Section: New Results
Imaging & Phenomics, Biostatistics
Statistical learning on large databases of heterogeneous imaging, cognitive and behavioral data
Participants : Luigi Antelmi [Correspondent] , Nicholas Ayache, Philippe Robert, Marco Lorenzi.
Supported by the French government, through the UCAJEDI Investments in the Future project managed by the National Research Agency (ANR) ref. num. ANR-15-IDEX-01, our research is within the MNC3 initiative (Médecine Numérique: Cerveau, Cognition, Comportement), in collaboration with the Institut Claude Pompidou (CHU of Nice). Computational facilities are funded by the grant AAP Santé 06 2017-260 DGA-DSH, and by the Inria Sophia Antipolis - Méditerranée, "NEF” computation cluster.
statistical learning, joint analysis, neuroimaging
The aim of our work is to build scalable learning models for the joint analysis of heterogeneous biomedical data, to be applied to the investigation of neurological and neuropsychiatric disorders from collections of brain imaging, body sensors, biological and clinical data available in current large-scale databases such as ADNI(http://adni.loni.usc.edu/) and local clinical cohorts.
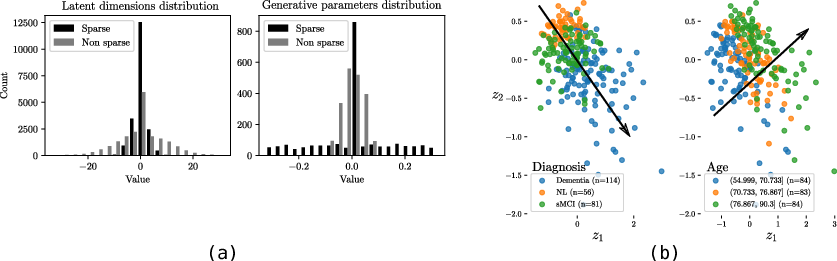
We developed a probabilistic latent variable model able to account for heterogeneous data modalities jointly [6]. In the latent space, this is achieved by constraining the variational distribution of each modality to a common target prior. Moreover, we added ad hoc prior distribution and parameterization for the latent space to induce sparsity (Fig. 7a). This approach is capable to highlight meaningful relationships among biomarkers in the context of Alzheimer's disease (Fig. 7b) that can be used to develop optimal strategies for disease quantification and prediction.
|
Joint Biological & Imaging markers for the Diagnosis of severe lung diseases
Participants : Benoit Audelan [Correspondant] , Hervé Delingette, Nicholas Ayache.
Lung cancer, Early detection, Biomarkers, Segmentation quality control
Lung cancer is among the most common cancer and is considered to be one of the most important public health problem. In recent years, immunotherapy has revolutionized cancer treatments but its efficiency is varying among patients. To prevent possible negative side effects there is a critical need in reliable biomarkers capable of predicting the response to immunotherapy treatments. We analyzed the performance of different biomarkers and studied their combination through logistic regression and decision tree models, as part of a joint project with the IRCAN laboratory (Pr P. Hofman, Dr S. Heeke) at Nice hospital [13].
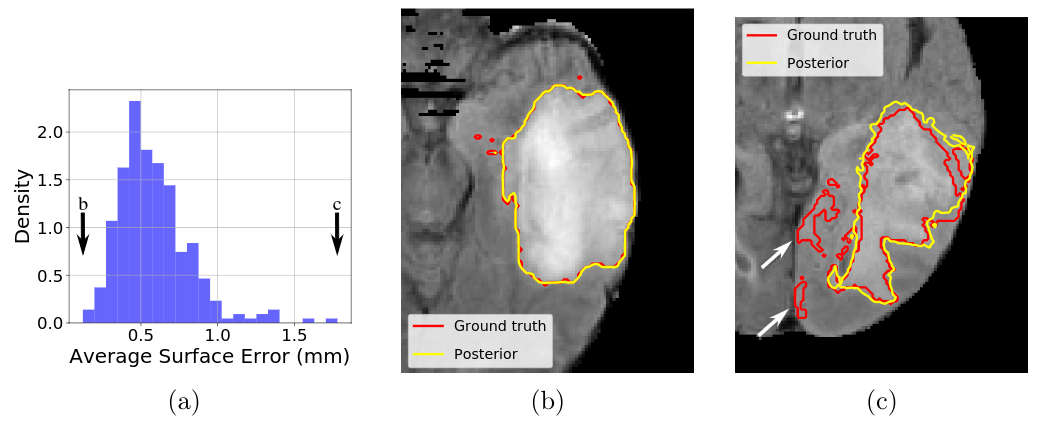
Furthermore, we investigated the issue of automated quality control assessment of image segmentations, which are a key point of medical image processing pipelines. We propose a novel unsupervised quality control approach based on simple intensity and smoothness assumptions [30]. We introduce a novel spatial prior which allows an automatic estimation of all parameters through Bayesian learning. The approach was tested on various medical imaging datasets (Fig. 8).
