Section: Research Program
Scientific Foundations
The scientific foundations of our team concern the design and development of new computational solutions for biological images, signals and measurements. Our objective is to develop a better understanding of the normal and pathological brain, at different scales.
This includes imaging brain pathologies in order to better understand pathological behavior from the organ level to the cellular level, and even to the molecular level (using molecule (e.g. through PET-MR imaging), as well as modeling with specific ligands/nanocarriers), and the modelling of normal and pathological large groups of individuals (cohorts) from image descriptors. It also includes the challenge of the discovery of episodic findings (i.e. rare events in large volumes of images and data), data mining and knowledge discovery from image descriptors, the validation and certification of new drugs from imaging features, and, more generally, the integration of neuroimaging into neuroinformatics through the promotion and support of virtual organizations of biomedical actors by means of e-health technologies.
|
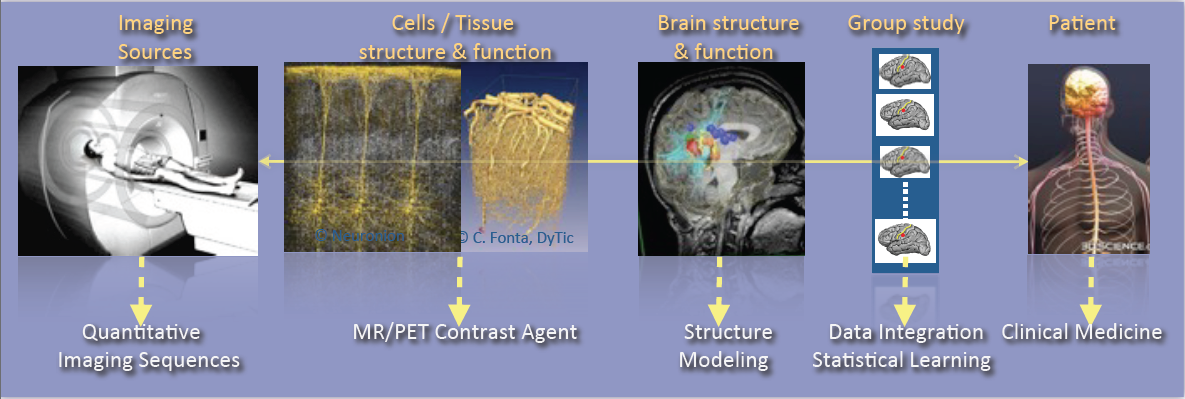
As shown in Fig. 1, the research activities of the Empenn team closely link observations and models through the integration of clinical and multiscale data, and phenotypes (cellular, and later molecular, with structural or connectivity patterns in the first stage). Our ambition is to build personalized models of central nervous system organs and pathologies, and to compare these models with clinical research studies in order to establish a quantitative diagnosis, prevent the progression of diseases and provide new digital recovery strategies, while combining all these research areas with clinical validation. This approach is developed within a translational framework, where the data integration process to build the models is informed by specific clinical studies, and where the models are assessed regarding prospective clinical trials for diagnosis and therapy planning. All of these research activities will be conducted in close collaboration with the Neurinfo platform, which benefited in 2018 from a new high-end 3T MRI system dedicated to research (3T Prisma™ system from Siemens), and through the development in the coming years of multimodal hybrid imaging (from the currently available EEG-MRI, to EEG-NIRS and PET-MRI in the future).
In this context, some of our major developments and newly arising issues and challenges will include:
-
The generation of new descriptors to study brain structure and function (e.g. the combination of variations in brain perfusion with and without a contrast agent; changes in brain structure in relation to normal, pathological, functional or connectivity patterns; or the modeling of brain state during cognitive stimulation using neurofeedback).
-
The integration of additional spatiotemporal and hybrid imaging sequences covering a larger range of observations, from the molecular level to the organ level, via the cellular level (arterial spin labeling, diffusion MRI, MR relaxometry, MR fingerprinting, MR cell labeling imaging, MR-PET molecular imaging, EEG-MRI functional imaging, EEG-NIRS-MRI, etc.).
-
The creation of computational models through the data fusion of molecular, cellular (i.e. through dedicated ligands or nanocarriers), structural and functional image descriptors from group studies of normal and/or pathological subjects.
-
The evaluation of these models in relation to acute pathologies, especially for the study of degenerative, psychiatric, traumatic or developmental brain diseases (primarily multiple sclerosis, stroke, traumatic brain injury (TBI) and depression, but applicable with a potential additional impact to epilepsy, Parkinson’s disease, dementia, Posttraumatic stress disorder, etc.) within a translational framework.
In terms of new major methodological challenges, we will address the development of models and algorithms to reconstruct, analyze and transform the images, and to manage the mass of data to store, distribute and “semanticize” (i.e. provide a logical division of the model’s components according to their meaning). As such, we expect to make methodological contributions in the fields of model inference; statistical analysis and modeling; the application of sparse representation (compressed sensing and dictionary learning) and machine learning (supervised/unsupervised classification and discrete model learning); data fusion (multimodal integration, registration, patch analysis, etc.); high-dimensional optimization; data integration; and brain-computer interfaces. As a team at the frontier between the digital sciences and clinical research in neuroscience, we do not claim to provide theoretical breakthroughs in these domains but rather to provide significant advances in using these algorithms through to the advanced applications we intend to address. In addition, we believe that by providing these significant advances using this set of algorithms, we will also contribute to exhibiting new theoretical problems that will fuel the domains of theoretical computer sciences and applied mathematics.
In summary, we expect to address the following major challenges:
-
Developing new information processing methods able to detect imaging biomarkers in the context of mental, neurological, and substance use disorders.
-
Providing new computational solutions for our target applications, allowing a more appropriate representation of data for image analysis and the detection of biomarkers specific to a form or grade of pathology, or specific to a population of subjects.
-
Providing, for our target applications, new patient-adapted connectivity atlases for the study and characterization of diseases from quantitative MRI.
-
Providing, for our target applications, new analytical models of dynamic regional perfusion, and deriving indices of dynamic brain local perfusion from normal and pathological populations.
-
Investigating whether the theragnostics paradigm of rehabilitation from hybrid neurofeedback can be effective in some behavioral and disability pathologies.
These major advances will be primarily developed and validated in the context of several priority applications in which we expect to play a leading role: multiple sclerosis, stroke rehabilitation, and the study and treatment of depression.