Section: New Results
Model-based Image Registration
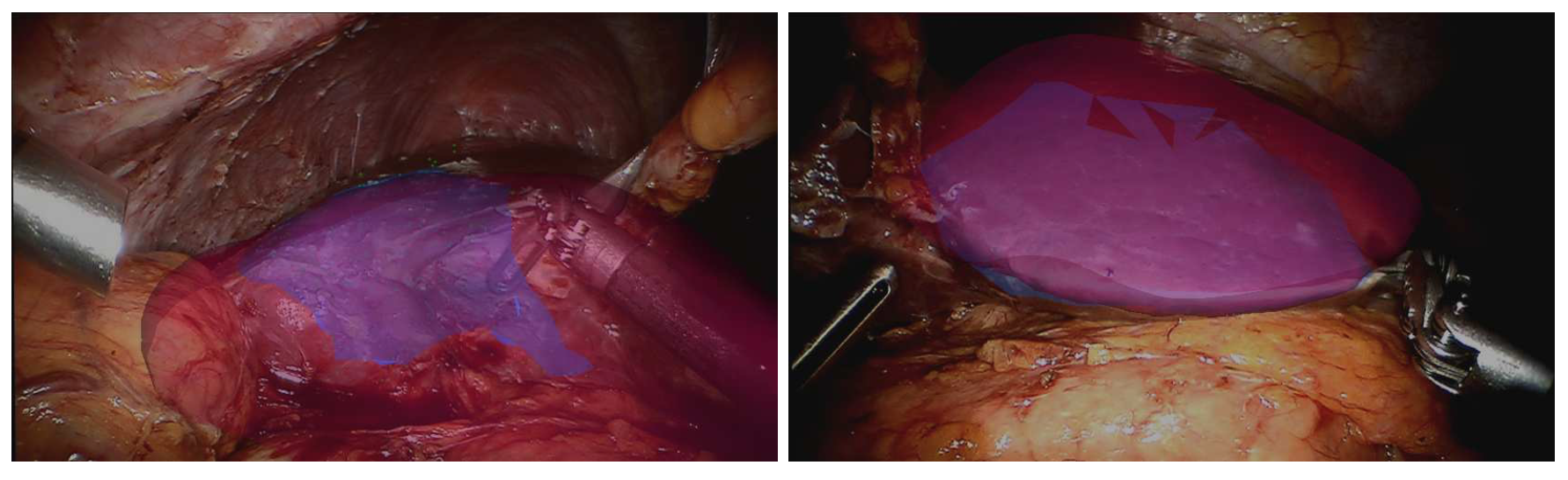
Intraoperative Biomechanical Registration of the Liver
Participants: R. Plantefève, I. Peterlik, S. Cotin
Different aspects of model-based registration in the context of surgical navigation employing the augmented reality were analyzed in an invited contribution [17] published in the context of the attributed Prix de thèse de former Ph.D. student Rosalie Plantefève. Preoperative images such as computed tomography scans or magnetic resonance imaging contain lots of valuable information that are not easily available for surgeons during an operation. To help the clinicians better target the structures of interest during an intervention, many registration methods that align preoperative images onto the intra-operative view of the organs have been developed. For important organ deformation, biomechanical model-based registration has proven to be a method of choice. Using an existing model-based registration algorithm for laparoscopic liver surgery we investigated the influence of the heterogeneity of the liver on the registration result. It was found that the use of an heterogeneous model does not improve significantly the registration result but increases the computation time necessary to perform the registration.
|
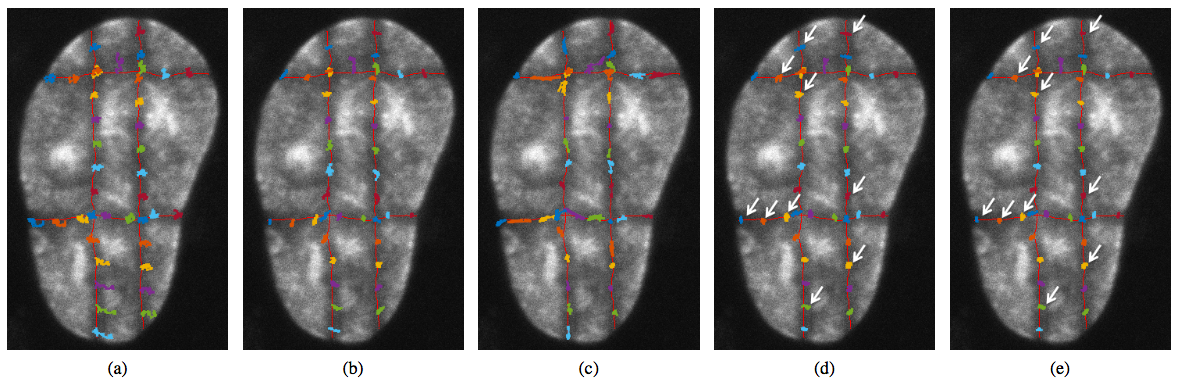
Registration of Cell Nuclei in Cell Microscopy
Participants: I. Peterlik
A contribution Registration of Cell Nuclei in 2D Live Cell Microscopy was published in a collaboration with Centre of Biomedical Image Analysis at Masaryk University, Czech Republic [18]. The analysis of the pure motion of sub-nuclear structures without influence of the cell nucleus motion and deformation is essential in live cell imaging. We proposed a 2D contour-based image registration approach for compensation of nucleus motion and deformation in fluorescence microscopy time-lapse sequences. The proposed approach extends our previous approach which uses a static elasticity model to register cell images. Compared to that scheme, the new approach employs a dynamic elasticity model for forward simulation of nucleus motion and deformation based on the motion of its contours. The contour matching process is embedded as a constraint into the system of equations describing the elastic behavior of the nucleus. This results in better performance in terms of the registration accuracy. Our approach was successfully applied to real live cell microscopy image sequences of different types of cells including image data that was specifically designed and acquired for evaluation of cell image registration methods.
|