
- PAMM-EORTC 44th Winter Meeting, February 2024 Marseille France
- 22nd International Congress of Therapeutic Drug Monitoring and Clinical Toxicology (IATDMCT), Sept 2024 Banff, Canada.
- GPCO-Unicancer Workshop, October 2024 Paris, France.
- F. Gattacceca
- PK national Masters Class (Every Friday, October-December 2024)
11.1.2 Scientific events: selection
Chair of conference program committees
- J. Ciccolini: PAMM-EORTC (Marseille France), GPCO-Unicancer (Paris France)
Member of the conference program committees
- J. Ciccolini: IATDMCT (Bannf Canada)
Reviewer
- A. Rodallec: CRS Bologna (Italy), CRS BNFL Ghent (Belgium)
11.1.3 Journal
Member of the editorial boards
- J. Ciccolini: Cancer chemotherapy and Pharmacology; Frontiers in Pharmacology
Reviewer - reviewing activities
- A. Rodallec: Pharmaceutics
- S. Benzekry: The Oncologist, Journal of Cancer Research and Clinical Oncology
- J. Ciccolini: Cancer Chemotherapy and Pharmacology, Clinical Cancer Research, British Journal of Clinical Pharmacology, Fundamental and Clinical Pharmacology, British Journal of Cancer, Journal of Clinical Oncology Oncology Practice, Clinical Pharmacology and Therapeutics, Clinical Pharmacology and Therapeutics Pharmacometrics and System Pharmacology, Critical Review in Oncology Hematology, Frontiers in Pharmacology, Clinical Pharmacokinetics, Expert Opinion On Drug Metabolism and Toxicology, Journal of Clinical Oncology Precision Oncology, Cancers, Pharmaceutics, International Journal of Nanomedicine, Nature Communications.
11.1.4 Invited talks
- S. Benzekry
- September, 2024. Twelfth International Workshop on Pharmacodynamics of Anticancer Agents. "Use of machine learning for predicting cancer outcomes", Ponta Delgada, Portugal.
- July, 2024. ECMTB congress (European Conference on Mathematical and Theoretical Biology). "Mechanistic learning for metastasis modeling and prediction", talk within the mini-symposium "Mechanistic learning in mathematical oncology" organized by A. Kohn-Luque and S. Haupt, Toledo, Spain.
- July, 2024. ECMTB congress (European Conference on Mathematical and Theoretical Biology). "Integrating kinetics and machine learning modeling for prediction of outcome following immunotherapy in lung cancer", talk within the mini-symposium "Digital twins for clinical oncology and cancer research" organized by G. Lorenzo, Toledo, Spain.
- July, 2024. SMB congress (Society of Mathematical Biology). "Integrative kinetics and machine learning modeling for prediction of outcome following immunotherapy in lung cancer". Talk within the mini-symposium "Mathematical modeling of cancer treatment" organized by E. Kim, Seoul, Korea.
- June, 2024. BIOMAT summer school. Mini-course "Mechanistic learning to predict response and survival in immuno-oncology.", Granada, Spain.
- March, 2024. Mechanistic learning for clinical cancer research. CANSEARCH, Geneva University, Switzerland.
- March, 2024. Keynote invited speaker in the session "Machine Learning for Rare Disease Applications: from Limited to In Silico Trials" of the Applied machine learning days (AMLD2024), "Mechanistic learning to predict the results of clinical trials", EPFL, Lausanne.
- January, 2024. EORTC-PAMM, The PIONeeR trial, Marseille.
- A. Rodallec
- September, 2024. Pharmacometric days, Paris, France.
- L. Nguyen Phuong
- October, 2024. Groupe de Métabolisme et Pharmacocinétique. "Computational modeling approaches for circulating cell-free DNA in oncology" within the symposium "Biomarker strategy for dose selection/optimization", Lyon, France.
- J. Ciccolini
- December, 2024. Groupe Français de Pneumo-Cancérologie. "E-R relationships with immunotherapy?", Paris, France.
