Section: New Results
Organoid growth tracking
Participants : Cédric Girard Riboulleau, Xavier Descombes.
This work is a collaboration with F.-R. Roustan, S. Torrino, S. Clavel and F. Bost from C3M. It was partially supported by the UCA Jedi Idex.
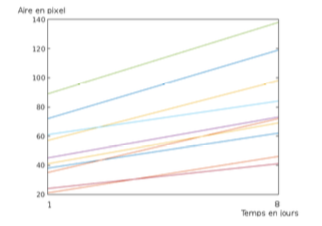
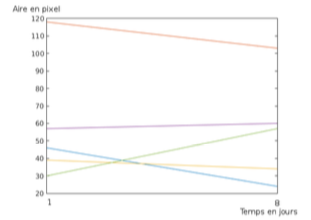
Organoid culture is a major challenge toward personalized medicine. It is now possible to partially reconstruct the structure of organs from a single biopsy. This new technology named organoid for sane cells or tumoroid for cancer cells allows the test of different molecules on cells withdrawn on a patient to retrieve the most efficient one for this patient. In this context, the main goal of this work is to develop a numerical scheme to automatically assess the effect of a given treatment on the organoid growth. We consider a time sequence of 2D confocal microscopy images of a population of organoids. We have considered different approaches to detect the organoids in the images. These approaches include edge detection using Canny filter, thresholding combined with mathematical morphology tools, texture analysis through Markov Random Fields, marked point processes. We have also modified several times the imaging protocol in order to simplify the object detection. To evaluate the growth of each organoid we have to match the objects detected in two consecutive frames. We have developed a matching algorithm based on a majority voting. We compute the vector between every pair of detected objects on both frames. Assuming that there is only a translation between the two frames we estimate it as the most represented vector. With this framework we have shown that the studied treatment has stopped the organoid growth (see figure 7).