|
Modelling and inference of protein dynamics in neurodegenerative diseases across brain networks
Participants : Sara Garbarino [Correspondant] , Marco Lorenzi.
Sara Garbarino acknowledges financial support from the French government managed by L’Agence Nationale de la Recherche under Investissements d’Avenir UCA JEDI (ANR-15-IDEX-01) through the project “AtroProDem: A data-driven model of mechanistic brain Atrophy Propagation in Dementia”.
Gaussian Processes, Bayesian non–parametric modelling, neuroimaging data, protein dynamics, brain network
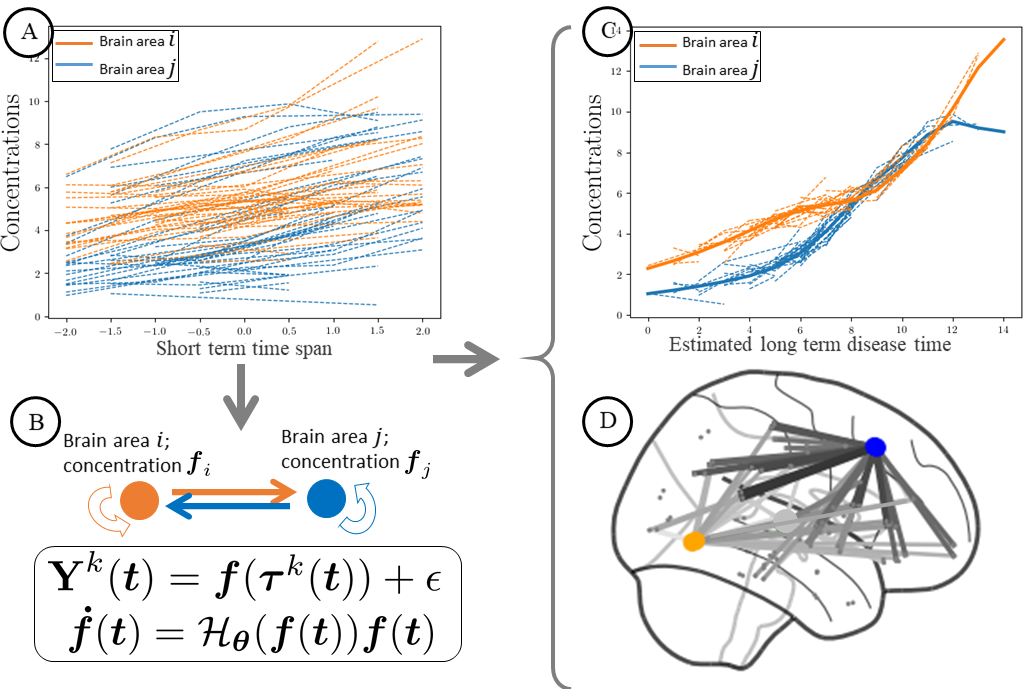
In this project we propose the first unified framework for the joint estimation of long term neurodegenerative disease progression and kinetic parameters describing pathological protein dynamics across brain networks [48]. The model is expressed within a constrained Gaussian Process regression setting. We use stochastic variational inference for scalable inference and uncertainty quantification. Experiments on simulated data and on AV45-PET brain imaging data measuring topographic amyloid deposition in Alzheimer's disease show that our model accurately recovers prescribed rates along graph dynamics and precisely reconstructs the underlying progression.
|