- November, 2024. GPCO-Unicancer Workshop"Place des anticorps conjugués dans le carcinome urothélial?" Paris, France.
- November, 2024. Cours Saint-Paul en Oncologie Digestive"PK/PD relationships with immune checkpoint inhibitors: is the earth flat?" Nice, France.
- October, 2024. Symposium Clinique Waters"Bioanalyse des biothérapies: application au STP en oncologie clinique de routine" Paris, France.
- October, 2024. 61th Oncology congress "Clinical pharmacology of Antibody Drug conjugates" Belgrade, Serbia.
- October, 2024. Management du MTXHD - Partage d'expérience sur les hémopathies en région PACA "Retards d'élimination du MTX: Aspects pharmacométriques" Marseille, France.
- September, 2024. Twelfth International Workshop on Pharmacodynamics of Anticancer Agents. "Pharmacometrics as a decision-making tool with immune checkpoint inhibitors: finding the perfect blend?", Ponta Delgada, Portugal.
- June, 2024. Science, Society and Values Symposium "Is biology soluble into ideology?" Bordeaux, France.
- June, 2024. Imagining the future of cancer treatments Mini-Symposium "ADC in oncology, reality beyond the myth?" Marseille, France.
- June, 2024. 7e édition des IFODS (Journées Franco-Internationales d'Oncologie) "Nanoparticules en Oncologie: enjeux & Perspectives" Paris, France.
- March, 2024. Fondazione Pisana per la Scienza "Anti-Body Drug Conjugates: why «Magic Bullet» is not «Magic Pharmacology»", Pisa, Italy.
- February, 2024. CISAM/RDV Innovation Santé "Individualisation des posologies en oncologie: apport de la modélisation mathématique en pratique clinique de routine à l'APHM" Marseille, France.
- February, 2024. PAMM-EORTC 44th Winter Meeting "BRAF inhibitors: pharmacology and pharmacokinetics considerations" Marseille, France.
- January, 2024. Triple Negative Breast Cancer : A dialog between basic science and clinical practice Workshop "Antibody-Drug Conjugates ; beyond the hype, which pharmacology in triple negative breast cancer?" Nantes, France.
11.1.5 Scientific expertise
- J. Ciccolini: Expert at Institut National du Cancer (INCA) PHRC-K, CIVIS, Ligue Régionale contre le Cancer, Cancéropôle CLARA, Cancéropôle GSO, Cancéropôle IdF, GIRCI-MED, Groupe Français des Myelodysplasies (GFM-ONUVEN-MDS), University of Peshawar, Pakistan, University of Sydney, Australia.
- R. Fanciullino: Expert at Association pour la Recherche contre le Cancer (ARC), CN5.
- F. Gattacceca: Scientific expert at ANSM (Agence Nationale de Sécurité du Médicament, national drug agency), member of the permanent scientific committee "Quality and safety of drugs"
11.1.6 Research administration
- A. Rodallec: Website and Social Media coordinator for the CRS benelux Local Chapter
- J. Ciccolini: Member of the Scientific Committee of the School of Pharmacy, Marseille France.
- J. Ciccolini: Member of the Steering Committee & Copil H&N of the Head and Neck Cancer Taskforce, Unicancer Paris France.
- J. Ciccolini: Member of the Scientific Committee of the Immuno-Oncology Group, Unicancer Paris France.
- J. Ciccolini: Member of the ADC TaskForce at EORTC, Brussels Belgium.
- J. Ciccolini: Co-Director of the TRANSLATE-IT Department at CRCM, Inserm U1068, Marseille France.
- J. Ciccolini: Member of the PETRA (PrEclinical and TRAnslational Network neuro-oncology research) Network, PACA France.
