Section: New Results
Parallel algorithms for adaptive molecular dynamics simulations
Participants : Dmitriy Marin, Stephane Redon.
We have developed a parallel implementation of Adaptively Restrained Particle Simulations (ARPS) in LAMMPS Molecular Dynamics Simulator with the usage of Kokkos (The Kokkos package is based on Kokkos library, which is a templated C++ library that provides two key abstractions: it allows a single implementation of an application kernel to run efficiently on different hardware, such as a many-core CPU, GPU, or MIC; it provides data abstractions to adjust (at compile time) the memory layout of basic data structures — like 2d and 3d arrays — for performance optimization on different platforms. These abstractions are set at build time (during compilation of LAMMPS).) package. The main idea of the ARPS method [22] is to speed up particle simulations by adaptively switching on and off positional degrees of freedom, while letting momenta evolve; this is done by using adaptively restrained Hamiltonian. The developed parallel implementation allows us to run LAMMPS with ARPS integrator on central processing units (CPU), graphics processing units (GPU), or many integrated core architecture (MIC). We modified the ARPS algorithm for efficient usage of GPU and many-core CPU, e.g. all computations were parallelized for efficient calculations on computational device; communications between host and device were decreased.
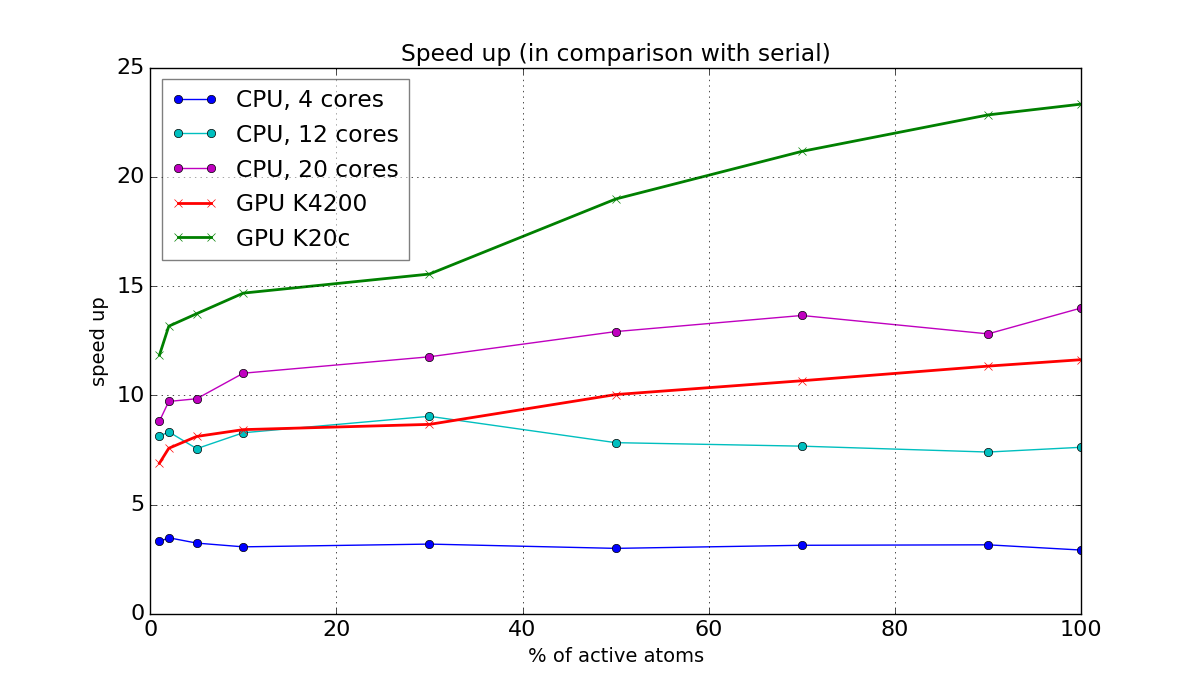
To measure speed up of the developed parallel implementation we used several benchmarks and heterogeneous computational systems with next parameters: 2x CPU Intel Xeon E5-2680 v3 (24 cores in total), GPU Nvidia Quadro K4200, GPU Nvidia Tesla K20c. Results on the speed up in comparison with serial ARPS code for one of the benchmarks (Lennard–Jones liquid, 515K atoms, 1% of particles switches their state at each timestep from active to restrained or from restrained to active) are shown in Figure 4. It can be seen, that for small number of CPU cores the speed up is almost constant for all the percentage of active atoms in the system. But for large number of CPU cores and for GPUs the speed up is decreasing with decreasing percentage of active atoms, because of divergence of threads and limited occupancy. The achieved speed up on 20 CPU-cores is up to 14 times, on GPU Nvidia Tesla K20c is up to 24 times.