Section: Overall Objectives
Overall Objectives
Diversity, evolution, and inheritance form the heart of modern biological thought. Modeling the complexity of biological systems has been a challenge of theoretical biology for over a century [29] and flourished with the evolution of data for describing biological diversity, most recently with the transformative development of high-throughput sequencing. However, most concepts and tools in ecology and population genetics for capitalizing on this wealth of data are still not adapted to high throughput data production. A better connection between high-throughput data production and tool evolution is highly needed: computational biodiversity.
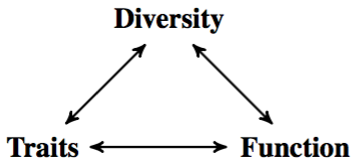
Paradoxically, diversity emphasizes differences between biological objects, while modeling aims at unifying them under a common framework. This means that there is a limit beyond which some components of diversity cannot be mastered by modeling. We need efficient methods for recognizing patterns in diversity, and linking them to patterns in function. It is important to realize that diversity in function is not the same as coupling observed diversity with function. Diversity informs both the study of traits, and the study of biological functions (Figure 1). The double challenge is to measure these links quickly and precisely with pattern recognition, and to explore the relations between diversity in traits and diversity in function through modeling
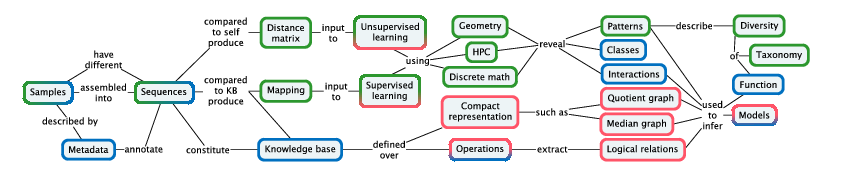
Pleiade links recognition of patterns, classes, and interactions with applications in biodiversity studies and biotechnology. We develop distance methods for NGS datasets at different levels of organization: between genomes, between individual organisms, and between communities; and develop high-performance pattern recognition and statistical learning techniques for analyzing the resulting point clouds. We refine inferential methods for building hierarchical models of networks of cellular functions, exploiting the mathematical relations that are revealed by large-scale comparison of related genomes and their models. We combine these methods into integrated e-Science solutions to place these tools directly in the hands of biologists.
Our methodology (Figure 2) is designed pragmatically to advance the state of the art in applications from biodiversity and biotechnology: molecular based systematics and community ecology, annotation and modeling for biotechnology.