- F. Gattacceca: Member of the scientific committee of the school of pharmacy
11.2 Teaching - Supervision - Juries
11.2.1 Teaching
- S. Benzekry: M2 Biologie Santé – Parcours IA biomarqueurs (6h). M2 "Pharmacokinetics" (6h).
- S. Benzekry: M2 PK "Fundamentals for modeling and simulation in pharmacokinetics/pharmacodynamics" (6h).
- A. Rodallec: lectures in MSc in Pharmacokinetics, MSc in Digipharm, MSc in Innovative Diagnostic and therapeutic Drug Products, DESU "Advances courses in pharmacometrics", DES in animal experiments, Pharm.D. studies (2nd, 3rd, 4th and 6th year), odonthology studies -> 190 h a year + additional lectures at University of Paris Saclay and Wuhan University (China).
- J. Ciccolini Lectures at Aix Marseille Univ in: MSc (2nd year) in Oncology, MSc (2nd year) in Oncogenetics, MSc (2nd year) in Pharmacokinetics, MSc (2nd year) in Digipharm, MSc (1st year) in Drugs & Health Products, D.U. in Animal Experiments, D.U. in Genetic Councelling, Master Class in Lung Cancer, Pharm.D. studies (2nd, 3rd, 4th and 6th year) -> 216 h a year.
- J. Ciccolini Teaching outside of AMU: additional lectures in pharmacokinetics at Université Catholique de Lyon, University of Amsterdam NL, University of Pisa Italy, School of Pharmacy of Belgrade, Serbia, and the International School of Metronomics. Lectures for Cours National Approfondissement DES Oncologie Médicale and Phase d'Approfondissement Docteur Junior, Paris Saclay University.
- J. Ciccolini Founder and co-Chair of the "Digital Tools for Pharmaceutical Sciences (Digipharm)"" Master Degree, Aix Marseille Univ.
- R. Fanciullino: Lectures in: MSc (2nd year) in Pharmacokinetics, MSc (2nd year) in Digipharm, CESU in Oncogeriatry, DES in PK Variability, Pharm.D. studies (3rd, 4th and 5th year) -> 190 h a year .
- F. Gattacceca: lectures in pharmacokinetics and pharmacometrics at Aix-Marseille University school of pharmacy (305h), teaching in other universities (50h: Nîmes, Angers, Montpellier): 90% at a post-graduate level.
- F. Gattacceca: Director of the master program "Pharmacokinetics". Director of two international post-graduate university diplomas: "Modeling and simulation: population approaches in pharmacokinetics/pharmacodynamics" and "Modeling and simulation: physiologically-based pharmacokinetic modeling for pharmacology and toxicology". Member of the national reflection committee for the industry pharmacy studies, of the national and local committees for a research option in pharmacy studies. Member of the training steering committee of ICI (Immunology Cancer Institute)
- F. Gattacceca: tutor in the first (2024) edition of the "Pharmacometrics Africa" training in French
- F. Gattacceca: CIVIS International Summer School "Drug Design and Discovery", Tübingen, Germany, July 2024 (Lectures and hands-ons)
- L. Greillier: M2 Recherche clinique et Simulation en Santé.
- L. Greillier: oncology and pulmonology for 3rd – 11th year medical students.
- X. Muracciole: DIU radio-urology for resident medical student.
- X. Muracciole: DCIU radio surgery for resident medical student
- S. Salas: Medical study/initial training. Seminary: palliative care Therapeutic module: pains Oncodigestive module Cancerology (44h), for 3rd - 6th year medical students
- S. Salas: Medical, paramedical study. DU Supportive care in oncology and palliative medicine. DU Wounds and healing. DIU Supportive care in oncology and palliative medicine. Master's in Advance Practice Nursing: Cancerology module, General module, pains. CEU Service providers at home, Home-based cancer care. DU Ambulatory shift, Oncology module. (37h)
- L. Nguyen Phuong: CESU "Fundamentals for modeling and simulation in pharmacokinetics/pharmacodynamics". Introduction to R for pharmacokinetic modeling. (9h).
