Section: New Results
Spatial statistics, point patterns, and colocalization in fluorescence imaging
Participants : Frédéric Lavancier, Charles Kervrann.
|
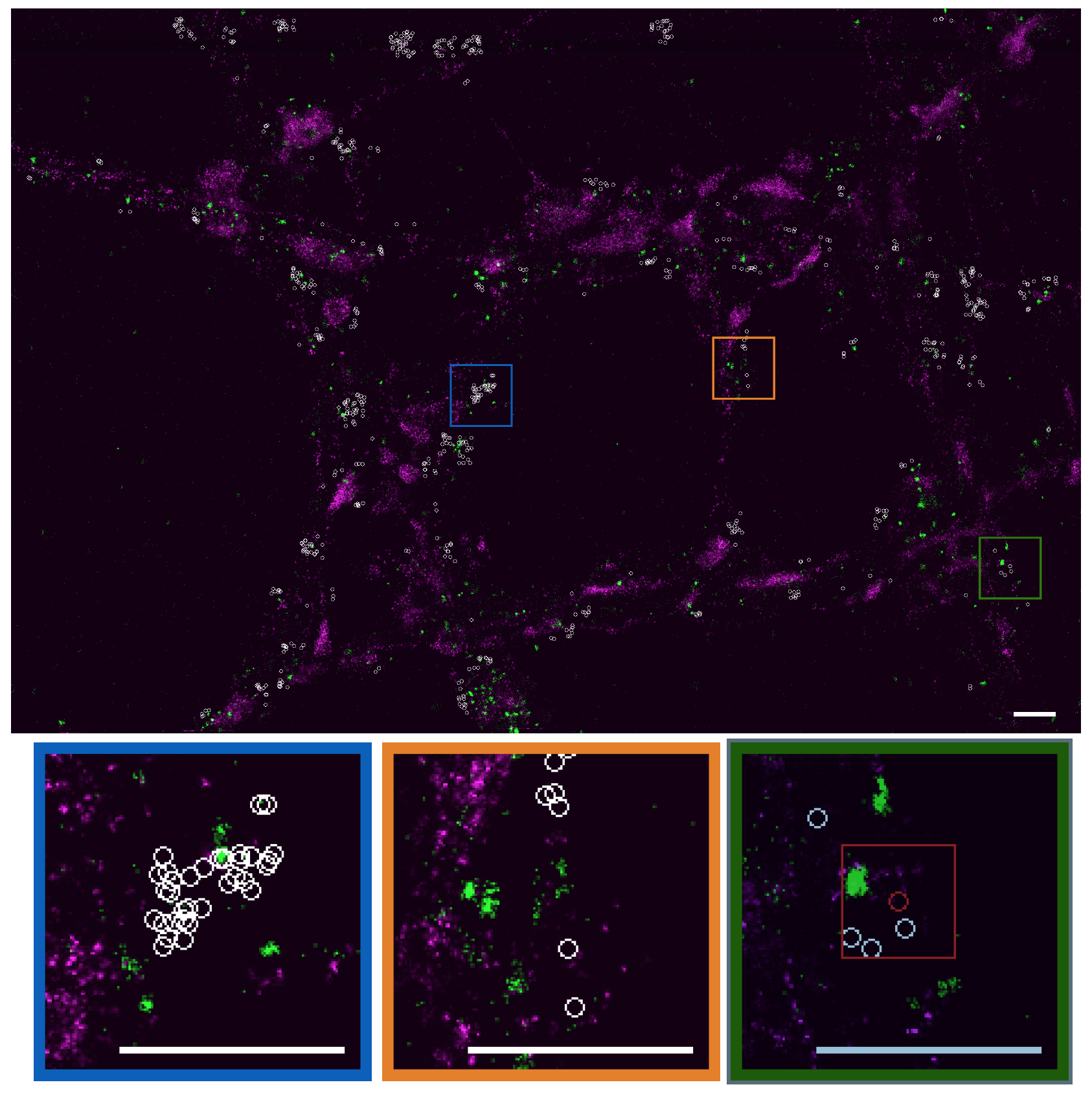
In the context of bioimaging, colocalization refers to the detection of emissions from two or more fluorescent molecules within the same pixel of the image. This approach enables to quantify the protein-protein interactions inside the cell, just at the resolution limit of the microscope. It refers to the detection of emissions from two or more fluorescent molecules within the same pixel of the image. Colocalization is an open problem for which no satisfying solution has been found up to now. Accordingly, we proposed an objective, robust-to-noise colocalization method (GcoPS – Geo-coPositioning System)) which only requires the adjustment of a p-value that guarantees more reproducibility and more objective interpretation. It is based on the statistical analysis of the intersection (area/volume) between the two 2D or 3D binary segmented images. GcoPS handles 2D and 3D images, variable signal-to-noise ratios and any fluorescence image pair acquired with conventional or super-resolution microscopy (see Fig. 6). To our knowledge, no existing method offers the same robustness and precision level with such an easy control of the algorithm. In a recent study (internships 2017), we started to adapt this framework to analyze the spatiotemporal molecular interactions from set of 3D computed trajectories or motion vector fields (e.g., co-alignment), and then to fully quantify specific molecular machineries.
More generally, analysis of molecule and protein localization, of interactions and spatial distributions in living cells is helpful to understand functions in the cell and to compare spatialized phenotypes. This is also true with the emergence of single-molecule localization microscopy techniques (e.g., PALM), relying on the cumulative spatial localization of fluorescently tagged markers, and whose outputs are sets of spatial coordinates of single molecules. Accordingly, we were interested in the spatial distribution of single molecules that exhibit some randomness, regularity and spatial clustering (or aggregation) at large scales, while having a minimal distance between them. In that context, we theoretically studied several point processes able to represent the spatial organization of points. We focused on determinantal point processes (DDP), since they are able to describe spatial point patterns where nearby points repel or repulse each other. We also partly solved a 30 years old conjecture by proving the consistency of the likelihood procedure for a large class of Gibbs models (e.g., Strauss model, area-interaction model) which are commonly used models in practice. We extended the pseudo-likelihood procedure to infinite range Gibbs interactions, and we proved its consistency and its asymptotic normality. All these models are now well understood and will be used in future works to analyse point patterns in cell imaging, generally described by Poisson point processes.
Collaborators: Jean Salamero and Liu Zengzhen (UMR 144 CNRS-Institut Curie),
David Dereudre (Laboratoire Paul Painlevé (UMR 8524), University of Lille 1),
Jean-François Coeurjolly (Laboratoire Jean Kutzmann, University of Grenoble).