Section: New Results
Mitochondrial network detection and classification
Participants : Guillaume Lavisse, Xavier Descombes.
This work is a collaboration with C. Badot and M. Chami from IPMC and A. Charezac, S. Clavel and F. Bost from C3M. It was partially supported by the UCA Jedi Idex.
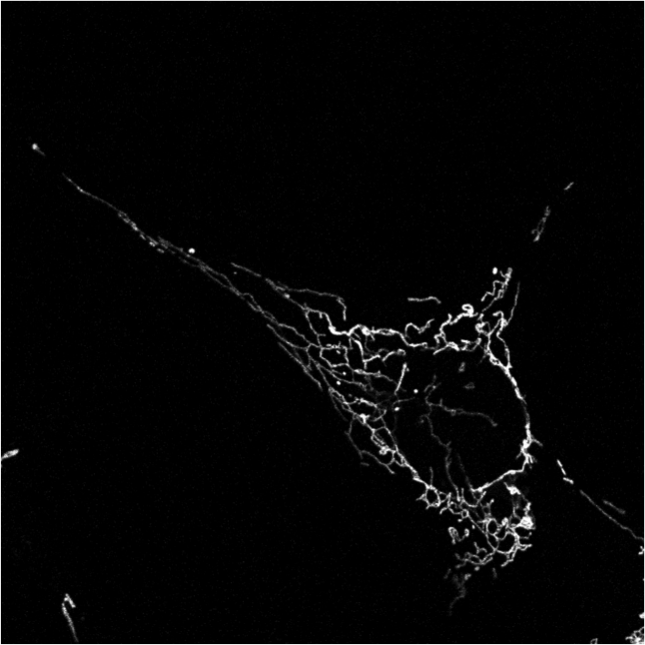
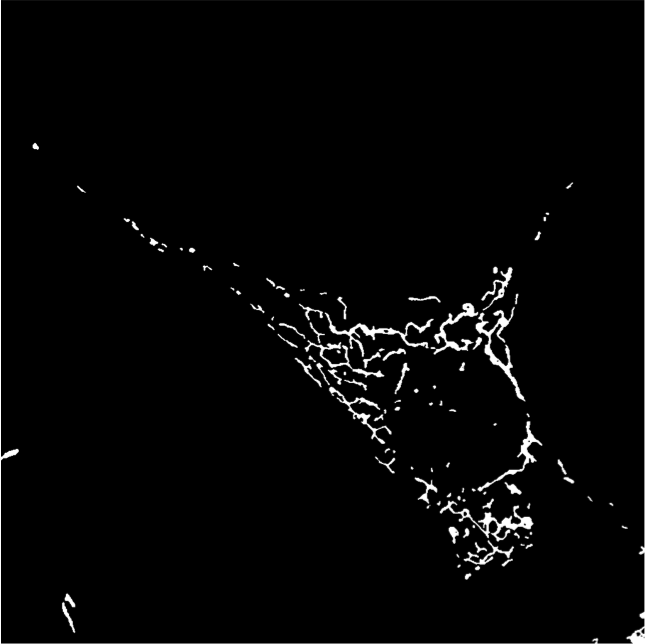
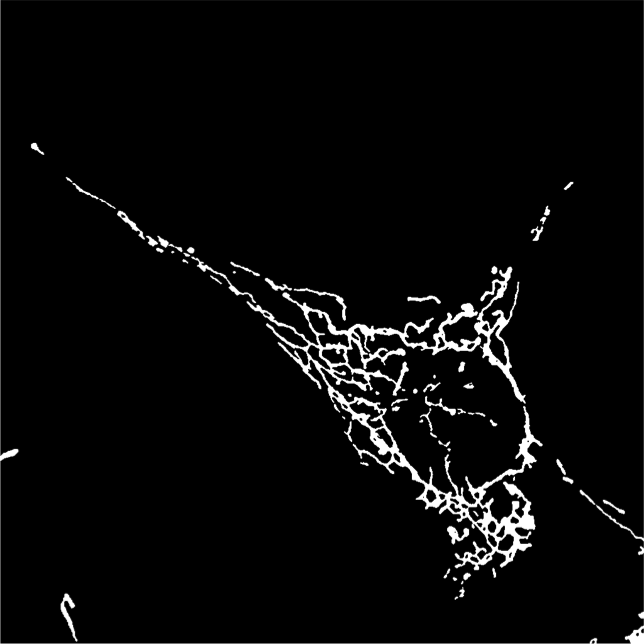
Last year we had developed a framework to classify mitochondrial networks. In this framework, mitchondrial networks are first binarized using our algorithm ATOLS. Some geometrical features are then computed for each connected component providing a clustering at the object level in the feature space. A signature of a given image is then defined by the ratio of objects in the different classes. A second classification, performed by an SVM on this signature, provides a global class for the image. The different classes are defined as fragmented, tubular and filamentous. This year, we have validated this framework on two other databases, one consisting of cultured cells, the other being constituted of Alzheimer neuronal cells. The results were not satisfactory compared to those obtained on the first database last year. This is mainly due to the signal heterogeneity within an image. To compensate this heterogeneity we have applied a local normalization (see figure 10). We then have recovered classification performances comparable to those obtained by an expert. The next step consists in following in time the mitochondria of Alzheimer neuron. To this aim, we have developed a matching algorithm between two sets of mitochondria based on geometrical features and location of detected objects at two different instants.