- L. Nguyen Phuong: M2 PK. Introduction to R for pharmacokinetic modeling. (20h).
11.2.2 Supervision
Postdoc
- S. Benzekry
- Paul Dufossé (Institut Laennec, AMU) : “Quantitative modeling and machine learning for prediction of the response to immunotherapy in lung cancer”.
- A. Rodallec
- P. Piris, (Thermonano)
Engineers
- S. Benzekry
- R. Zakrasjek (SChISM)
- A. Vaglio (RHU PIONeeR)
- J. Ciccolini
- C. Perez (RHU PIONeeR)
PhD students
- S. Benzekry
- A. Bakhmach (PEPR Santé Numérique DIGPHAT), 2023 - 2026: “Modelling and statistical learning for pharmacology in oncology”, co-supervision with R. Fanciullino (COMPO, APHM) and S. Garciaz (IPC)
- M. Boussena (Institut Laënnec, AMU), 2023 - 2026: “Machine learning methods for clinical oncology data: application to the prediction of immunotherapy response in lung cancer”, co-supervision with J. Josse (Premedical, Inria) and L. Greillier (COMPO, APHM)
- C. Bigarré, 2020 - 2024: "Mathematical modeling for prediction of metastatic relapse in breast cancer", co-supervision X. Muracciole, funding Inria – Inserm
- L. Nguyen Phuong, 2022 - 2025, SChISM: "Mechanistic modeling of circulating DNA combined to machine learning for prediction of response and survival following immunotherapy", co-supervision S. Salas, funding Amidex ICI (Institute for Cancer Immunotherapy)
- H. Hamdache, 2023 - 2026: "Improving therapeutic efficacy and managing side effects and sequelae in pediatric cancer through improved and personalized nutritional programs using computer simulations", co-supervision V. Pancaldi (IUCT, Inserm, Toulouse), funding Inria–Inserm.
- E. Ventre
- C. Berthaud, 2024-2027: "Bioinformatic approach to the impact of modulation of the respiratory chain SDH complex on cell states dynamics in pediatric gliomas and rhabdomyosarcomas", co-supervision M. Castets (Inserm, CRCL), funding Ligue contre le cancer
- D. Boulate
- C. Buton (Institut Laënnec, AMU), 2024-2027: "Development of interactive expert software stratifying the risk of lung cancer diagnosis in the setting of lung cancer screening based on mathematical modeling and machine learning approaches", co-supervision with D. Barbolosi (COMPO)
- A. Todesco (CRCM - E19 - SMARTc), 2023-2026: "Study of the maintenance of pulmonary and systemic vascular permeability: from physiology to pathology", co-supervision with P. Habert (APHM)
- E. Armand (CRCM), 2023-2026: "French screening programme: development of risk stratification tools"
- J. Ciccolini
- C. Marin, CetuxIMAX, funding APHM & Merck Serono
- A. Ronda, Pembro Monitoring in real-world patients, funding APHM
- G. Kallee, DPD status and clinical outcome, funding APHM
- D. Protzenko, PK-guided dosing in HCT conditioning, funding APHM
- F. Gattacceca
- A. Deschamps, 2019-2024, funding APHM, co-supervision R. Guilhaumou (APHM)
- J. Ou, 2021-2024, funding AMU doctoral school 62, co-supervision P. Garrigue (AMU)
- S. Benamara, 2023-2026, funding CIFRE, co-supervision D. Teutonico (Sanofi)
- R. Fanciullino
- M. Dacos, Nanimmuno funding APHM & Institut Roche
- L. Osanno, Venaza project funding APHM
Interns (Master 2)
