Section: Application Domains
Modeling and analysis of membrane transport and molecule trafficking at the single cell scale
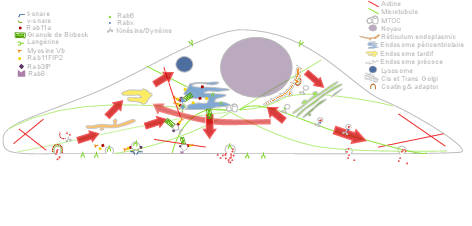
In the past recent years, research carried out out together with the “Space Time imaging of Endomembranes and organelles Dynamics” team at CNRS-UMR 144Institut Curie contributed to a better understanding of the intracellular compartmentation, particularly in specialized model cells such as melanocytes and Langerhans cells of the epidermis, of the components and structural events involved in the biogenesis of their specialized organelles: melanosomes and Birbeck granules, respectively and to the understanding on how the dynamics of those structures relate to their physiological functions. These studies have started to highlight: i/ the measurement of multiple sorting and structural events involved in the biogenesis of these organelles; ii/ complexity of the endo-melanosomal network of these highly specialized cells; iii/ complex molecular architecture organizing and coordinating their dynamics; iv/ intracellular transport steps affected in genetic diseases, among which the Hermansky Pudlak syndrome (HPS) or involved in viral infection (HIV and Langerin in Langerhans cells).
In this context, the central aim of serpico is to understand how the different machineries of molecular components involved are interconnected and coordinated to generate such specialized structures, an issue that become more and more accessible, thanks to improvement in all domains related to live imaging. We need to address the following topics:
-
developing new bioimaging approaches to observe and statistically analyze such coordinated dynamics in live material;
-
correlating this statistically relevant spatiotemporal organization of protein networks with the biological architectures and at the ultrastructural level;
-
modeling intracellular transport of those reference biological complex systems and proposing new experimental plans in an iterative and virtuous circle;
-
managing and analyzing the workflow of image data obtained along different multidimensional microscopy modalities.
These studies are essential to unravel the complexity of the endomembrane system, how different machineries evolve together and thus coordinate (e.g. see Fig. 1). They help to decipher cell organization and function at different scales through an integrative workflow of methodological and technological developments. New approaches, such as optogenetics may even help controlling cell functions.
At long term, these studies will shed light on the cellular and molecular mechanisms underlying antigen presentation, viral infection or defense mechanisms, skin pigmentation, the pathogenesis of hereditary genetic disorders (lysosomal diseases, immune disorders) and on the mechanisms underlying cell differentiation and cell transformation. Our methodological goal is also to link dynamics information obtained through diffraction limited light microscopy, at a time regime compatible with live cell imaging and close to biochemical molecular interactions. The overview of ultrastructural organization will be achieved by complementary electron microscopy methods which have also undergone a revolutionary improvement over the last decade. Image visualization and quantitative analysis are of course essential issues in this context.