Section: New Results
Fast optical flow methods for 3D fluorescence microscopy
Participants : Sandeep Manandhar, Patrick Bouthemy, Charles Kervrann.
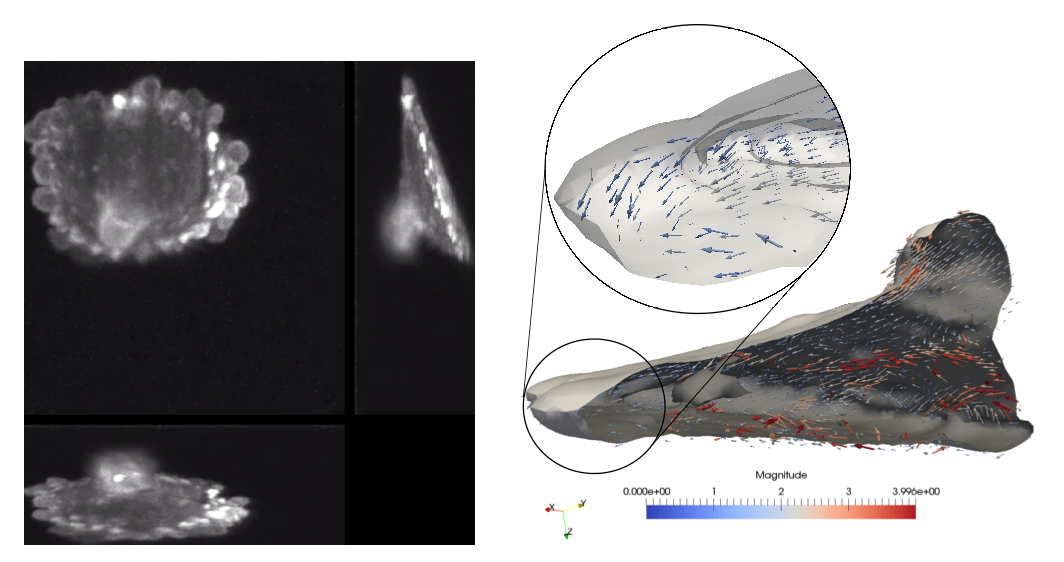
Estimating motion of cells and of subcellular particles is crucial for deciphering cell mechanisms and understanding cell behaviors. Modern 3D light microscopy (LM) for cell biology enables to observe cell dynamics at a good resolution, in both space and time, motivating the development of 3D optical flow methods. However, the acquired 3D LM image sequences exhibit several specificities making 3D motion computation a difficult problem. We have defined an original and efficient two-stage estimation method for light microscopy image volumes. The method, developed in the frame of S. Manandhar PhD thesis, takes a pair of LM image volumes as input, segments the 2D slices of the source volume in super-pixels, and first estimates the 3D displacement vectors of the super-pixel centers. To this end, we have extended the well-known PatchMatch method to 3D volumes, where correspondences act between voxels. Both the propagation and the random search steps were adapted to 3D volumes. Then, a weighted interpolation has been designed to recover the dense 3D flow field for all the voxels, from the sparse 3D displacement field. The super-pixel segmentation is exploited to define the neighborhood for interpolation, and the interpolation weights take into account intensity edges and local motion differences to preserve flow discontinuities. The experimental results show good gain in execution speed, and accuracy evaluated in computer-generated 3D data with ground-truth. The results were promising on two real 3D LM image sequences supplied by USTW. The sequences depict blebbing in a MV3 cell (see Fig. 9). The cell membrane protrudes increasing the surface area of the cell. These protrusions, referred to as blebs, appear and disappear in interval of minutes, the bleb appearance corresponding to the stretching of a local region of the cell membrane. The total computation time was for the first sequence 163 seconds (resp. 101s for the second sequence), with 19 (resp. 49), 120 (resp. 44) and 24 (resp. 8) seconds for super-pixel generation, 3D patch matching, and interpolation respectively, on a computer with 2.8 GHz Intel i7 processor and 16 GB of RAM.
Collaborators: Philippe Roudot and Gaudenz Danuser (UTSW, Dallas, USA).