- S. Benzekry
- A. Boniffay (M2 AI4PH AMU, funding Laënnec)
- A. Daumas (radiotherapist, M2 AI and biomarkers AMU)
- A. Ouloum (Centrale Marseille, sup. S. Benzekry)
- R. Taïeb (M2, ENS Lyon)
- R. Zakrasjek (M2 PK AMU, funding ICI, sup. S. Benzekry)
- A. Rodallec
- O. Jaonino, immunomodulation of nanoparticles, University of Bonn
- J. Ciccolini, R. Fanciullino
- E. Diroff, nanoparticles in pancreatic cancer, Aix Marseille Univ funding Inserm
- J. Ciccolini
- Q. Gerbault, PK/PD of lirilumab in paediatric cancers, funding APHM
- F. Gattacceca
- M. Hammami, Development of a physiologically-based pharmacokinetic model of fludarabine in cancer patients, funding COMPO
11.2.3 Juries
- S. Benzekry: reviewer of the PhDs of Virginie Montalibet (MONC, Inria Bordeaux), Florian Jeanneret (CEA Grenoble) and Beatriz Ocaña-Tienda (Universidad de Castilla La Mancha, Spain)
- A. Rodallec: Jury of Pharm.D. Thesis and DES Hospital Pharmacist thesis, School of Pharmacy of Marseille (approx. 5 thesis/year)
- J. Ciccolini:
- Reviewer for PhD Defense: Thao Nguyen Pham, Approches biomathématiques sur les effets immunitaires systémiques induits par les radiations dans les cancers du cerveau et de la tête & cou en utilisant des modèles précliniques et cliniques. Université de Caen Normandie. October 2024
- President for PhD Defense: Arthur Millet, Dosage d’anticorps médicaments utilisés en cancérologie par spectrométrie de masse. Application à une étude clinique. Université Claude Bernard Lyon 1. January 2024
- Jury of Pharm.D. Thesis and DES Hospital Pharmacist thesis, School of Pharmacy of Marseille (approx. 30 thesis/year)
- R. Fanciullino:
- Jury of Pharm.D. Thesis and DES Hospital Pharmacist thesis, School of Pharmacy of Marseille (approx. 15 thesis/year)
- S. Salas
- President for PhD defense: Clémence Marin.
- Reviewer for Pharm.D. defense: Clara Boeri, Relation PK/PD du Nivolumab dans les cancers ORL.
- F. Gattacceca
- Reviewer for PhD Defense: Blaise Pasquier, Apport de la modélisation PKPD dans l’extrapolation interespèce et dans l’adaptation de posologie en pratique clinique des anticorps monoclonaux thérapeutiques. Université Paris Cité. October 2024
- Member of PhD scientific evaluation committees: I. Benbouziane (Jagiellonian University in Krakow, Poland), B. Cardozo (Aix-Marseille Université), A. Baillod (Université de Poitiers), Céline Delansay (Université Paris-Saclay), Magali Godard (Université de Montpellier)
- Jury of Pharm.D. thesis: Aura Seroussi.
11.3 Popularization
- Joseph Ciccolini:
- March 2024: Rencontres Enseignants-IPC "Comment les mathématiques peuvent guider les traitements" Marseille, France.
- Joseph Ciccolini, Sébastien Benzekry, Dominique Barbolosi, Xavier Muracciole, David Boulate:
- June 2024: Rencontres Maths et Santé, Université Pascal Paoli, Corté, Corse.
11.3.1 Productions (articles, videos, podcasts, serious games, ...)
- Joseph Ciccolini:
- October 2024: Serious Game TRANSFORM-O, Société Française du Cancer et Association Enseignement Recherche Internes en Oncologie (AERIO) "Développement clinique en oncologie" Montpellier, France.
