Section: New Results
Colocalization for live cell and super-resolution imaging
Participants : Frédéric Lavancier, Thierry Pécot, Charles Kervrann.
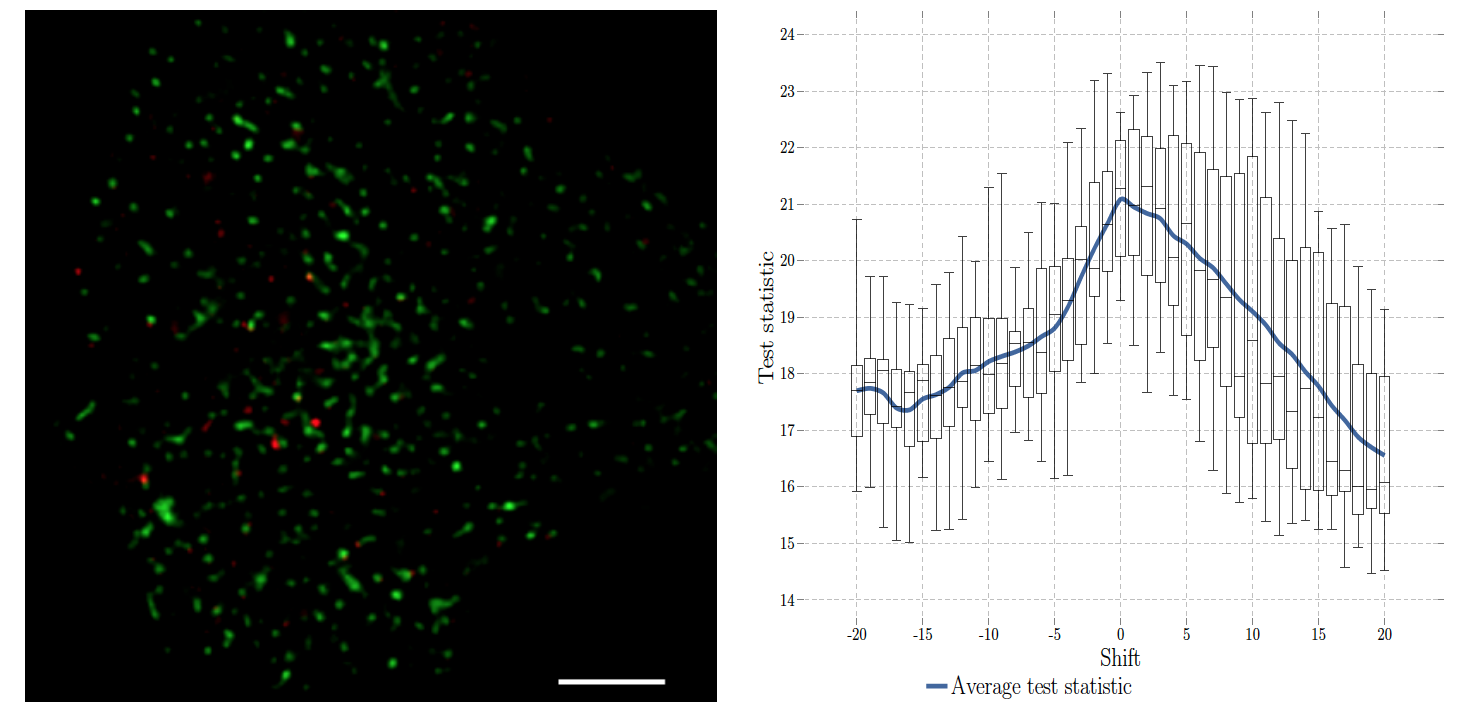
In the context of bioimaging, colocalization refers to the detection of emissions from two or more fluorescent molecules within the same pixel of the image. This approach enables to quantify the protein-protein interactions inside the cell, just at the resolution limit of the microscope. In statistics, this amounts to characterizing the joint spatial repartition and the spatial overlap between different fluorescent labels. Two distinct categories of colocalization approaches are considered to address this issue: intensity-based methods and object-based methods. The popular (intensity-based) Pearson's correlation method, which returns values between -1 and +1, is known to be sensitive to high intensity backgrounds and provides errors if the signal-to-noise ratio (SNR) is typically low. The object-based method, recommended in single molecule imaging, analyses the spatial distribution of the two sets of detected spots by using point process statistics.
Accordingly, we developed an original, fast, robust-to-noise and versatile approach that reconciles intensity-based and object-based methods for both conventional diffraction-limited microscopy and sub-resolved microscopy. The procedure is only controlled by a p-value and tests whether the Pearson correlation between two binary images is significantly positive. This amount to quantifying the interaction strength by the area/volume of the intersection between the two binary images viewed as random distributions of geometrical objects. Under mild assumptions, it turns out that the appropriately normalized Pearson correlation follows a standard normal distribution under the null hypothesis if the number of image pixels is large. Unlike previous methods, the method handles 2D and 3D images, variable SNRs and any kind of cell shapes. It is able to colocalize large regions with small dots, as it is the case in TIRF-PALM experiments and to detect negative colocalization. The typical processing time is two milliseconds per image pair in 2D and a few seconds in 3D, with no dependence on the number of objects per image. Finally, the method provides maps to geocolocalize molecule interactions in specific image regions.
Collaborators: Jean Salamero and Liu Zengzhen (UMR 144 CNRS-Institut Curie).
|