Section: New Results
Image analysis for the characterization of cell mobility
Participant : Igor Peterlik.
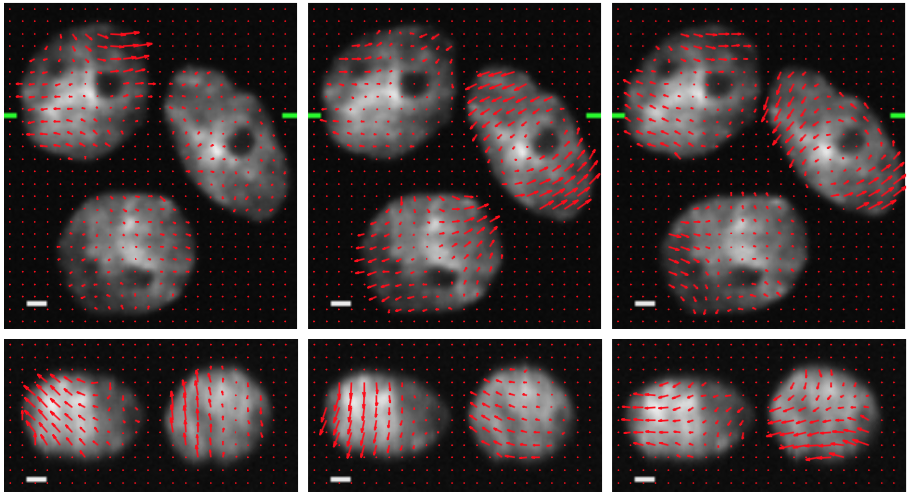
The complex behaviour of motile cells plays a crucial role in biological processes such as tissue growth and tumorigenesis. Biomedical research that focuses on understanding the mechanisms of cell motility generates large amounts of multidimensional image data acquired by fully automated optical microscopes. Manual analysis of such data is extremely laborious and therefore, it is necessary to develop reliable automatic methods of image analysis. However, evaluation and assessment of such methods remains a challenging task, since in the case of real data, no ground truth is available to establish simple and robust metrics. Therefore, an important task in development of automatic methods of image analysis is the synthetic generation of realistic images allowing for quantitative assessment based on the ground truth.
We collaborate with the Centre of Biomedical Image Analysis (CBIA) at Masaryk University, Czech Republic on the development of reliable image analysis methods for quantitative characterization of cell motility driven by cellular protrusions at the leading edge of crawling cells. In particular, we develop physics-based models of living cells which are used to generate synthetic time-lapse 3D image series that realistically mimic the motile cells with protrusions (Figure 10). Although modeling of living cells has many specificities, we successfully exploit the modeling algorithms originally designed and developed for the simulation soft tissues. We have already demonstrated that realistic simulation of living cells can be achieved using SOFA and are working toward more complex models and scenarios, involving interactions among the cells and mitosis.