12 Scientific production
12.1 Major publications
- 1 articleComputational oncology — mathematical modelling of drug regimens for precision medicine.Nature Reviews Clinical Oncology134April 2016, 242-254HALDOI
- 2 inproceedings3MO - Comprehensive biomarkers (BMs) analysis to predict efficacy of PD1/L1 immune checkpoint inhibitors (ICIs) in combination with chemotherapy: a subgroup analysis of the Precision Immuno-Oncology for advanced Non-Small CEll Lung CancER (PIONeeR) trial.Annals of Oncology2022 ESMO Immuno-Oncology Congress16suppl_1Geneva, Switzerland2022HAL
- 3 articleArtificial Intelligence and Mechanistic Modeling for Clinical Decision Making in Oncology.Clinical Pharmacology and TherapeuticsJune 2020HALDOI
- 4 articlePredicting Survival in Patients with Advanced NSCLC Treated with Atezolizumab Using Pre‐ and on‐Treatment Prognostic Biomarkers.Clinical Pharmacology and Therapeutics1164July 2024, 1110–1120HALDOIback to text
- 5 articleMechanistic modeling of metastatic relapse in early breast cancer to investigate the biological impact of prognostic biomarkers.Computer Methods and Programs in Biomedicine231April 2023, 107401HALDOI
- 6 articlePharmacokinetics and Pharmacodynamics-Based Mathematical Modeling Identifies an Optimal Protocol for Metronomic Chemotherapy.Cancer Research77172017, 4723-4733HALDOI
- 7 articleCDA as a predictive marker for life-threatening toxicities in patients with AML treated with cytarabine.Blood Advances25February 2018, 462 - 469HALDOI
- 8 articleTowards Rational Cancer Therapeutics: Optimizing Dosing, Delivery, Scheduling, and Combinations.Clinical Pharmacology and Therapeutics1083August 2020, 458-470HALDOI
- 9 articleRevisiting bevacizumab + cytotoxics scheduling using mathematical modeling: proof of concept study in experimental non-small cell lung carcinoma.CPT: Pharmacometrics and Systems Pharmacology2018, 1-9HALDOI
12.2 Publications of the year
International journals
Invited conferences
International peer-reviewed conferences
Conferences without proceedings
Doctoral dissertations and habilitation theses
Reports & preprints
Other scientific publications
12.3 Cited publications
- 97 articleExploiting Cancer’s Tactics to Make Cancer a Manageable Chronic Disease.Cancers1266Jun 2020, 1649DOIback to text
- 98 articleFrom patient-specific mathematical neuro-oncology to precision medicine..Front Oncol3AprilJan 2013, 62–62back to text
- 99 articleComputational Modelling of Metastasis Development in Renal Cell Carcinoma.PLoS Comput Biol111100007Nov 2015, e1004626DOIback to text
- 100 articleDetermination of the unmetabolized 18F-FDG fraction by using an extension of simplified kinetic analysis method: clinical evaluation in paragangliomas.Medical & biological engineering & computing541Jan 2016, 103–111DOIback to text
- 101 articleOncotarget829Jul 2017, 47161–47166DOIback to text
- 102 articleGlobal Dormancy of Metastases Due to Systemic Inhibition of Angiogenesis.PLoS ONE9100019Jan 2014, e84249-11DOIback to text
- 103 articleMathematical Modeling of Tumor-Tumor Distant Interactions Supports a Systemic Control of Tumor Growth..Cancer Research7718Sep 2017, 5183–5193DOIback to text
- 104 articleModeling Spontaneous Metastasis following Surgery: An In Vivo-In Silico Approach.Cancer Research76300000Feb 2016, 535–547DOIback to text
- 105 articleProgress and Opportunities to Advance Clinical Cancer Therapeutics Using Tumor Dynamic Models..Clinical cancer research : an official journal of the American Association for Cancer ResearchDec 2019, 1–22DOIback to text
- 106 articleStan: A Probabilistic Programming Language.Journal of Statistical Software761Jan 2017, 1–32DOIback to text
- 107 articleEmergence of drug tolerance in cancer cell populations: an evolutionary outcome of selection, nongenetic instability, and stress-induced adaptation.Cancer Research756Mar 2015, 930–939DOIback to text
- 108 articleMechanistic Learning for Combinatorial Strategies With Immuno-oncology Drugs: Can Model-Informed Designs Help Investigators?JCO Precision Oncology4May 2020, 486–491DOIback to textback to text
- 109 articleCombinatorial immunotherapy strategies: most gods throw dice, but fate plays chess.Annals of Oncology3011Nov 2019, 1690–1691DOIback to text
- 110 articleRealistic simulation of the 3-D growth of brain tumors in MR images coupling diffusion with biomechanical deformation.IEEE Transactions on Medical Imaging2410Oct 2005, 1334–1346DOIback to text
- 111 articleA new methodology to derive 3D kinetic parametric FDG PET images based on a mathematical approach integrating an error model of measurement.EJNMMI Research8Nov 2018, URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6238015/DOIback to text
- 112 articlePrediction of the Evolution of Thyroidal Lung Nodules Using a Mathematical Model..82Jan 2010, 37–38back to text
- 113 articleA multi-layered systems approach for renal cell carcinoma.bioRxivJan 2020, 2020.01.13.904235DOIback to text
- 114 articleConvergence of a stochastic approximation version of the EM algorithm.The Annals of Statistics271Zbl: 0932.62094Mar 1999, 94–128DOIback to text
- 115 articleA 2/1 Sunitinib Dosing Schedule Provides Superior Antitumor Effectiveness and Less Toxicity Than a 4/2 Schedule for Metastatic Renal Cell Carcinoma: A Systematic Review and Meta-Analysis.Frontiers in Oncology102020, 313DOIback to text
- 116 phdthesisApproche pharmacocinétique de population centrée sur le patient : Preuve de concept et application en soins critiques.Embargo jusqu'au 19/12/2025Aix Marseille UniversitéDecember 2024HALback to text
- 117 articleUsing the SAEM algorithm for mechanistic joint models characterizing the relationship between nonlinear PSA kinetics and survival in prostate cancer patients.Biometrics731Mar 2017, 305–312DOIback to text
- 118 articleNumerical solution of an inverse problem in size-structured population dynamics.Inverse Problems254Apr 2009, 045008–045008back to text
- 119 articleA phase Ia/Ib clinical trial of metronomic chemotherapy based on a mathematical model of oral vinorelbine in metastatic non-small cell lung cancer and malignant pleural mesothelioma: rationale and study protocol.BMC cancer161000002016, 278DOIback to text
- 120 articleIn vitro and in vivo reversal of resistance to 5-fluorouracil in colorectal cancer cells with a novel stealth double-liposomal formulation.British Journal of Cancer977Oct 2007, 919–926DOIback to text
- 121 articleMethodological aspects and pharmacological applications of three-dimensional cancer cell cultures and organoids.Life Sciences254Aug 2020, 117784DOIback to text
- 122 articlePembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung.Cancer. N Engl J Med3782078–20922018back to text
- 123 articleThe evolutionary history of lethal metastatic prostate cancer.Nature5207547Apr 2015, 353–357DOIback to text
- 124 articleThe elements of statistical learning: data mining, inference, and prediction, Springer Series in Statistics.2009back to text
- 125 articleRevisiting dosing regimen using PK/PD modeling: the MODEL1 phase I/II trial of docetaxel plus epirubicin in metastatic breast cancer patients.Breast Cancer Research and Treatment1562Mar 2016, 331–341DOIback to text
- 126 articleDevelopment of ipilimumab: contribution to a new paradigm for cancer immunotherapy.Seminars in Oncology375Oct 2010, 533–546DOIback to text
- 127 articleCalibrating a Predictive Model of Tumor Growth and Angiogenesis with Quantitative MRI.Annals of Biomedical Engineering477Jul 2019, 1539–1551DOIback to text
- 128 articleSingle-agent paclitaxel for the treatment of breast cancer: an overview.Seminars in Oncology231 Suppl 1Feb 1996, 4–9back to text
- 129 articleQuantitative evidence for early metastatic seeding in colorectal cancer.Nature Genetics5177Jul 2019, 1113–1122DOIback to text
- 130 articleRevisiting Bevacizumab + Cytotoxics Scheduling Using Mathematical Modeling: Proof of Concept Study in Experimental Non-Small Cell Lung Carcinoma..CPT: Pharmacometrics & Systems Pharmacology71Jan 2018, 42–50DOIback to text
- 131 articleA Dynamical Model for the Growth and Size Distribution of Multiple Metastatic Tumors.Journal of Theoretical Biology2032Mar 2000, 177–186DOIback to text
- 132 articleDynamic Risk Profiling Using Serial Tumor Biomarkers for Personalized Outcome Prediction.Cell1783Jul 2019, 699-713.e19DOIback to text
- 133 bookMixed Effects Models for the Population Approach.CRC PressJul 2014back to text
- 134 articleNonlinear mixed effects models for repeated measures data..Biometrics463Sep 1990, 673–687back to text
- 135 articleA phase II randomized trial of nivolumab with stereotactic body radiotherapy (SBRT) versus nivolumab alone in metastatic (M1) head and neck squamous cell carcinoma (HNSCC)..Journal of Clinical Oncology3615_supplMay 2018, 6009–6009DOIback to text
- 136 articleRevisiting Dosing Regimen Using Pharmacokinetic/Pharmacodynamic Mathematical Modeling: Densification and Intensification of Combination Cancer Therapy.Clinical Pharmacokinetics55800000Aug 2016, 1015–1025DOIback to text
- 137 articleModel driven optimization of antiangiogenics + cytotoxics combination: application to breast cancer mice treated with bevacizumab + paclitaxel doublet leads to reduced tumor growth and fewer metastasis.Oncotarget814Feb 2017, 23087–23098DOIback to text
- 138 articleIn Vivo Bioluminescence Tomography for Monitoring Breast Tumor Growth and Metastatic Spreading: Comparative Study and Mathematical Modeling.Scientific Reports600000Nov 2016, 36173DOIback to text
- 139 articleIntegrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas.Nature Biotechnology3833Mar 2020, 333–342DOIback to text
- 140 articleAvelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma.New England Journal of Medicine38012Mar 2019, 1103–1115DOIback to text
- 141 phdthesisMathematical modelling of neoadjuvant antiangiogenic therapy and prediction of post-surgical metastatic relapse in breast cancer patients.Université de BordeauxOct 2019, URL: https://tel.archives-ouvertes.fr/tel-02560543back to text
- 142 articleMatrix metalloproteinases at cancer tumor–host interface.Seminars in Cell & Developmental Biology191Feb 2008, 52–60DOIback to text
- 143 articleAn alternative parameter for early forecasting clinical response in NSCLC patients during radiotherapy: proof of concept study.The British Journal of Radiology891062Jun 2016, 20160061DOIback to text
- 144 articleFrom 3D spheroids to tumor bearing mice: efficacy and distribution studies of trastuzumab-docetaxel immunoliposome in breast cancer.International Journal of Nanomedicine132018, 6677–6688DOIback to text
- 145 articleDefining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies.Annals of Oncology: Official Journal of the European Society for Medical Oncology2772016, 1190–1198DOIback to text
- 146 articleOptimal Scheduling of Bevacizumab and Pemetrexed/cisplatin Dosing in Non-Small Cell Lung Cancer.CPT: pharmacometrics & systems pharmacologyApr 2019DOIback to text
- 147 articleAtezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC..New England Journal of Medicine37824Jun 2018, 2288–2301DOIback to text
- 148 articleComprehensive analysis of the clinical immuno-oncology landscape..Ann. Oncol.291Jan 2018, 84–91DOIback to text
- 149 phdthesisMathematical modeling of antibody nanoconjugates transport in tumors.Bordeaux UniversityDec 2020back to text
- 150 articlePREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer..Breast cancer research121Jan 2010, R1DOIback to text

